文章信息
- 基于TCGA数据库乳头状甲状腺癌miRNA预后风险模型的建立与分析
- Establishment and Analysis of MicroRNA Prognostic Risk Model of Papillary Thyroid Carcinoma Based on TCGA Database
- 肿瘤防治研究, 2020, 47(4): 262-267
- Cancer Research on Prevention and Treatment, 2020, 47(4): 262-267
- http://www.zlfzyj.com/CN/10.3971/j.issn.1000-8578.2020.19.0947
- 收稿日期: 2019-07-24
- 修回日期: 2019-11-01
甲状腺癌是人类内分泌系统和头颈部最常见的恶性肿瘤之一,其发病率在国内外均呈上升趋势[1]。许多危险因素与甲状腺癌的发病相关,包括电离辐射、雌激素、碘摄入和遗传因素等。研究表明,手术联合放化疗以及分子靶向治疗有助于控制甲状腺癌进展,延长患者的无进展生存期,提高总体生存率[2-4]。然而由于早期缺乏典型的临床表现,甲状腺癌很容易被患者自身忽略,从而错过最佳治疗时间[5]。分化型甲状腺癌一般预后较好,但当其失分化为碘难治性甲状腺癌时便进展迅速,病死率较高,预后较差[6]。乳头状甲状腺癌(papillary thyroid carcinoma, PTC)是最常见的甲状腺恶性肿瘤,约占甲状腺癌的80%[7]。据报道,在发达国家PTC的发病率呈上升趋势[8-10],PTC的不良预后因素包括年龄、性别、肿瘤大小等[7]。Grogan等[11]对269例PTC患者进行平均27年的长期随访,结果发现超过25%的患者复发,并且在11%的病例中,PTC复发发生在治疗后20多年。因此,评估患者的疾病进展情况,寻找影响PTC患者预后的相关因素值得重视[12]。
微小RNA(miRNA)是一类短的非编码RNA分子,仅含有19~22个核苷酸。它们是高度保守的分子,通过以完全或不完全互补的方式与靶基因的3’端非翻译区(3’-UTR)结合,参与靶基因的调节[13]。越来越多的研究证实,miRNA广泛参与人类癌症的发生和进展,包括甲状腺癌。miRNA在肿瘤发生和发展过程中参与细胞增殖、凋亡和肿瘤侵袭等多种恶性生物学行为[14]。虽有研究报道某些miRNA可以作为PTC的分子标志物,但目前仍缺乏敏感的miRNA组合标志物以评估PTC患者的预后。因此,本研究通过使用TCGA数据库,结合生物信息学分析对PTC组织中差异表达的miRNA进行筛选,利用Cox和Lasso回归分析出与PTC患者预后相关的miRNA,构建基于miRNA表达的预测PTC患者预后的生存模型。
