文章信息
- 超级增强子在肿瘤转移中的作用机制研究进展
- Role and Mechanism of Super-enhancers in Tumor Metastasis
- 肿瘤防治研究, 2023, 50(5): 518-524
- Cancer Research on Prevention and Treatment, 2023, 50(5): 518-524
- http://www.zlfzyj.com/CN/10.3971/j.issn.1000-8578.2023.22.1167
- 收稿日期: 2022-10-08
- 修回日期: 2022-11-29
2. 341000 赣州,赣南医学院药学院
2. College of Pharmacy, Gannan Medical University, Ganzhou 341000, China
上世纪80年代,Banerji等发现SV40病毒上一段长度为100~300 bp的DNA序列可促进家兔β珠蛋白转录,并将这段DNA片段定义为增强子[1]。2013年,Young团队在胚胎干细胞中鉴定出超级增强子(super-enhancers, SEs)[2]。研究发现SEs具有组织及细胞特异性,是决定细胞身份的主要因素[3]。SEs调控功能广泛,与多种生物进程密切相关,例如视网膜发育[4]、促进心肌成熟[5]、维持角膜上皮细胞稳态[6]等。SEs功能紊乱与多种疾病发生发展相关,例如促进血栓形成[7]、加剧动脉粥样硬化的炎性反应[8]、上调CXCL1、CXCL6、CXCL8等趋化因子转录水平从而增强其表达、加重肝中性粒细胞浸润而导致酒精性肝炎发生[9]等。并且,新的研究还发现一种B细胞特异性SEs与系统性红斑狼疮发病机制有关[10]。
研究表明SEs与肿瘤进程及患者预后密切相关[11]。在肿瘤细胞中,SEs通过驱动关键性致癌因子及成瘾性癌基因活跃转录,从而促进肿瘤的发生发展[12]。SEs驱动的核心转录程序对肿瘤细胞转移极其重要。而转移是肿瘤患者死亡的主要原因,因此,了解肿瘤转移的过程及机制并根据转移过程涉及的调控分子开发出新的药物靶点显得尤为重要[13]。上皮-间充质转化(epithelial-mesenchymal transition, EMT)是肿瘤转移的重要过程,可以使得癌细胞骨架发生重构从而获得迁移性和侵袭性。在肿瘤转移过程中,只有小部分细胞能获得干性特征从而克服失巢凋亡、免疫杀伤和剪切应力得以存活,这部分细胞称之为肿瘤干细胞(cancer stem cells, CSCs)。肿瘤细胞突破、溶解基底膜进入基质,在基质中有成纤维细胞、肌成纤维细胞、内皮细胞、脂肪细胞和各种骨髓源性细胞,如间充质干细胞、巨噬细胞和其他免疫细胞等基质细胞[14],这些基质细胞参与构成肿瘤微环境(tumor microenvironment, TME)。研究证实SEs可通过调控EMT、CSCs、重塑TME等过程促进肿瘤转移[15-17]。本文即对SEs的基本特点及其调控肿瘤转移进程的研究现状进行总结。
1 超级增强子的结构特征及功能增强子是位于启动子上游或下游区域的一段长度约为100~300 bp的DNA调节区域,具有增强基因转录的功能[1, 18]。而SEs可招募大量转录因子、辅因子、Mediator、RNA聚合酶Ⅱ(RNA polymeraseⅡ, RNA PolⅡ)等组成相互作用的SEs启动子环,在空间上通过内聚缩小了SEs和启动子距离,强力驱动靶基因的转录。SEs相对于增强子具有以下特点:(1)具有特异性,不同的组织或细胞,SEs种类与数量不一样;(2)跨越的DNA区域比普通增强子长一个数量级,通常为1~12.5 kb;(3)募集更多的主要转录因子,如Oct4、Sox2、Nanog及c-Myc等;(4)高度富集转录激活物及相关组蛋白,如RNA PolⅡ、转录增强子位点的RNA、Mediator、H3K4me1、H3K27ac、组蛋白乙酰转移酶p300、CREB结合蛋白(CREB-binding protein, CBP)等;(5)SEs驱动的靶基因转录活性更高;(6)SEs调控的基因表达对转录抑制剂更敏感[2, 19-20]。SEs结构特征和功能见图 1。目前,主要通过增强子上组蛋白修饰水平、转录活性标志分子结合强度及跨越DNA区域长度,采用H3K27ac、H3K4me1、p300和Mediator等进行ChIP-Seq技术鉴定SEs[21]。此外,也可通过染色质开放区域检测技术(如DNase-Seq、ATAC-Seq等)CRISPR基因编辑技术、转录组测序技术等探究SEs作用及功能[12]。
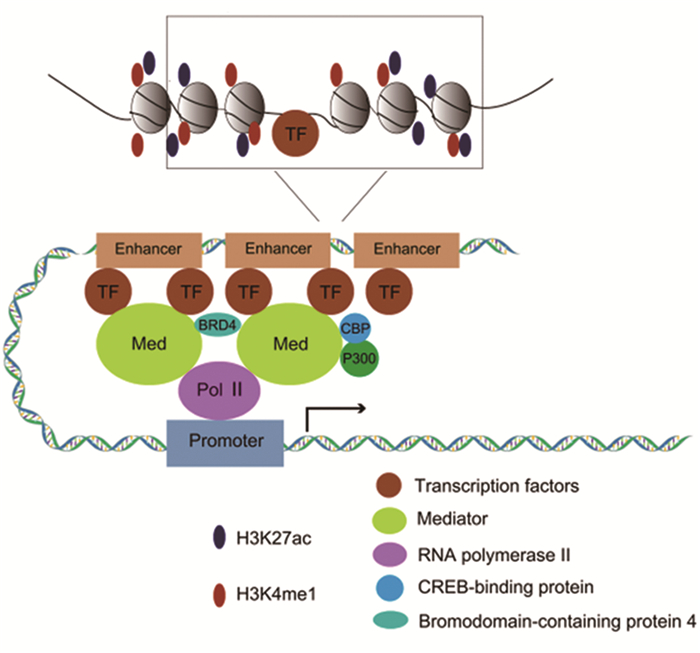
|
| 图 1 超级增强子的结构和功能 Figure 1 Structure and function of super-enhancers (SEs) |
SEs驱动基因转录主要依赖于细胞周期依赖蛋白激酶(cyclin-dependent kinases, CDKs)、溴结构域和末端外家族蛋白(bromodomain and extra-terminal domains, BETs)、RNA PolⅡ等多种转录复合物。其中,与SEs促进转录相关的CDKs主要有CDK7、CDK9、CDK12/CDK13。RNA PolⅡ羧基末端结构域(carboxy-terminal domain, CTD)磷酸化是激活转录的关键,CDK7主要通过磷酸化RNA PolⅡ第五位丝氨酸(RNA PolⅡ CTD phosphor Ser5)和第七位丝氨酸(Ser7)来启动转录过程,也可促进RNA PolⅡ CTD的第二位丝氨酸(Ser2)促进转录延伸。CDK9通过磷酸化RNA PolⅡ CTD Ser2促进基因转录延伸[22]。CDK12与CDK13结构类似,均可磷酸化RNA PolⅡ CTD Ser2调控基因转录延伸[23]。溴结构域(Bromodomain, BRD)可以识别并结合乙酰化赖氨酸。BETs是BRD蛋白中的一种,主要包括BRD2、BRD3、BRD4以及结构域睾丸特异蛋白激酶(bromodomain testis specific protein, BEDT)。大多数BRD蛋白只含有一个BRD,此外还含有一个外结构域(extra-terminal domains, ET),其可串联两个BRD。BETs主要通过与乙酰化赖氨酸位点结合,充当转录因子和基本转录复合物的支架[24]。其中,BRD4在SEs驱动的基因转录中起主要作用。BRD4首先通过识别SEs区域中H3K27ac位点从而促进基因活跃转录。此外,BRD4结构中还含有一个CTD,可通过招募阳性转录延伸因子b(positive transcription elongation factor b, P-TEFb)促进靶基因转录激活和延伸[24]。研究表明BRD4可在TP63、MET和FOSL1等肿瘤干性基因上招募Mediator和NF-κB、p65形成SEs复合物[17]。BRDT在睾丸中特异性表达。有研究发现BRDT在食管鳞状细胞癌中也存在异常表达,与转录因子ΔNp63结合共同调控SEs来驱动靶基因表达[25]。
