文章信息
- 胡蕴菲,何鹏,吴宇杰,金长文
- HU Yun-fei, HE Peng, WU Yu-jie, JIN Chang-wen
- 枯草芽孢杆菌双精氨酸转运系统TatAy蛋白的溶液结构
- Solution Structure of Bacillus subtilis Twin-Arginine Translocation TatAy Protein
- 波谱学杂志, 2015, 32(2): 291-307
- Chinese Journal of Magnetic Resonance, 2015, 32(2): 291-307
- http://dx.doi.org/10.11938/cjmR20150212
-
文章历史
- 收稿日期:2015-02-09
- 收修改稿日期:2015-05-09
2. 北京大学 化学与分子工程学院,北京1000871;
3. 北京大学 生命科学学院,北京1000871;
4. 北京大学 北京分子国家实验室,北京1000871
2. College of Chemistry and Molecular Engineering, Peking University, Beijing 100871;
3. College of Life Sciences, Peking University, Beijing 100871;
4. Beijing National Laboratory for Molecular Sciences, Peking University, Beijing 100871
Protein translocation across cellular membranes requires specific machineries. Besides the general Sec system which transports proteins in their unfolded forms via a ‘threading’ mechanism,the novel twin-arginine transport (Tat) system identified in bacteria and plant chloroplasts has attracted much interest due to its ability to transport proteins in their folded states[1, 2]. Some of these proteins are transported with bound co-factors,or in homo-/hetero-oligomeric states. Therefore,the Tat system is challenged with the unique task to transport substrates of varies sizes while at the same time preserve the membrane permeability barrier to small molecules.
The Tat translocases identified thus far can be divided into two types,namely the TatABC and TatAC systems. The ABC-type Tat system in Gram-negative bacteria and plant chloroplasts contains three functional components TatA,TatB and TatC (Tha4,Hcf106 and cpTatC in chloroplasts) while the AC-type Tat systems found in most Gram-positive bacteria is only comprised of TatA and TatC subunits[3, 4, 5, 6, 7]. TatA and TatB are sequentially related single-pass transmembrane proteins,and TatC contains six transmembrane segments. For the ABC-type system,it is believed that the TatA component forms the protein translocation channel via self-oligomerization,whereas the TatBC complex functions as the receptor for cargo proteins[8, 9, 10, 11, 12, 13]. In the frequently proposed model,the TatBC complex binds to cargo proteins and subsequently recruits the TatA component to form the protein translocation channel[9, 12, 13]. The translocation process is dependent on the pH gradient via a yet unknown mechanism[14]. Recent studies on the AC-type Tat system strongly suggested dual roles for its TatA component,which may act not only as a channel-forming subunit,but also as a receptor analogous to the TatB component of the ABC-type system[15, 16]. Moreover,evidence for the existence of soluble TatA forming a micelle-like structure and its ability to bind cargo proteins in the cytosol indicated that the TatAC system might operate via a distinct mechanism from the one proposed for the TatABC system[17]. Despite recent advances in structural elucidations of the ABC-type system[18, 19, 20, 21, 22],structural information on the AC-type system is scarce and its functional mechanism remains elusive[23, 24, 25].
The Gram-positive bacterium Bacillus subtilis harbours three TatA (TatAy,TatAd and TatAc) and two TatC proteins (TatCy and TatCd). While the function of TatAc protein remains unknown,the TatAy-TatCy and TatAd-TatCd pairs form two autonomous systems targeting different protein substrates[7, 26]. The TatAy-TatCy system specifically transports the YwbN protein,whereas the TatAd-TatCd system specifically transports the phosphodiesterase PhoD. Therefore,systematic structural studies of the two parallel Tat systems in B. subtilis are essential for elucidating the common molecular basis for protein translocation by the TatAc systems,as well as identifying the structural determinants underlying substrate specificity. Herein,we report the solution NMR structure of the B. subtilis TatAy protein,which,in combination with previously reported TatAd structures[23, 24],provides insights into the mechanism of channel complex formation.
1 Experimental proceduress 1.1 Expression of TatAy proteinThe B. subtilis TatAy gene was cloned into the pET21a(+) vector (Novagen) and expressed in Escherichia coli BL21(DE3) strain (Novagen) with a C-terminal fused His-tag. The cell culture was grown overnight in 4 mL of Luria-Bertani media with ampicillin at 35 ℃,centrifuged and resuspended in 200 mL M9 minimal medium. For the production of 13C/15N-labeled samples,15NH4Cl and 13C6-glucose were used in the M9 medium. When OD600 reached 0.6,protein expression was induced by adding isopropyl-β-D- thiogalactoside (IPTG) at a final concentration of 200 mg/L. The cells were harvested after 6 hr induction at 35 ℃ and frozen at -80 ℃.
1.2 Protein purification and sample preparationThe protein purification procedure was similar to that previously reported[23]. Briefly,cells containing over-expressed TatAy protein were sonicated and treated with lysozyme for 1 h at 4 ℃. After centrifugation (17 000 g for 20 min at 4 ℃),the soluble fraction was resolved in buffer A (20 mmol/L Tris,200 mmol/L NaCl,pH 8.0) containing 0.5 % AZ314 (Anatrace,Affymetrix) for 1 h. The AZ314 solubilized sample was mixed with 2 mL Ni-NTA resin (Qiagen),and sequentially washed with 20 mL buffer A containing 0.1 % AZ314,15 mL buffer A containing 0.1 % AZ314 and 20 mmol/L imidazole,10 mL buffer A containing 0.2 % dodecylphosphocholine (DPC) and 20 mmol/L imidazole. Finally,TatAy protein was eluted using 10 mL buffer A containing 0.5 % DPC (or deuterated DPC) and 200 mmol/L imidazole. Purified TatAy in DPC was concentrated and exchanged into 50 mmol/L sodium phosphate buffer (pH 5.5 or 7.0). 10% of D2O was added for preparations of the NMR samples,and 2,2-dimethyl-2-silapentanesulfonic acid was added as the internal chemical shift reference.