1 资料与方法 1.1 原始数据的下载与处理在TCGA官方网站上下载PTC患者的miRNA测序数据和患者的临床数据。其中癌组织514例,正常甲状腺组织59例。使用Perl5.24.3软件将原始miRNA测序数据转换成miRNA表达矩阵。在分析过程中,若有多个探针检测同一miRNA表达量,则取该miRNA表达量的平均值作为该miRNA的表达值。对于患者临床资料的分析,删除生存时间未知和生存时间为0的患者临床信息。
1.2 差异miRNA的筛选以|log foldchange|≥2,错误发现率FDR < 0.05为筛选条件,使用R3.6.0软件中的edgeR包筛选出在甲状腺癌中差异表达的miRNA。使用ggplot2和heatmap软件程序包绘制差异基因的火山图和热图。
1.3 Cox/Lasso回归分析结合患者的生存信息,首先对上述得到的差异miRNA进行单因素Cox回归分析,计算每个miRNA与甲状腺癌患者生存的风险比(hazard ratio, HR)和P值,以P < 0.05的标准筛选出与甲状腺癌患者预后显著相关的miRNA。将这些miRNA进一步行Lasso回归分析,目的减少基因之间共线性的影响,防止后续构建的风险模型变量过度拟合。Lasso回归使用交叉验证以确定参数,得到合适的模型。再将Lasso回归得到的miRNA进行多因素Cox回归分析,计算每个miRNA的多因素回归系数,构建风险评分方程。
1.4 风险预后模型的建立与分析根据上述多因素Cox回归分析的结果,构建基于miRNA表达的风险评分方程risk score。按照文献报道使用公式:Risk score=β1×miRNA1EXP+β2×miRNA2EXP+......+βn×miRNAnEXP [15-16]。式中β为相应miRNA的多因素回归系数,miRNAEXP为相应miRNA的表达量。根据risk score数值的中位值,将PTC患者分为高风险评分组和低风险评分组。利用R3.6.0软件绘制模型预测预后的列线图,并比较两组患者之间生存的差异。利用R3.6.0软件绘制模型的ROC曲线和校准曲线以评价模型的敏感度和特异性。使用Survival ROC软件程序包计算受试者工作特征曲线(ROC)下面积(AUC)的数值。
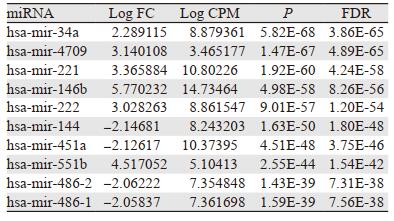