除了上述核心的转录调节蛋白,最新研究也鉴定出某些肿瘤中关键的致癌性SEs及核心转录网络以进行靶向治疗。如在急性髓细胞白血病(acute myeloid leukemia, AML)中,McKeown等鉴定出超强活性SEs相关基因视黄酸受体α(retinoic acid receptor alpha, RARα),RARα抑制剂可显著诱导AML分化及抑制细胞增殖[26]。在胰腺导管腺癌(pancreatic ductal adenocarcinoma, PDAC)中,FOXA1可驱动SEs发生重构以促进肿瘤肝转移[27]。此外,组蛋白乙酰转移酶p300和CBP在启动子、增强子、SEs处招募H3K27ac,从而促进BRD4募集并上调相关癌基因转录[28]。
SEs还可通过激活非编码RNA(non-coding RNAs, ncRNAs)表达间接调节生物过程。SEs调控的ncRNAs在肿瘤细胞增殖、转移、耐药等过程中发挥重要的作用[29-30]。这些ncRNAs主要包括微小RNA(microRNAs, miRNAs)、长链非编码RNA(long non-coding RNAs, lncRNAs)、环状RNA(circular RNAs, circRNAs)和增强子RNA(enhancer RNAs, eRNAs)等。SEs相关lncRNAs通过在SEs与启动子之间形成的loop环状结构有利于SEs活性维持及其转录功能调控[31-33]。
2 肿瘤转移过程肿瘤转移是指肿瘤细胞离开原发灶,通过血液循环或淋巴循环到达远端组织形成新的转移灶并不断生长的过程。由于肿瘤的异质性及转移过程的多步骤复杂性,肿瘤转移患者的治疗存在较大困难[34]。肿瘤转移是“种子”与“土壤”不断相互作用的过程(肿瘤细胞即“种子”,特定转移靶器官的微环境为“土壤”)[35]。首先,单个或小群肿瘤细胞通过神经纤维、胶原纤维或细胞外基质发生迁移或侵袭,从而脱离原发灶组织。在TGF-β及相关信号蛋白的作用下,肿瘤细胞发生EMT。随后,肿瘤细胞壁通过MMPs酶及组织蛋白酶水解细胞外基质,内渗进入血管或淋巴管,随着血液或淋巴循环转移到远端靶器官,成为循环肿瘤细胞(circulating tumor cells, CTCs)。肿瘤细胞进入血液循环24小时内,由于失巢凋亡、免疫杀伤和剪切应力作用,只有不到0.1%的肿瘤细胞可以存活,能够发生转移的细胞数不足0.01%,因此,肿瘤转移成功率不高[14, 34]。有研究表明SEs在驱动肿瘤干细胞基因和转移相关基因转录过程中发挥关键作用,从而调控肿瘤转移发生的各阶段,进而促进肿瘤转移[17]。因此,此文旨在论述SEs在肿瘤转移阶段中的作用及其潜在的治疗意义。
3 SEs在肿瘤转移中的作用 3.1 SEs通过调控细胞EMT促进肿瘤转移肿瘤细胞具有迁移和侵袭能力,这使得肿瘤细胞可以进入到血管或(和)淋巴管从而扩散到循环系统中,进一步在远处的靶器官中进行定植生长[36]。细胞外基质(extracellular matrix, ECM)为肿瘤细胞迁移提供了基础,为前进的细胞体提供了屏障。细胞在组织中的迁移是一个循环反复、相互依赖的过程。首先,细胞发生骨架重构,向前缘延伸形成伪足,附着在ECM上。随后,细胞前缘或整个细胞体通过收缩产生牵引力,使细胞体及其后缘逐渐向前滑动。肿瘤细胞可通过EMT获得迁移和侵袭特性,并克服衰老和失巢凋亡[37]。在细胞EMT过程中,Slug、Snail、Twist、ETS2、ZEB1、ZEB2等EMT相关的转录因子可通过与E-钙黏蛋白(E-cadherin)启动子上的E盒结合从而抑制E-钙黏蛋白的转录,促进上皮来源的细胞失去极性向间充质表型转化[14, 38]。近年来,越来越多的研究发现,SEs可通过强力驱动EMT相关转录因子转录促进EMT过程[4, 9]。在非小细胞肺癌中,SEs通过增强EMT相关转录因子ETS2、HNF4A和JUNB的表达促进TGF-β诱导的EMT过程,而BRD4特异性抑制剂JQ1可抑制SEs驱动的ETS2、HNF4A和JUNB等基因转录,导致EMT过程受阻[15]。在肝细胞癌中,研究报道SEs通过调控靶基因AJUBA转录,从而激活Akt/GSK-3β/Snail信号通路,诱导肝癌细胞EMT,最终促进肝癌细胞侵袭和转移[39]。SEs还可通过驱动ncRNAs表达进而加速EMT过程。Li团队在肝细胞癌中鉴定了一种新型SEs驱动的lncRNA-HCCL5,可以通过上调EMT相关的转录因子Snail、Twist1、ZEB1、Slug等来促进EMT,从而增强肝细胞癌的迁移和侵袭能力。此外,转录因子ZEB1又可以结合lncRNA-HCCL5的SEs区域及启动子区域反过来驱动lncRNA-HCCL5的转录,进一步促进肝细胞癌的转移[40]。Quaking(QKI)是促进circRNA生成的主要剪切因子,Han等发现QKI的SEs与YY1结合促进circRNA生成,从而诱导肝细胞癌发生EMT,增强肝细胞癌细胞转移能力[41-42]。
3.2 SEs通过调控肿瘤微环境促进肿瘤转移肿瘤微环境(TME)主要由间质细胞、内皮细胞、免疫细胞以及大量的信号分子构成[43-44],在肿瘤中具有促进血管新生、耐药、免疫调节及转移的作用[43]。Baghel等发现肿瘤相关巨噬细胞可通过上调乳腺癌细胞MYO3A基因转录增强细胞的侵袭和转移能力[45]。成纤维细胞是间质细胞的主要类型,通过分泌细胞因子及产生细胞外基质促进肿瘤细胞迁移、侵袭及转移[46]。多项研究表明SEs可通过调控TME促进肿瘤转移。在晚期肾透明细胞癌中,有研究发现肿瘤细胞通过SEs促进大量趋化因子表达,募集中性粒细胞,从而促进肾透明细胞癌肺转移。使用BRD4抑制剂JQ1可显著降低SEs活性,抑制中性粒细胞介导的肾癌肺转移[47]。研究表明,在胰腺导管腺癌细胞及其肿瘤相关成纤维细胞(cancer-associated fibroblasts, CAFs)中存在不同的SEs。天然化合物雷公藤内酯醇(triptolide, TPL)显著抑制PDAC细胞及CAFs中SE相关的基因,如MYC、Collagen 1的转录水平,最后抑制PDAC中促肿瘤生长、增殖和侵袭等相关信号通路[48]。大数据挖掘发现胃腺癌中,SE调控的lncRNA与免疫细胞浸润密切相关。其中,lncRNA TM4SF1-AS1在胃腺癌中高表达,抑制TME中CD8+ T杀伤作用,促进肿瘤进程且影响患者预后[49]。