1.3 NMR spectroscopyThe triple-resonance NMR experiments were performed at 35 ℃ on Bruker Avance 600 and 800 MHz spectrometers equipped with four RF channels and triple-resonance cryo-probes with pulsed-field gradients. The 2D 15N-edited HSQC spectra of TatAy sample prepared at pH 5.5 and 7.0 were essentially similar,with slight peak shifts for water-exposed residues located at the C-terminal part of the protein sequence. The sample condition at pH 5.5 allowed for the observation of additional side chain NH signals and was therefore used for the collection of triple-resonance and NOESY experiments. The chemical-shift assignments of backbone and side-chain atoms were obtained by 2D 15N- and 13C-edited HSQC experiments and 3D HNCA,HNCACB,HNCO,HN(CA)CO,CBCACONH,HBHACONH,(H)CC(CO)NH,(H)CCH-COSY,and (H)CCH-TOCSY experiments[27] using 13C/15N labeled TatAy sample in DPC micelles. 3D 15N- and 13C-edited NOESY-HSQC spectra as well as the 13C-edited NOESY-HSQC spectrum optimized for aromatic resonances (mixing times of 100 ms) for TatAy in deuterated DPC micelles were collected to confirm the chemical-shift assignments and generate distance restraints for structure calculations. An additional 15N-edited NOESY-HSQC experiment was collected at pH 7.0 to confirm that the NOE pattern of TatAy remains unchanged under the two different pH conditions. The NMR spectra were processed using NMRPipe[28] and analyzed by NMRView[29].
1.4 Residual dipolar coupling (RDC) measurementsThe backbone N-H RDC measurements of TatAy was performed using the liquid crystalline phase of G-tetrad DNA[30]. The 15N-labeled TatAy protein sample was exchanged to a buffer containing 50 mmol/L potassium phosphate (pH 7.0) and 150 mmol/L potassium chloride. To prepare weakly aligned sample,approximately 9 mg d(GpG) was solubilized in 250 μL water (containing 50 μL D2O) and mixed with 250 μL of the above TatAy sample. The reference sample was similarly prepared in which the d(GpG) solution was substituted by pure water (containing 50 μL D2O). The RDCs were extracted from the difference in 1H-15N splitting measured by 1H-15N IPAP-HSQC spectra[31] between the weakly aligned and the isotropic samples at 35 ℃ on a Bruker Avance 800 MHz spectrometer. The data were analyzed using software packages PALES[32] and MODULE[33].
1.5 Structure calculationsThe structure calculation procedure of TatAy was similar to that previously reported[23]. Inspection of the NOESY spectra revealed that the C-terminal tail (Glu51-Gln57) is unstructured and prone to degradation. Thus,the structure calculation was performed on the Met1-Glu50 fragment of TatAy protein based on dihedral angle restraints predicted by TALOS[34] and inter-proton NOE-derived distance restraints. The structures were calculated using simulated annealing approach in torsion angle space with the program Xplor-NIH[35]. In each round,200 structures were calculated,and 100 conformers with the lowest energy were subsequently subjected to validation by the RDC data. Fitting of the backbone N-H RDC values with calculated structures were performed using the single value decomposition (SVD) method by the program PALES[32]. Residues located at the flexible termini and linker regions were excluded during the fitting. Conformers with correlation coefficient R value higher than 0.85 were subjected to further structural and violation analysis. In the final round of calculation,89 out of the 100 lowest-energy conformers showed R values higher than 0.88. Therefore,40 conformers among those with the lowest backbone root mean square deviation (RMSD) to mean structure were selected as representative structures for the TatAy protein.
1.6 Backbone relaxation measurementsThe backbone 15N relaxation parameters of the longitudinal relaxation rates (R1),transverse relaxation rates (R2),and steady-state heteronuclear {1H}-15N NOE values of both B. subtilis TatAy and TatAd proteins were measured on a Bruker Avance 800 MHz NMR spectrometer equipped with a cryoprobe at 35 ℃[36]. The delays used for the R1 experiments were 10 (×2),100,200,300,400,600,800,1 000,1 600,2 400,3 200 and 4 000 ms,and those used for the R2 experiments were 7.424 (×2),14.848,22.272,29.696,37.120,51.968,66.816,84.088,111.360 and 133.632 ms. The relaxation rate constants were obtained by fitting the peak intensities to a single exponential function using the nonlinear least squares method. The {1H}-15N NOE experiments were performed in the presence and absence of a 3-s proton pre-saturation period prior to the 15N excitation pulse.
2 Results 2.1 Overall structure of B. subtilis TatAy proteinWe obtained near complete backbone and side-chain chemical shift assignments for the 57-residue B. subtilis TatAy protein (Fig. 1). The backbone NH signal of residue Leu27 was missing in the 15N-edited heteronuclear single quantum coherence (HSQC) spectrum,while its side chain atoms were successfully assigned. Chemical shift and NOE pattern analysis demonstrated that the C-terminal region of TatAy starting from residue Ser47 is largely unstructured,which is consistent with the secondary structure prediction. Therefore,the solution structure of B. subtilis TatAy protein was calculated using the fragment Met1-Glu50,while the C-terminal residues were removed. A total of 2 108 NOE derived distance restraints and 60 chemical shifts derived dihedral angle restraints were used in the structure calculations. Since NOE data only reflects local conformation details and is intrinsically insensitive to long-range structural information,such as helix orientations,we utilized the backbone N-H RDC data for structure validation and analysis during the calculation procedure (see Experimental Procedures). The majority of the conformers calculated based on NOE and dihedral angle restraints showed good correlation with the RDC data,with correlation coefficient R values larger than 0.85 (and Q factors less than 0.45) for residues located in the regular helical segments. Fig. 2(b) and 2(c) show the ensemble of 40 representative structures of TatAy protein best satisfying both NOE and RDC data and the ribbon diagram of one representative structure,respectively. The structural statistics are summarized in Table 1.