2 结果 2.1 差异miRNA的筛选利用|logfoldchange|≥2,FDR < 0.05为筛选条件,在PTC组织中共筛选到差异表达的miRNA75个。其中,上调表达的有70个、下调表达的有5个。根据FDR数值的排序前十位差异表达的miRNA,见表 1。图 1为差异miRNA相应的火山图和热图。图中红色表示与正常甲状腺组织相比,该基因在PTC组织中表达上调;绿色表示与正常甲状腺组织相比,该基因在PTC组织中表达下调。
|
|
| 图 1 PTC组织中差异表达的miRNA火山图(A)和热图(B) Figure 1 Volcano plot(A) and heat map(B) of differentially-expressed miRNA in PTC tissues |
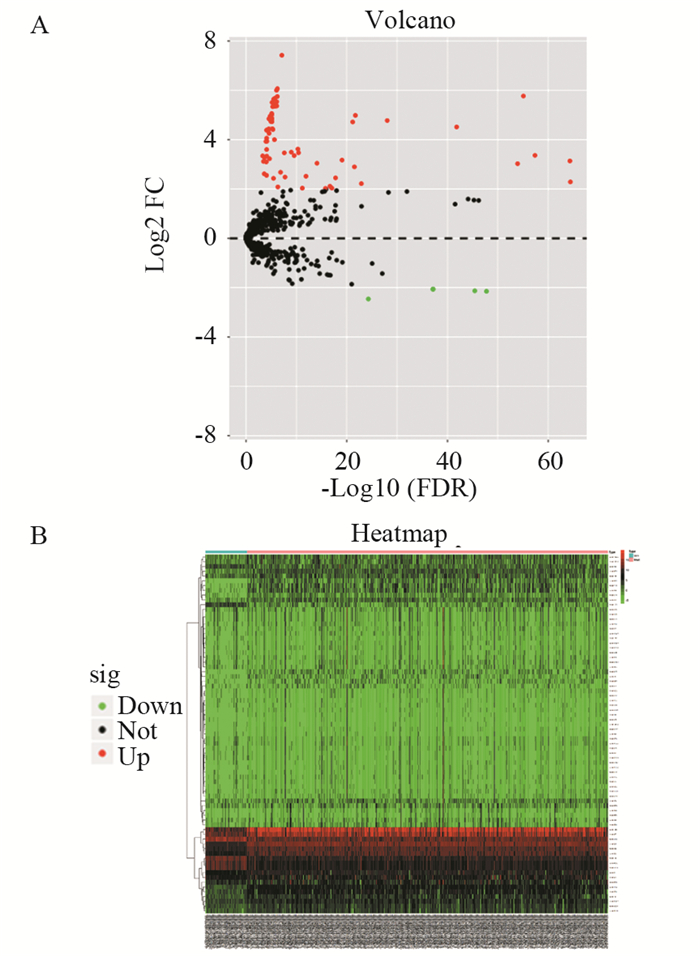
对上述差异表达的miRNA进行单因素Cox回归分析,计算相应miRNA与甲状腺癌患者的HR和P值,结果共有9个miRNA与甲状腺癌患者生存相关(P < 0.05):hsa-mir-6730、hsa-mir-4709、hsa-mir-196a-2、hsa-mir-146b、hsa-mir-6860、hsa-mir-509-3、hsa-mir-513c、hsa-mir-515-1、hsa-mir-551b。进一步使用lasso回归对这些miRNA进行筛选,见图 2A,并使用交叉验证建立模型,见图 2B。结果共有8个miRNA(hsa-mir-6730, hsa-mir-4709, hsa-mir-196a-2, hsa-mir-146b, hsa-mir-6860, hsa-mir-509-3, hsa-mir-513c, hsa-mir-515-1)纳入分析模型。
|
| A: the Lasso regression model and cross validation method were utilized to screen miRNA. When the number of variables was 8, the partial likelihood deviation was the minimum, corresponding to the minimum λ value; B: the regression coefficient map of miRNA in lasso model, and double dashed lines showed the 1-fold standard error of the minimum partial likelihood deviation. 图 2 Lasso回归分析筛选miRNA(A)和交叉验证结果(B) Figure 2 miRNA screened by Lasso regression analysis(A) and cross validation results(B) |