3.3 SEs通过调控肿瘤干细胞干性促进肿瘤转移肿瘤干细胞(CSCs)是肿瘤细胞中一小部分具有自我更新、高度异质性及无限增殖分化能力的细胞。在肿瘤转移过程中,CTCs和MICs均可获得CSCs样特性从而在靶器官内定植形成转移灶[50]。CSCs是肿瘤转移的根源这个学说已得到多数研究者的认同。因此清除CSCs可能是抑制肿瘤转移的有效手段。越来越多的研究证实SEs在干细胞命运中起重要作用。在胚胎干细胞中,SEs驱动干性调控关键转录因子Oct4、Sox2、Klf4、Nanog活跃转录,维持干细胞更新及促进胚胎发育[2]。在毛囊干细胞中,SEs促进转录因子NFIB、NFIX高表达来维持毛囊干细胞的干性[51]。在白血病干细胞中,敲除MYC基因启动子下游的SE区域,显著抑制干/祖细胞自我更新、增殖及分化能力[52]。在恶性骨肉瘤中,SE通过驱动LIF基因转录从而激活Notch1信号通路维持肿瘤干细胞干性,进而增强肿瘤细胞侵袭及转移能力[53]。在三阴性乳腺癌中,SEs驱动干性相关分子TCOF1表达维持肿瘤干细胞干性[54]。组蛋白去乙酰化酶1(histone deacetylases 1, HDAC1)维持着乳腺癌CSCs,研究表明HDAC1和HDAC3共同调控HDAC7表达,维持SEs附近H3K27ac活性,促进乳腺癌干细胞中SEs驱动的癌基因表达[55]。Dai等发现在结直肠癌中CDK12通过维持SE相关靶基因CCDC137活跃转录,促进结直肠癌中CSCs自我更新,从而促进肿瘤肝转移[56]。
4 SEs的抑制剂与肿瘤进程肿瘤细胞可以通过染色体易位、基因扩增、突变等方式获得超级增强子。而在许多肿瘤中,关键性癌基因的高表达依赖于SEs驱动的活跃转录能力,如神经母细胞瘤中的MYCN基因、黑色素瘤中MITF基因、骨肉瘤细胞中的F3基因[57]。干预这些基因的表达将显著影响肿瘤细胞的生存、增殖及转移,而在正常细胞中不存在这种依赖性,表明肿瘤细胞对SE活跃转录能力的高度依赖将可能导致转录成瘾性癌基因的产生,针对这一特征可以开发靶向超级增强子转录复合物关键组分的小分子抑制剂以消除癌基因成瘾性从而干预肿瘤进程。
SEs相关的抑制剂目前主要包括CDKs相关抑制剂、BETs相关抑制剂等。其中,CDKs相关抑制剂主要有THZ1、THZ2及SR-4835。THZ1是CDK7的共价抑制剂,通过其丙烯酰胺键竞争性地与CDK7中第312位半胱氨酸的ATP结构域结合,进而抑制CDK7激酶活性[58]。THZ2是THZ1的类似物,是将THZ1丙烯酰胺键上的4-丙烯酰胺-苯甲酰胺结构修饰为3-丙烯酰胺-苯甲酰胺的产物。THZ2与THZ1相比,药代动力学得以显著改善,体内半衰期提高了5倍[59]。研究表明在骨肉瘤中,THZ2可以抑制SEs驱动的成瘾性癌基因转录,在体内外都具有强大的抗骨肉瘤活性[60]。SR-4835是CDK12/13的共价抑制剂,在三阴性乳腺癌中,SR-4835触发内含子多聚腺苷酸化切割,抑制DNA损伤翻译蛋白表达,与PARP抑制剂和破坏DNA的化疗药物均具有协同作用,可引起乳腺癌细胞凋亡[61]。而JQ1是研发的第一个BETs抑制剂,其通过竞争性地结合BRD4的乙酰赖氨酸识别区域,干扰BRD4识别SEs区域的H3K27ac位点,进而抑制SEs驱动的基因转录[62]。I-BET151是BETs抑制剂衍生物,属于3,5-二甲基异恶唑,最早应用于混合谱系白血病(mixed lineage leukaemia, MLL),其通过置换染色质中BRD2/3/4、聚合酶相关因子(polymerase associated factor, PAFc)、超延伸复合物(super elongation complex, SEC)来抑制关键基因BCL-2、C-MYC、CDK6转录,对MLL治疗具有巨大价值[63]。随后研究也发现其在卵巢癌[64]、胶质母细胞瘤[65]及多发性骨髓瘤[66]等肿瘤中起作用。A-485是CBP/P300抑制剂,通过与乙酰辅酶A竞争来抑制CBP/P300活性,在生长激素垂体腺瘤中抑制PI3K/Akt/mTOR通路,起到抗肿瘤作用[67]。Cortistatin A可选择性抑制Mediator,并在急性髓系白血病中上调抑癌基因CEBPA、IRF8、TRF1和ETV6表达[68]。SEs相关抑制剂总结见表 1。
肿瘤的异质性使肿瘤的治疗尤为困难,肿瘤转移是患者死亡的主要原因,癌基因存在转录成瘾性,而SEs具有强大的转录活性,可以促进肿瘤转移,是导致癌基因转录成瘾性的主要原因。因此,开发针对SEs的靶点,在肿瘤的治疗上具有很好的应用前景。CRISPR-Cas9基因编辑技术可以靶向敲除肿瘤细胞的SEs,为SEs的研究与治疗带来了新的策略。
利益冲突声明:
所有作者均声明不存在利益冲突。
作者贡献:
郭彩瑶:文献查阅、文章撰写
王宇:文献检索
戴伟:资料总结
刘圣兰:立题构思及论文指导
| [1] |
Banerji J, Rusconi S, Schaffner W. Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences[J]. Cell, 1981, 27(2 Pt 1): 299-308. |
| [2] |
Whyte WA, Orlando DA, Hnisz D, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes[J]. Cell, 2013, 153(2): 307-319. DOI:10.1016/j.cell.2013.03.035 |
| [3] |
Hnisz D, Abraham BJ, Lee TI, et al. Super-enhancers in the control of cell identity and disease[J]. Cell, 2013, 155(4): 934-947. DOI:10.1016/j.cell.2013.09.053 |
| [4] |
Honnell V, Norrie JL, Patel AG, et al. Identification of a modular super-enhancer in murine retinal development[J]. Nat Commun, 2022, 13(1): 253. DOI:10.1038/s41467-021-27924-y |
| [5] |
Sakamoto T, Batmanov K, Wan S, et al. The nuclear receptor ERR cooperates with the cardiogenic factor GATA4 to orchestrate cardiomyocyte maturation[J]. Nat Commun, 2022, 13(1): 1991. DOI:10.1038/s41467-022-29733-3 |
| [6] |