|
| Fig. 1 Backbone assignments of B. subtilisTatAy. The figure shows 2D 15N-edited HSQC spectrum of B.subtilits TatAy protein in DPC micelles collected at 35 ºC in asodium phosphate buffer (pH 5.5). The backbone assignments are annotated usingthe one-letter amino acid code and residue number. The peak corresponding toresidue Met1 is folded and labeled in blue. For clarity,peaks originating fromthe C-terminal His-tag are not labeled |

|
| Fig. 2 Solution structure of B. subtilisTatAy. (a) Primary structure of B. subtilis TatAywith the TMH and APH segments coloured in red and green,respectively. (b)Ensemble of the 40 representative conformers of TatAy (segment Met1-Glu50).(c) Ribbon diagram of the solution structure of TatAy viewed fromtwo angles. The C-terminal unstructured region (Glu51-Gln57)is schematically represented by a dashed line in the left panel. (d)Experimentally measured backbone N-H RDC values of TatAy. The curves(magenta) show the fitting of the RDC data to sinusoid functions with a fixedperiodicity of 3.6 using a MATLAB script (60) (e) surface representation ofTatAy. The positively charged,negatively charged,and polarresidues are coloured in blue,red and yellow,respectively. Hydrophilicresidues in the APH segment are selectively labeled. For clarity,only residuesMet1-Thr46 are shown. (f) Mapping of wateraccessibility onto the structure of TatAy. The water accessibilityof each residue was qualitatively analyzed by measuring the intensity of theNOE cross peaks between backbone amides and water in the 15N NOESYspectrum. Residues that show no cross peak to water signal,the prolineresidues,as well as residues difficult to analyze due to signal overlap arecoloured in grey. Residues that show cross peaks to water are divided into 4groups according to the peak intensities (I),and are coloured in red (I≥ 10),magenta (5 ≤ I <10),yellow (1 ≤ I < 5) andwheat (0 < I < 1),respectively. For clarity,only residues Met1-Thr46are shown |
| Experimental restraints | |
| Total NOE | 2108 |
| Total unambiguous NOE | 1537 |
| Intra-residue | 527 |
| Inter-residue | 1010 |
| Sequential (|i-j| = 1) | 525 |
| Medium-range (1 < |i-j| < 5) | 464 |
| Long-range ( |i-j|≥ 5) | 21 |
| Total ambiguous NOE | 571 |
| Total dihedral angle restraints (j + y) | 60 (30 + 30) |
| Restraint violations | |
| Distance restraint violations > 0.5 nm | 0 |
| Dihedral angle restraint violations > 5˚ | 0 |
| RMSD from mean structure (0.1 nm) | |
| All heavy atoms | 1.86 |
| All backbone atoms | 1.45 |
| Secondary structure heavy atoms | 0.92 |
| Secondary structure backbone atoms | 0.73 |
| Ramachandran statistics¶ (%) | |
| Most favored regions | 89.9 |
| Additional allowed regions | 9.3 |
| Generously allowed regions | 0.4 |
| Disallowed regions | 0.4 |
The monomeric TatAy protein adopts an L-shaped structure formed by the perpendicular packing of two helices. The transmembrane helix (TMH) is comprised of residues Pro5-Phe19,and the amphipathic helix (APH) contains approximately residues Leu24-Thr46,as demonstrated by NOE pattern analysis and chemical shift information. This is also evidenced by the backbone N-H RDC values [Fig. 2(d)],which show clear dipolar wave pattern of α-helices for both segments. Only residues Gly6-Leu8,Leu24-Pro25 and Leu45-Thr46 at the ends of the helices show RDC values deviating from the dipolar wave functions,presumably due to their higher structural flexibility (vide infra). Moreover,both segments can be fitted using a single sine function with no significant phase or amplitude changes,indicating the lack of notable kink or bend in these two helices.
The APH segment is oriented such that its hydrophobic surface points toward the TMH,while the hydrophilic residues,in particular the charged residues,are exposed to water [Fig. 2(e)]. This was further supported by the analysis of the NOE cross peaks to water [Fig. 2(f)]. For residues in the TMH region,neither their backbone amide groups nor side chain hydrogen atoms show NOE cross peaks to the water. For residues in the N-terminal half of the APH segment,water accessibility is dependent on the position of the residue. For example,the backbone amides of residues Gly28,Leu35,Arg36 and Phe38 have limited water accessibility,and these residues are located on the hydrophobic side of the APH. On the other hand,the backbone amides of residues Lys29,Gly32,Asp33,Thr34 and Asn40 show relatively strong NOE cross peaks to water,and these residues are distributed on the hydrophilic side of the APH. The observed cross peaks may arise from either cross-relaxation effect or chemical exchange process with water,both are indication of water accessibility. In contrast,the backbone amides of the C-terminal half of the APH segment (residues Asn40-Thr46) as well as their side chain hydrogen atoms all show strong NOE cross peaks to water in the NOESY spectra. This is also consistent with the fact that this region displays higher motional flexibility,which will be described in the ‘Backbone dynamics of B. subtilis TatAy protein’ section (vide infra).