Li M, Huang H, Li L, et al. Core transcription regulatory circuitry orchestrates corneal epithelial homeostasis[J]. Nat Commun, 2021, 12(1): 420. DOI:10.1038/s41467-020-20713-z |
| [7] |
Stefanucci L, Frontini M. Non-coding genetic variation in regulatory elements determines thrombosis and hemostasis phenotypes[J]. J Thromb Haemost, 2022, 20(8): 1759-1765. DOI:10.1111/jth.15754 |
| [8] |
Li X, Zhu R, Jiang H, et al. Autophagy enhanced by curcumin ameliorates inflammation in atherogenesis via the TFEB-P300-BRD4 axis[J]. Acta Pharm Sin B, 2022, 12(5): 2280-2299. DOI:10.1016/j.apsb.2021.12.014 |
| [9] |
Liu M, Cao S, He L, et al. Super enhancer regulation of cytokine-induced chemokine production in alcoholic hepatitis[J]. Nat Commun, 2021, 12(1): 4560. DOI:10.1038/s41467-021-24843-w |
| [10] |
Zhang Y, Day K, Absher DM. STAT3-mediated allelic imbalance of novel genetic variant Rs1047643 and B-cell-specific super-enhancer in association with systemic lupus erythematosus[J]. Elife, 2022, 11: e72837. DOI:10.7554/eLife.72837 |
| [11] |
Tang F, Yang Z, Tan Y, et al. Super-enhancer function and its application in cancer targeted therapy[J]. NPJ Precis Oncol, 2020, 4: 2. DOI:10.1038/s41698-020-0108-z |
| [12] |
Thandapani P. Super-enhancers in cancer[J]. Pharmacol Ther, 2019, 199: 129-138. DOI:10.1016/j.pharmthera.2019.02.014 |
| [13] |
Chambers AF, Groom AC, Macdonald IC. Dissemination and growth of cancer cells in metastatic sites[J]. Nat Rev Cancer, 2002, 2(8): 563-572. DOI:10.1038/nrc865 |
| [14] |
Valastyan S, Weinberg RA. Tumor metastasis: molecular insights and evolving paradigms[J]. Cell, 2011, 147(2): 275-292. DOI:10.1016/j.cell.2011.09.024 |
| [15] |
Chang H, Liu Y, Xue M, et al. Synergistic action of master transcription factors controls epithelial-to-mesenchymal transition[J]. Nucleic Acids Res, 2016, 44(6): 2514-2527. DOI:10.1093/nar/gkw126 |
| [16] |
Evan GI, Hah N, Littlewood TD, et al. Re-engineering the Pancreas Tumor Microenvironment: A "Regenerative Program" Hacked[J]. Clin Cancer Res, 2017, 23(7): 1647-1655. DOI:10.1158/1078-0432.CCR-16-3275 |
| [17] |
Dong J, Li J, Li Y, et al. Transcriptional super-enhancers control cancer stemness and metastasis genes in squamous cell carcinoma[J]. Nat Commun, 2021, 12(1): 3974. DOI:10.1038/s41467-021-24137-1 |
| [18] |
Banerji J, Olson L, Schaffner W. A lymphocyte-specific cellular enhancer is located downstream of the joining region in immunoglobulin heavy chain genes[J]. Cell, 1983, 33(3): 729-740. DOI:10.1016/0092-8674(83)90015-6 |
| [19] |
Pott S, Lieb JD. What are super-enhancers?[J]. Nat Genet, 2015, 47(1): 8-12. DOI:10.1038/ng.3167 |
| [20] |
Bulger M, Groudine M. Functional and mechanistic diversity of distal transcription enhancers[J]. Cell, 2011, 144(3): 327-339. DOI:10.1016/j.cell.2011.01.024 |
| [21] |
Sur I, Taipale J. The role of enhancers in cancer[J]. Nat Rev Cancer, 2016, 16(8): 483-493. DOI:10.1038/nrc.2016.62 |
| [22] |
Kolloch L, Kreinest T, Meisterernst M, et al. Control of Expression of Key Cell Cycle Enzymes Drives Cell Line-Specific Functions of CDK7 in Human PDAC Cells[J]. Int J Mol Sci, 2022, 23(2): 812. DOI:10.3390/ijms23020812 |
| [23] |
Paculová H, Kohoutek J. The emerging roles of CDK12 in tumorigenesis[J]. Cell Div, 2017, 12: 7. DOI:10.1186/s13008-017-0033-x |
| [24] |
Borck PC, Guo LW, Plutzky J. BET Epigenetic Reader Proteins in Cardiovascular Transcriptional Programs[J]. Circ Res, 2020, 126(9): 1190-1208. DOI:10.1161/CIRCRESAHA.120.315929 |
| [25] |
Wang X, Kutschat AP, Yamada M, et al. Bromodomain protein BRDT directs ΔNp63 function and super-enhancer activity in a subset of esophageal squamous cell carcinomas[J]. Cell Death Differ, 2021, 28(7): 2207-2220. DOI:10.1038/s41418-021-00751-w |
| [26] |
Mckeown MR, Corces MR, Eaton ML, et al. Superenhancer Analysis Defines Novel Epigenomic Subtypes of Non-APL AML, Including an RARα Dependency Targetable by SY-1425, a Potent and Selective RARα Agonist[J]. Cancer Discov, 2017, 7(10): 1136-1153. DOI:10.1158/2159-8290.CD-17-0399 |
| [27] |
Kim MP, Li X, Deng J, et al. Oncogenic KRAS Recruits an Expansive Transcriptional Network through Mutant p53 to Drive Pancreatic Cancer Metastasis[J]. Cancer Discov, 2021, 11(8): 2094-2111. DOI:10.1158/2159-8290.CD-20-1228 |
| [28] |
Chen Q, Yang B, Liu X, et al. Histone acetyltransferases CBP/p300 in tumorigenesis and CBP/p300 inhibitors as promising novel anticancer agents[J]. Theranostics, 2022, 12(11): 4935-4948. DOI:10.7150/thno.73223 |
| [29] |
Wang Y, Nie H, He X, et al. The emerging role of super enhancer-derived noncoding RNAs in human cancer[J]. Theranostics, 2020, 10(24): 11049-11062. DOI:10.7150/thno.49168 |
| [30] |
Suzuki HI, Young RA, Sharp PA. Super-Enhancer-Mediated RNA Processing Revealed by Integrative MicroRNA Network Analysis[J]. Cell, 2017, 168(6): 1000-1014. DOI:10.1016/j.cell.2017.02.015 |
| [31] |
Angrand PO, Vennin C, Le Bourhis X, et al. The role of long non-coding RNAs in genome formatting and expression[J]. Front Genet, 2015, 6: 165. |
| [32] |
Castro-Oropeza R, Melendez-Zajgla J, Maldonado V, et al. The emerging role of lncRNAs in the regulation of cancer stem cells[J]. Cell Oncol (Dordr), 2018, 41(6): 585-603. |
| [33] |
Xie JJ, Jiang YY, Jiang Y, et al. Super-Enhancer-Driven Long Non-Coding RNA LINC01503, Regulated by TP63, Is Over-Expressed and Oncogenic in Squamous Cell Carcinoma[J]. Gastroenterology, 2018, 154(8): 2137-2151. DOI:10.1053/j.gastro.2018.02.018 |
| [34] |
Fidler IJ. The pathogenesis of cancer metastasis: the 'seed and soil' hypothesis revisited[J]. Nat Rev Cancer, 2003, 3(6): 453-458. DOI:10.1038/nrc1098 |
| [35] |
Paget S. The distribution of secondary growths in cancer of the breast. 1889[J]. Cancer Metastasis Rev, 1989, 8(2): 98-101. |
| [36] |
Massagué J, Obenauf AC. Metastatic colonization by circulating tumour cells[J]. Nature, 2016, 529(7586): 298-306. DOI:10.1038/nature17038 |
| [37] |