2.2 Local conformation at the hinge regionThe overall structure of B. subtilis TatAy is similar to that of the TatAd protein,previously solved by our group[23]. Similar to TatAd,the L-shaped conformation of TatAy is also stabilized via extensive medium and long range NOE contacts among residues located in the short linker Gly20-Lys23,residues at the C-terminal of TMH and the N-terminal of APH. This region,known as the “hinge” region,contains a number of highly conserved residues that are essential to the functions of the TatA and TatB protein families[37, 38]. Interestingly,the hinge region of the TatA subunit in the AC-type system displays sequence similarity to both TatA and TatB components of the ABC-type system. The B. subtilis TatAy and TatAd proteins both harbour the invariant FG dipeptide characteristic of the TatA family,while they also contain the conserved GPxxLP (x corresponds to any amino acids) motif of the TatB family.
Fig. 3 shows the local conformation at the hinge region of B. subtilis TatAy,together with that of TatAd protein for comparison. In TatAy,residues Leu16,Pro21,Leu24 and Pro25 form relayed interactions on one side of the hinge [Fig. 3(a)],while residues Phe19,Lys23 and Leu27 form relayed interactions on the other side [Fig. 3(c)]. In TatAd,a highly similar mode of interactions is present. As shown in Fig. 3(b),residues Leu18,Pro23,Leu26 and Pro27 in TatAd are sequentially equivalent to residues Leu16,Pro21,Leu24 and Pro25 in TatAy,and are structurally located at similar positions to form a relatively stable interacting network which may be essential to the maintenance of the L-shaped conformation. On the other hand,residues Phe21,Lys25 and Ile19 in TatAd are sequentially equivalent to residues Phe19,Lys23 and Leu27 in TatAy,and also adopt similar positions in the three-dimensional structure. Though the detailed side chain conformations between the two proteins differ,particularly for residue Lys23 of TatAy and Lys25 of TatAd,the general interaction pattern is essentially the same for these two proteins. Sequence alignments of TatA proteins from different sources [Fig. 4(a)] show that residues at positions Phe19 and Lys23 (as numbered in TatAy) are strictly conserved,while residues at positions Leu16,Leu24 and Leu27 (as numbered in TatAy) are generally restricted to hydrophobic residues like Leu,Ile and Val. In addition,residues Pro21,Leu24 and Pro25 in the GP21xxL24P25 motif (as numbered in TatAy) are invariant among TatB family proteins and the TatA proteins of the AC-type system.
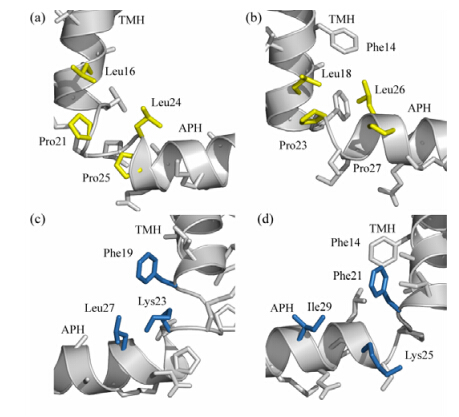
|
| Fig. Local conformation at the hinge region of B. subtilis TatA proteins. (a),(c) Local conformation at the hinge region of B. subtilis TatAy protein viewed from two sides. In (a),residues Leu16,Pro21,Leu24 and Pro25 that form relayed interactions on one side of the structure are coloured in yellow. In (c),residues Phe19,Lys23 and Leu27 that form relayed interactions on the other side of the structure are coloured in blue. (b),(d) Local conformation at the hinge region of B. subtilis TatAd protein viewed from two sides. The residues in TatAd protein equivalent to those of TatAy protein labeled in (a) and (c) are coloured in yellow (b) and blue (d),correspondingly. In addition,residue Phe14 of TatAd,which was found to contribute to the interaction network,is also labeled |
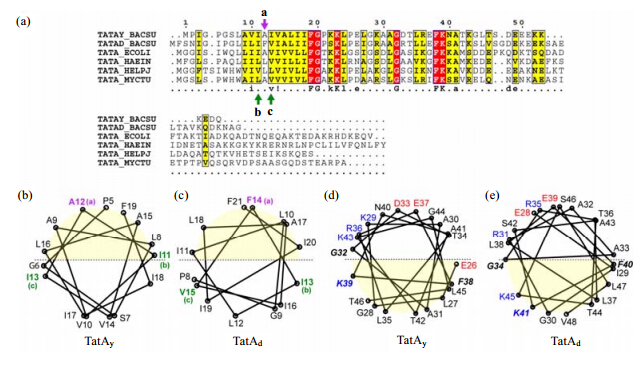
|
| Fig. Sequence and structural conservations among TatA proteins. (a) Multiple sequence alignment of TatA proteins from different sources. The sequences used in the alignment are TatAy (TatAy_BACSU) and TatAd (TatAd_BACSU) proteins from B. subtilis,TatA proteins from E. coli (TATA_ECOLI),Haemophilus influenzae (TATA_HAEIN),Helicobacter pylori (TATA_HALPJ),and Mycobacterium tuberculosis (TATA_MYCTU). The sequence alignment was performed using ClustalW (61) and drawn using ESPript (62). The sequences are numbered according to B. subtilis TatAy,and the consensus residues are illustrated. (b),(c) Structure-based helical wheel diagrams of the TMH segment of B. subtilis TatAy (b) and TatAd (c) proteins. The upper half of the wheel with light yellow background colour depicts the side of TMH that orients toward the APH segment,while the lower half of the wheel represents the side of TMH that points away from the APH. Residue Ala12 in TatAy (Phe14 in TatAd) located toward the TMH-APH interface is labeled in purple and designated as position a. The residues Ile11 and Ile13 in TatAy (Ile13 and Val15 in TatAd) located at the sides of the TMH potential for forming inter-subunit contacts are labeled in green and designated as positions b and c. These three positions are marked by arrows correspondingly in the sequence alignment in (a). (d),(e) Structure-based helical wheel representations of the APH segment of B. subtilis TatAy (d) and TatAd (e) proteins. The lower half of the wheel with light yellow background color depicts the side of APH that orients toward the TMH segment (thus the hydrophobic lipid environment),while the upper half of the wheel represents the side of APH that is faces the aqueous solution. Positively and negatively charged residues are labeled in blue and red,respectively. The strictly conserved residues Gly32,Phe38 and Lys39 (as numbered in TatAy) are labeled in bold italic characters. The helical wheel diagrams (b)-(e) were generated by an in-house written script based on the normalized coordinates of Cα atoms in the 3D solution structure |