Pastushenko I, Blanpain C. EMT Transition States during Tumor Progression and Metastasis[J]. Trends Cell Biol, 2019, 29(3): 212-226. DOI:10.1016/j.tcb.2018.12.001 |
| [38] |
Ye X, Tam WL, Shibue T, et al. Distinct EMT programs control normal mammary stem cells and tumour-initiating cells[J]. Nature, 2015, 525(7568): 256-260. DOI:10.1038/nature14897 |
| [39] |
Zhang C, Wei S, Sun WP, et al. Super-enhancer-driven AJUBA is activated by TCF4 and involved in epithelial-mesenchymal transition in the progression of Hepatocellular Carcinoma[J]. Theranostics, 2020, 10(20): 9066-9082. DOI:10.7150/thno.45349 |
| [40] |
Peng L, Jiang B, Yuan X, et al. Super-Enhancer-Associated Long Noncoding RNA HCCL5 Is Activated by ZEB1 and Promotes the Malignancy of Hepatocellular Carcinoma[J]. Cancer Res, 2019, 79(3): 572-584. DOI:10.1158/0008-5472.CAN-18-0367 |
| [41] |
Han J, Meng J, Chen S, et al. YY1 Complex Promotes Quaking Expression via Super-Enhancer Binding during EMT of Hepatocellular Carcinoma[J]. Cancer Res, 2019, 79(7): 1451-1464. DOI:10.1158/0008-5472.CAN-18-2238 |
| [42] |
Conn SJ, Pillman KA, Toubia J, et al. The RNA binding protein quaking regulates formation of circRNAs[J]. Cell, 2015, 160(6): 1125-1134. DOI:10.1016/j.cell.2015.02.014 |
| [43] |
Gysler SM, Drapkin R. Tumor innervation: peripheral nerves take control of the tumor microenvironment[J]. J Clin Invest, 2021, 131(11): e147276. DOI:10.1172/JCI147276 |
| [44] |
Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis[J]. Nat Med, 2013, 19(11): 1423-1437. DOI:10.1038/nm.3394 |
| [45] |
Baghel KS, Tewari BN, Shrivastava R, et al. Macrophages promote matrix protrusive and invasive function of breast cancer cells via MIP-1β dependent upregulation of MYO3A gene in breast cancer cells[J]. Oncoimmunology, 2016, 5(7): e1196299. DOI:10.1080/2162402X.2016.1196299 |
| [46] |
Erdogan B, Webb DJ. Cancer-associated fibroblasts modulate growth factor signaling and extracellular matrix remodeling to regulate tumor metastasis[J]. Biochem Soc Trans, 2017, 45(1): 229-236. DOI:10.1042/BST20160387 |
| [47] |
Wang M. Epigenetic control of inflammatory cells[J]. Nat Rev Nephrol, 2020, 16(6): 316. |
| [48] |
Noel P, Hussein S, Ng S, et al. Triptolide targets super-enhancer networks in pancreatic cancer cells and cancer-associated fibroblasts[J]. Oncogenesis, 2020, 9(11): 100. DOI:10.1038/s41389-020-00285-9 |
| [49] |
Peng L, Peng JY, Cai DK, et al. Immune Infiltration and Clinical Outcome of Super-Enhancer-Associated lncRNAs in Stomach Adenocarcinoma[J]. Front Oncol, 2022, 12: 780493. DOI:10.3389/fonc.2022.780493 |
| [50] |
Chaffer CL, Weinberg RA. A perspective on cancer cell metastasis[J]. Science, 2011, 331(6024): 1559-1564. DOI:10.1126/science.1203543 |
| [51] |
Adam RC, Yang H, Ge Y, et al. NFI transcription factors provide chromatin access to maintain stem cell identity while preventing unintended lineage fate choices[J]. Nat Cell Biol, 2020, 22(6): 640-650. DOI:10.1038/s41556-020-0513-0 |