In the TatAd protein sequence,an aromatic Phe14 residue is located in the TMH segment and was observed to have NOE contacts with residues Leu18 and Leu26 in the hinge region. These interactions appear to pull the APH of TatAd closer to TMH as compared to TatAy. A similar Phe residue was found at the same position in the E. coli TatB sequence,but not in other TatA sequences. In TatAy,the corresponding position is occupied by a small residue Ala12,and we were not able to identify long-range contacts between this residue and the hinge region. Therefore,the interaction between Phe14 and the hinge region observed in TatAd may be non-essential,whereas the conserved interactions among residues at the hinge region are sufficient for the maintenance of the L-shaped conformation. 2.3 Conserved structural features among TatA proteins
As demonstrated above and in our previous work,both TatAy and TatAd proteins adopt an L-shaped structure containing a TMH that is embedded in the detergent micelles,and an APH that lies at the water-detergent interface. To further analyze the conserved structural features between these two proteins and to provide more general insights for the TatA protein family,we present the multiple sequence alignment of the TatA proteins from different bacterial species [Fig. 4(a)],together with the helical wheel pattern of both TMH and APH segments of TatAy and TatAd proteins drawn based on the 3D structures [Fig. 4(b) -(e)].
The relative orientation of the two helices against each other is essentially identical between TatAy and TatAd proteins. The hydrophobic TMH segment can be approximately divided into two halves,one that faces the APH [the upper half in Fig. 4(b),(c)],and the other that orients away from the APH [the lower half in Fig. 4(b),(c)]. In the TatAd structure,the side chain of Phe14 points toward the direction of the APH segment and involves in the interaction with the hinge region. As shown by the multiple sequence alignment,the equivalent residue at this position is Ala12 in TatAy [position a as labeled in Fig. 4(a)]. Although residue Ala12 in B. subtilis TatAy does not participate in the interactions of the hinge region,it is also pointed toward the APH. Both residues locate at the central region of the TMH side orienting toward the APH.
Moreover,residues Ile12 and Val14 of E. coli TatA are equivalents to residues Ile11 and Ile13 of B. subtilis TatAy (or Ile13 and Val15 of B. subtilis TatAd),respectively [positions b and c as labeled in Fig. 4(a)]. Structural study of E. coli TatA using EPR suggested that Ile12 and Val14 residues may form the inter-subunit contact site between adjacent subunits in the TatA homo-oligomeric complex[39]. The structures of both B. subtilis TatAy and TatAd show that residues at these two sites are well positioned at the two sides of TMH,and are therefore competent for forming the inter-subunit contact surface [positions b and c as labeled in Fig. 4(b),(c)].
As shown in Fig. 4(d),(e),the APH segment can be easily divided into the hydrophobic and hydrophilic sides. The hydrophobic side is oriented toward the TMH [the lower half in Fig. 4(d),(e)]. The hydrophilic side is oriented away from the TMH [the upper half in Fig. 4(d),(e)]. The dissection of these two halves was supported by the presence or absence of cross peaks to water in the NOESY spectra [Fig. 2(f)]. As expected,the charged and polar residues are mostly positioned on the hydrophilic side of the APH,with the exception of residues in the C-terminal end of the helix. For example,residues Thr42 and Thr46 in TatAy,are located in the lower (hydrophobic) half of the APH,while residues Asn40 and Lys43 are located at the upper (hydrophilic) half. A similar pattern is also found for TatAd protein. Therefore,the C-terminal region of the APH segment does not show clear amphipathic characteristics,which is consistent with the observation that this region is largely accessible to water.
Multiple sequence alignment of TatA proteins [Fig. 4(a)] highlights three strictly conserved residues in the APH region,residues Gly32,Phe38 and Lys39 (as numbered in TatAy). Mutagenesis studies of E. coli TatA demonstrated that point mutations at these three positions (Gly33,Phe39 and Lys40 in E. coli TatA) caused complete inactivation of the Tat system[37, 38, 40, 41, 42]. Comparison of the TatAy and TatAd structures indicates that all three residues are positioned sideways [Fig. 4(d),(e)],thus lying at the interface between the hydrophobic and hydrophilic halves of the helix. In particular,the charged Lys residue in the strictly conserved FK dipeptide is positioned in the hydrophobic half of the helix,the possible biological relevance of which will be discussed below (see Discussion).
2.4 Backbone dynamics of B. subtilis TatAy proteinProtein dynamics play critical roles in its biological functions. The Tat system,in particular,shows highly dynamic behaviour as demonstrated by various biochemical observations. For example,the E. coli TatA protein may adopt dual topology[43, 44],and the E. coli TatC protein was suggested to have two of its transmembrane segments shuttling in and out of the membrane[45, 46]. As representatives of the AC-type Tat system,both TatAy and TatAd proteins from B. subtilis were shown to exist in two distinct forms: the membrane bound form and the soluble form[15],which also suggested that this class of proteins has certain intrinsic flexibility to adapt to different environments. Since the dynamic behaviour of the Tat components might be related to the protein translocation process,we used NMR relaxation experiments to investigate the backbone dynamics of B. subtilis TatAy protein.