| [52] |
Bahr C, Von Paleske L, Uslu VV, et al. A Myc enhancer cluster regulates normal and leukaemic haematopoietic stem cell hierarchies[J]. Nature, 2018, 553(7689): 515-520. DOI:10.1038/nature25193 |
| [53] |
Lu B, He Y, He J, et al. Epigenetic Profiling Identifies LIF as a Super-enhancer-Controlled Regulator of Stem Cell-like Properties in Osteosarcoma[J]. Mol Cancer Res, 2020, 18(1): 57-67. DOI:10.1158/1541-7786.MCR-19-0470 |
| [54] |
Hu J, Lai Y, Huang H, et al. TCOF1 upregulation in triple-negative breast cancer promotes stemness and tumour growth and correlates with poor prognosis[J]. Br J Cancer, 2022, 126(1): 57-71. DOI:10.1038/s41416-021-01596-3 |
| [55] |
Caslini C, Hong S, Ban YJ, et al. HDAC7 regulates histone 3 lysine 27 acetylation and transcriptional activity at super-enhancer-associated genes in breast cancer stem cells[J]. Oncogene, 2019, 38(39): 6599-6614. DOI:10.1038/s41388-019-0897-0 |
| [56] |
Dai W, Wu J, Peng X, et al. CDK12 orchestrates super-enhancer-associated CCDC137 transcription to direct hepatic metastasis in colorectal cancer[J]. Clin Transl Med, 2022, 12(10): e1087. |
| [57] |
Morrow JJ, Bayles I, Funnell APW, et al. Corrigendum: Positively selected enhancer elements endow osteosarcoma cells with metastatic competence[J]. Nat Med, 2018, 24(4): 525. |
| [58] |
Kwiatkowski N, Zhang T, Rahl PB, et al. Targeting transcription regulation in cancer with a covalent CDK7 inhibitor[J]. Nature, 2014, 511(7511): 616-620. DOI:10.1038/nature13393 |
| [59] |
Wang Y, Zhang T, Kwiatkowski N, et al. CDK7-dependent transcriptional addiction in triple-negative breast cancer[J]. Cell, 2015, 163(1): 174-186. DOI:10.1016/j.cell.2015.08.063 |
| [60] |
Zhang J, Liu W, Zou C, et al. Targeting Super-Enhancer-Associated Oncogenes in Osteosarcoma with THZ2, a Covalent CDK7 Inhibitor[J]. Clin Cancer Res, 2020, 26(11): 2681-2692. DOI:10.1158/1078-0432.CCR-19-1418 |
| [61] |
Quereda V, Bayle S, Vena F, et al. Therapeutic Targeting of CDK12/CDK13 in Triple-Negative Breast Cancer[J]. Cancer Cell, 2019, 36(5): 545-558. DOI:10.1016/j.ccell.2019.09.004 |
| [62] |
Liu Z, Wang P, Chen H, et al. Drug Discovery Targeting Bromodomain-Containing Protein 4[J]. J Med Chem, 2017, 60(11): 4533-4558. DOI:10.1021/acs.jmedchem.6b01761 |
| [63] |
Dawson MA, Prinjha RK, Dittmann A, et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia[J]. Nature, 2011, 478(7370): 529-533. DOI:10.1038/nature10509 |
| [64] |
Zhang Z, Ma P, Jing Y, et al. BET Bromodomain Inhibition as a Therapeutic Strategy in Ovarian Cancer by Downregulating FoxM1[J]. Theranostics, 2016, 6(2): 219-230. DOI:10.7150/thno.13178 |
| [65] |
Pastori C, Kapranov P, Penas C, et al. The Bromodomain protein BRD4 controls HOTAIR, a long noncoding RNA essential for glioblastoma proliferation[J]. Proc Natl Acad Sci U S A, 2015, 112(27): 8326-8331. DOI:10.1073/pnas.1424220112 |
| [66] |