The backbone relaxation parameters of B. subtilis TatAy including the steady-state heteronuclear {1H}-15N NOE values,the longitudinal relaxation rate R1,and the transverse relaxation rate R2 measured on an 800 MHz spectrometer are shown in Fig. 5,together with the values of R2/R1 and R1*R2. The N- and C-terminal residues are highly flexible as judged from the low {1H}-15N NOE and R2/R1 values. The TMH segment,which is fully buried in DPC micelles,shows the highest structural rigidity,with {1H}-15N NOE values around 0.75. The central part of the APH region (residues Gly32-Lys39) has slightly increased flexibility on the ps-ns timescales,with {1H}-15N NOE values around 0.65. Residues in the linker region and the N-terminal part of the APH segment (residues Lys23-Ala30) show higher structural flexibility,with {1H}-15N NOE values close to 0.60. The C-terminal end of the APH (starting from residue Asn40) displays a sharp decrease of {1H}-15N NOE values and hence significant increase of motional flexibility in the fast timescales. The relaxation data also suggested that the motional properties of the two helices are slightly different: the TMH segment is the most rigid part of the structure,whereas the APH has a certain degree of flexibility to sample more conformational spaces.
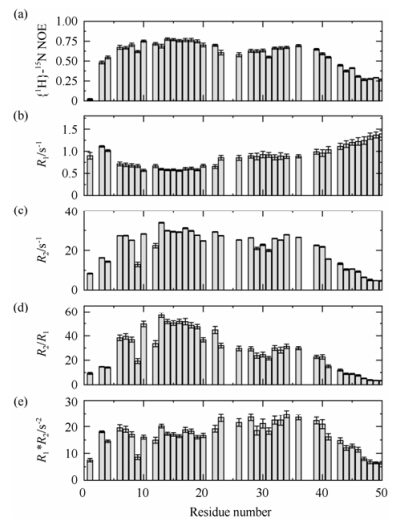
|
| Fig. 5 Backbone dynamics of B. subtilis TatAy. (a)-(c) The backbone 15N relaxation parameters of TatAy,including the steady-state heteronuclear {1H}-15N NOE values (a),the longitudinal relaxation rates R1 (b),and the transverse relaxation rates R2 (c). (d) The R2/R1 values as a function of residue number. (e) The R1*R2 values as a function of residue number |
Fig. 5(d),(e) show the R2/R1 and R1*R2 values as a function of residue number. The R1*R2 value is independent of rotational anisotropy,and thus a better indicator of possible chemical/conformational exchanges than the R2/R1 value[47]. The data shows that residues in the APH region display elevated R1*R2 values as compared to those in the TMH region [Fig. 5(e)]. We also collected and analyzed the backbone relaxation data of the B. subtilis TatAd protein,and observed a similar increase of R1*R2 values in the APH segment. These results suggest that the APH segment may be involved in conformational exchanges on the μs-ms timescales,which could result from on-off interaction with the detergent micelles,or intrinsic conformational exchanges of the APH. Taken together,the above data suggest that the TMH region of the TatAy protein is relatively rigid,while the APH segment displays higher structural flexibility on the ps-ns timescales,and possibly involves in s-ms timescale conformational exchanges.
3 DiscussionOur analysis of NOE cross peaks with water in the APH segment suggested that the amphipathic nature is most prominent at the N-terminal half,while the C-terminal region (starting from residue Asn40 in TatAy and Ser42 in TatAd) is largely exposed to water. In addition,backbone dynamics of both proteins show that this region displays significantly increased motional flexibility on the ps-ns timescales. It is therefore possible that in a lipid bilayer,the N-terminal half of the APH lie at the water-lipid interface,while the C-terminal region protrudes into the aqueous environment. This was supported by the data from solid-state NMR study of B. subtilis TatAd,which showed that in membrane mimicking bicelles,the APH segment does not lie flat on the membrane surface,but show a relatively steep tilt angle with the membrane[24]. This orientation allows the N-terminal half of APH to be submerged in the hydrophobic lipid bilayer,while the C-terminal half sticked out of the membrane and exposed to water.
In addition,three strictly conserved residues (Gly32,Phe38 and Lys39 in TatAy) in the APH segment locate at the interface between the hydrophobic and hydrophilic halves of the helix (Fig. 4),with the invariant FK dipeptide more oriented to the hydrophobic side. The positioning of the Phe residue on the hydrophobic side of the helix,and the observation that its backbone amide group and side chain hydrogen atoms show limited access to water,together suggest that this residue may insert into the lipid. It is interesting to notice that the charged side chain of the Lys is also located on the same side of the APH. As previous studies suggested that the TatA protein may preferably bind to negatively charged membranes[48, 49, 50],it is possible that this conserved Lys residue contributes to the protein-lipid binding via electrostatic interaction. Therefore,the conserved Phe and Lys residues in the APH segment may both play important roles in the protein-lipid interactions. Further,the sideway positions of all the three residues may also implicate their possible contributions to inter-subunit contacts during oligomerization,which is supported by the biochemical observations that F39A mutation of E. coli TatA caused aberrant TatA complex assembly[41] and the recently reported E. coli TatA dimer structure[21].
4 ConclusionIn this study,we determined the solution structure of B. subtilis TatAy protein in DPC micelles. It adopts an L-shaped conformation similar to the B. subtilis TatAd protein as previously reported by our group. Comparison of the two structures,together with sequence analysis and other published biochemical evidences,highlights the conserved residues at the hinge region for maintenance of the L-shaped conformation,and further suggests a number of common structural and dynamic features for the TatA protein family. The results provide new insights into the channel assembly of the Tat protein translocation system.