Abruzzese MP, Bilotta MT, Fionda C, et al. Inhibition of bromodomain and extra-terminal (BET) proteins increases NKG2D ligand MICA expression and sensitivity to NK cell-mediated cytotoxicity in multiple myeloma cells: role of cMYC-IRF4-miR-125b interplay[J]. J Hematol Oncol, 2016, 9(1): 134. DOI:10.1186/s13045-016-0362-2 |
| [67] |
Ji C, Xu W, Ding H, et al. The p300 Inhibitor A-485 Exerts Antitumor Activity in Growth Hormone Pituitary Adenoma[J]. J Clin Endocrinol Metab, 2022, 107(6): e2291-e2300. DOI:10.1210/clinem/dgac128 |
| [68] |
Pelish HE, Liau BB, Nitulescu Ⅱ, et al. Mediator kinase inhibition further activates super-enhancer-associated genes in AML[J]. Nature, 2015, 526(7572): 273-276. DOI:10.1038/nature14904 |
| [69] |
Wang J, Zhang R, Lin Z, et al. CDK7 inhibitor THZ1 enhances antiPD-1 therapy efficacy via the p38α/MYC/PD-L1 signaling in non-small cell lung cancer[J]. J Hematol Oncol, 2020, 13(1): 99. DOI:10.1186/s13045-020-00926-x |
| [70] |
Jiang YY, Lin DC, Mayakonda A, et al. Targeting super-enhancer-associated oncogenes in oesophageal squamous cell carcinoma[J]. Gut, 2017, 66(8): 1358-1368. DOI:10.1136/gutjnl-2016-311818 |
| [71] |
Rasool RU, Natesan R, Deng Q, et al. CDK7 Inhibition Suppresses Castration-Resistant Prostate Cancer through MED1 Inactivation[J]. Cancer Discov, 2019, 9(11): 1538-1555. DOI:10.1158/2159-8290.CD-19-0189 |
| [72] |
Lovén J, Hoke HA, Lin CY, et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers[J]. Cell, 2013, 153(2): 320-334. DOI:10.1016/j.cell.2013.03.036 |
| [73] |
Ni M, Li J, Zhao H, et al. BRD4 inhibition sensitizes cervical cancer to radiotherapy by attenuating DNA repair[J]. Oncogene, 2021, 40(15): 2711-2724. DOI:10.1038/s41388-021-01735-3 |
| [74] |
Dai X, Gan W, Li X, et al. Prostate cancer-associated SPOP mutations confer resistance to BET inhibitors through stabilization of BRD4[J]. Nat Med, 2017, 23(9): 1063-1071. DOI:10.1038/nm.4378 |
| [75] |
Shu S, Lin CY, He HH, et al. Response and resistance to BET bromodomain inhibitors in triple-negative breast cancer[J]. Nature, 2016, 529(7586): 413-417. DOI:10.1038/nature16508 |
| [76] |
Zuber J, Shi J, Wang E, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia[J]. Nature, 2011, 478(7370): 524-528. DOI:10.1038/nature10334 |
| [77] |
Yoshino S, Yokoyama T, Sunami Y, et al. Trib1 promotes acute myeloid leukemia progression by modulating the transcriptional programs of Hoxa9[J]. Blood, 2021, 137(1): 75-88. DOI:10.1182/blood.2019004586 |
| [78] |
Berthon C, Raffoux E, Thomas X, et al. Bromodomain inhibitor OTX015 in patients with acute leukaemia: a dose-escalation, phase 1 study[J]. Lancet Haematol, 2016, 3(4): e186-e195. DOI:10.1016/S2352-3026(15)00247-1 |
| [79] |
Chen J, Nelson C, Wong M, et al. Targeted Therapy of TERT-Rearranged Neuroblastoma with BET Bromodomain Inhibitor and Proteasome Inhibitor Combination Therapy[J]. Clin Cancer Res, 2021, 27(5): 1438-1451. DOI:10.1158/1078-0432.CCR-20-3044 |
 2023, Vol. 50
2023, Vol. 50