Acknowledgement: All NMR experiments were performed at the Beijing NMR Center. The MATLAB script used for the fitting of dipolar waves was kindly provided by Prof. Stanley Opella,University of California San Diego. This work was supported by Grant 2010IM030700 from the Ministry of Science and Technology of China,and Grant 31070649 from the National Natural Science Foundation of China to C. J. Y. H acknowledges a special support from China Postdoctoral Foundation.| [1] | Palmer T, Berks B C. The twin-arginine translocation (Tat) protein export pathway[J]. Nat Rev Micro, 2012, 10(7):483-496. |
| [2] | Sargent F. The twin-arginine transport system:moving folded proteins across membranes[J]. Biochem Soc Trans, 2007, 35(Pt 5):835-847. |
| [3] | Weiner J H, Bilous P T, Shaw G M, et al. A novel and ubiquitous system for membrane targeting and secretion of cofactor-containing proteins[J]. Cell, 1998, 93(1):93-101. |
| [4] | Sargent F, Bogsch E G, Stanley N R, et al. Overlapping functions of components of a bacterial Sec-independent protein export pathway[J]. EMBO J, 1998, 17(13):3 640-3 650. |
| [5] | Bogsch E G, Sargent F, Stanley N R, et al. An essential component of a novel bacterial protein export system with homologues in plastids and mitochondria[J]. J Biol Chem, 1998, 273(29):18 003-18 006. |
| [6] | Sargent F, Stanley N R, Berks, B C, et al. Sec-independent protein translocation in Escherichia coli. A distinct and pivotal role for the TatB protein[J]. J Biol Chem, 1999, 274(51):36 073-36 082. |
| [7] | Jongbloed J D, Grieger U, Antelmann H, et al. Two minimal Tat translocases in Bacillus[J]. Mol Microbiol, 2004, 54(5):1 319-1 325. |
| [8] | Gohlke U, Pullan L, McDevitt C A, et al. The TatA component of the twin-arginine protein transport system forms channel complexes of variable diameter[J]. Proc Natl Acad Sci USA, 2005, 102(30):10 482-10 486. |
| [9] | Dabney-Smith C, Mori H, Cline K. Oligomers of Tha4 organize at the thylakoid Tat translocase during protein transport[J]. J Biol Chem, 2006, 281(9):5 476-5 483. |
| [10] | Leake M C, Greene N P, Godun R M, et al. Variable stoichiometry of the TatA component of the twin-arginine protein transport system observed by in vivo single-molecule imaging[J]. Proc Natl Acad Sci USA, 2008, 105(40):15 376- 15 381. |
| [11] | de Leeuw E, Granjon T, Porcelli I, et al. Oligomeric properties and signal peptide binding by Escherichia coli Tat protein transport complexes[J]. J Mol Biol, 2002, 322(5):1 135-1 146. |
| [12] | Cline K, Mori H. Thylakoid deltapH-dependent precursor proteins bind to a cpTatC-Hcf106 complex before Tha4-dependent transport[J]. J Cell Biol, 2001, 154(4):719-729. |
| [13] | Alami M, Lüke I, Deitermann S, et al. Differential interactions between a twin-arginine signal peptide and its translocase in Escherichia coli[J]. Mol Cell, 2003, 12(4):937-946. |
| [14] | Yahr T L, Wickner W T. Functional reconstitution of bacterial Tat translocation in vitro[J]. EMBO J, 2001, 20(10):2 472-2 479. |
| [15] | Westermann M, Pop O I, Gerlach R, et al. The TatAd component of the Bacillus subtilis twin-arginine protein transport system forms homo-multimeric complexes in its cytosolic and membrane embedded localization[J]. Biochim Biophys Acta, 2006, 1 758(4):443-451. |
| [16] | Barnett J P, Eijlander R T, Kuipers O P, et al. A minimal Tat system from a gram-positive organism:a bifunctional TatA subunit participates in discrete TatAC and TatA complexes[J]. J Biol Chem, 2006, 283(5):2 534-2 542. |
| [17] | Pop O I, Westermann M, Volkmer-Engert R, et al. Sequence-specific binding of prePhoD to soluble TatAd indicates protein-mediated targeting of the Tat export in Bacillus subtilis[J]. J Biol Chem, 2003, 278(40):38 428-38 436. |
| [18] | Rollauer S E, Tarry M J, Graham J E, et al. Structure of the TatC core of the twin-arginine protein transport system[J]. Nature, 2012, 492(7428):210-214. |
| [19] | Ramasamy S, Abrol R, Suloway C J, et al. The glove-like structure of the conserved membrane protein TatC provides insight into signal sequence recognition in twin-arginine translocation[J]. Structure, 2013, 21(5):777-788. |
| [20] | Rodriguez F, Rouse S L, Tait C E, et al. Structural model for the protein-translocating element of the twin-arginine transport system[J]. Proc Natl Acad Sci USA, 2013, 110(12):E1 092-E1 101. |
| [21] | Zhang Y, Hu Y, Li H, et al. Structural basis for TatA oligomerization:an NMR study of Escherichia coli TatA dimeric structure[J]. PLoS One, 2014, 9(8):e103157 |
| [22] | Zhang Y, Wang L, Hu Y, et al. Solution structure of the TatB component of the twin-arginine translocation system[J]. Biochim Biophys Acta, 2014, 1838(7):1 881-1 888. |
| [23] | Hu Y, Zhao E, Li H, et al. Solution NMR structure of the TatA component of the twin-arginine protein transport system from gram-positive bacterium Bacillus subtilis[J]. J Am Chem Soc, 2010, 132(45):15 942-15 944. |
| [24] | Walther T H, Grage S L, Roth N, et al. Membrane alignment of the pore-forming component TatAd of the twin-arginine translocase from Bacillus subtilis resolved by solid-state NMR spectroscopy[J]. J Am Chem Soc, 2010, 132(45):15 945-15 956. |
| [25] | Walther T H, Gottselig C, Grage S L, et al. Folding and self-assembly of the TatA translocation pore based on a charge zipper mechanism[J]. Cell, 2013, 152(1-2):316-326. |
| [26] | Pop O, Martin U, Abel C, et al. The twin-arginine signal peptide of PhoD and the TatAd/Cd proteins of Bacillus subtilis form an autonomous Tat translocation system[J]. J Biol Chem, 2002, 277(5):3 268-3 273. |
| [27] | Sattler M, Schleucher J, Griesinger C. Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulse field gradients[J]. Prog Nucl Magn Reson Spect, 1999, 34:93-158. |
| [28] | Delaglio F, Grzesiek S, Vuister G W, et al. NMRPipe:a multidimensional spectral processing system based on UNIX pipes[J]. J Biomol NMR, 1995, 6(3):277-293. |
| [29] | Johnson B A. Using NMRView to visualize and analyze the NMR spectra of macromolecules[J]. Methods Mol Biol, 2004, 278:313-352. |
| [30] | Lorieau J, Yao L, Bax A. Liquid crystalline phase of G-tetrad DNA for NMR study of detergent-solubilized proteins[J]. J Am Chem Soc, 2008, 130(24):7 536-7 537. |
| [31] | Ottiger M, Delaglio F, Bax A. Measurement of J and dipolar couplings from simplified two-dimensional NMR spectra[J]. J. Magn. Reson. 1998, 131(2):373-378. |
| [32] | Zweckstetter M. NMR:prediction of molecular alignment from structure using the PALES software[J]. Nat Protoc, 2008, 3(4):679-690. |
| [33] | Dosset P, Hus J C, Marion D, et al. A novel interactive tool for rigid-body modeling of multi-domain macromolecules using residual dipolar couplings[J]. J Biomol NMR, 2001, 20(3):223-231. |
| [34] | Cornilescu G, Delaglio F, Bax, A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology[J]. J Biomol NMR, 1999, 13(3):289-302. |
| [35] | Schwieters C D, Kuszewski J J, Tjandra N, et al. The Xplor-NIH NMR molecular structure determination package[J]. J Magn Reson, 2003, 160(1):65-73. |
| [36] | Farrow N A, Muhandiram R, Singer A U, et al. Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation[J]. Biochemistry, 1994, 33(19):5 984-6 003. |
| [37] | Barrett C M, Mathers J E, Robinson C. Identification of key regions within the Escherichia coli TatAB subunits[J]. FEBS Lett, 2003, 537(1-3):42-46. |
| [38] | Hicks M G, de Leeuw E, Porcelli I, et al. The Escherichia coli twin-arginine translocase:conserved residues of TatA and TatB family components involved in protein transport[J]. FEBS Lett, 2003, 539(1-3):61-67. |
| [39] | White G F, Schermann S M, Bradley J, et al. Subunit organization in the TatA complex of the twin arginine protein translocase:a site-directed EPR spin labeling study[J]. J Biol Chem, 2010, 285(4):2 294-2 301. |
| [40] | Greene N P, Porcelli I, Buchanan G, et al. Cysteine scanning mutagenesis and disulfide mapping studies of the TatA component of the bacterial twin arginine translocase[J]. J Biol Chem, 2007, 282(33):23 937-23 945. |
| [41] | Barrett C M L, Mangels D, Robinson C. Mutations in subunits of the Escherichia coli twin-arginine translocase block function via differing effects on translocation activity or tat complex structure[J]. J Mol Biol, 2005, 347(2):453-463. |
| [42] | Hicks M G, Lee P A, Georgiou G, et al. Positive selection for loss-of-function tat mutations identifies critical residues required for TatA activity[J.] J Bacteriol, 2005, 187(8):2 920-2 925. |
| [43] | Gouffi K, Gérard F, Santini C L, et al. Dual topology of the Escherichia coli TatA protein[J]. J Biol Chem, 2004, 279(12):11 608-11 615. |
| [44] | Chan C S, Zlomislic M R, Tieleman D P, et al. The TatA subunit of Escherichia coli twin-arginine translocase has an N-in topology[J]. Biochemistry, 2007, 46(25):7 396-7 404. |
| [45] | Behrendt J, Standar K, Lindenstrauss U, et al. Topological studies on the twin-arginine translocase component TatC[J]]. FEMS Microbiol Lett, 2004, 234(2):303-308. |
| [46] | Gouffi K, Santini C L, Wu L F. Topology determination and functional analysis of the Escherichia coli TatC protein[J]. FEBS Lett, 2002, 525(1-3):65-70. |
| [47] | Kneller J M, Lu M, Bracken C. An effective method for the discrimination of motional anisotropy and chemical exchange[J]. J Am Chem Soc, 2002, 124(9):1 852-1 853. |
| [48] | Lange C, Müller S D, Walther T H, et al. Structure analysis of the protein translocating channel TatA in membranes using a multi-construct approach[J]. Biochim Biophys Acta, 2007, 1 768(10):2 627-2 634. |
| [49] | Porcelli I, de Leeuw E, Wallis R, et al. Characterization and membrane assembly of the TatA component of the Escherichia coli twin-arginine protein transport system[J]. Biochemistry, 2002, 41(46):13 690-13 697. |
| [50] | Mikhaleva N I, Santini C L, Giordano G, et al. Requirement for phospholipids of the translocation of the trimethylamine N-oxide reductase through the Tat pathway in Escherichia coli[J]. FEBS Lett, 1999, 463(3):331-335. |
 2015, Vol. 32
2015, Vol. 32