对上述8个miRNA进行多因素Cox回归分析,根据相应的回归系数,建立风险评分方程。Risk score=0.41×hsa-mir-196a-2EXP–0.14×hsa-mir-146bEXP–0.22×hsa-mir-4709EXP+0.83×hsa-mir-509-3EXP –0.03×hsa-mir-513cEXP +0.36×hsa-mir-515-1EXP – 0.33×hsa-mir-6730EXP –0.63×hsa-mir-6860 EXP。根据此方程,计算每位甲状腺癌患者的risk score数值,并根据risk score数值的中位值,将甲状腺癌患者分为高风险评分组和低风险评分组。
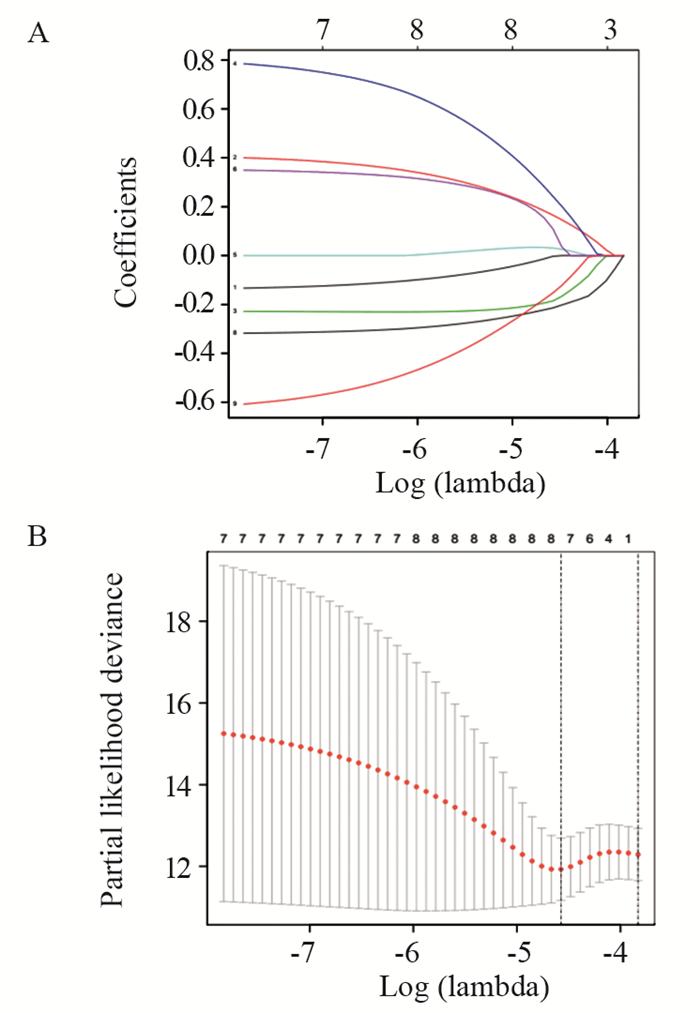
2.4 风险预后模型分析根据上述结果,利用R3.6.0软件绘制基于8个miRNA的组合预测甲状腺癌患者生存的列线图,见图 3。
|
| 图 3 基于8个miRNA表达预测甲状腺癌患者生存的列线图 Figure 3 Nomogram of thyroid cancer patients' survival predicted based on eight miRNA expression |
利用R3.6.0软件绘制模型的ROC曲线以评价模型的敏感度和特异性,见图 4。我们所构建的miRNA模型预测患者3年生存率和5年生存率的ROC曲线下面积数值AUC分别为0.860和0.896,这表明模型具有较好的敏感度和特异性。同时,校准曲线的结果也显示构建的模型可靠,这些结果表明我们构建的miRNA预后模型可以较准确的预测甲状腺癌患者的生存,见图 5。
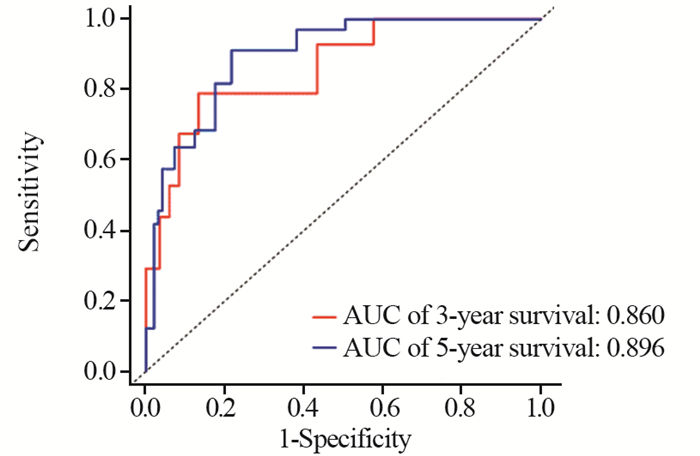
|
| 图 4 风险预后模型的ROC曲线 Figure 4 ROC curve of risk prognosis model |
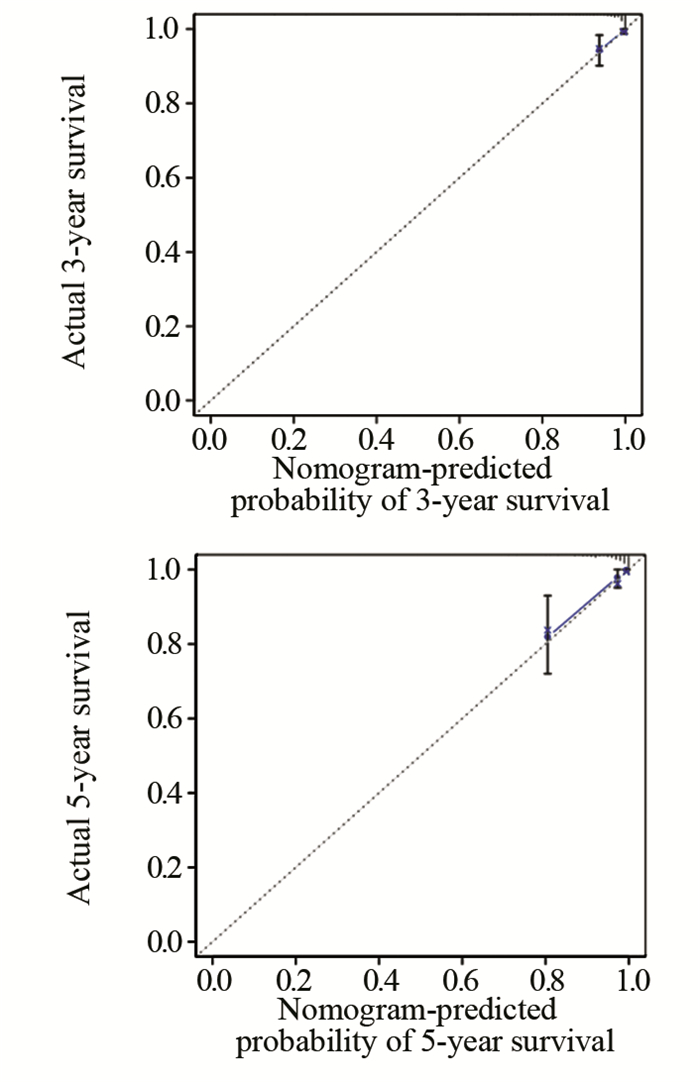
|
| 图 5 预测模型的校准曲线 Figure 5 Calibration curve of prediction model |
利用R3.6.0软件中的survival程序包分析高低风险评分组患者的生存差异,绘制生存点图,图中绿色点表示未死亡患者,红色点表示死亡患者。随着risk score数值的升高,高风险评分组患者的死亡人数显著多于低风险评分组,说明高风险评分组的患者生存率较差,见图 6。
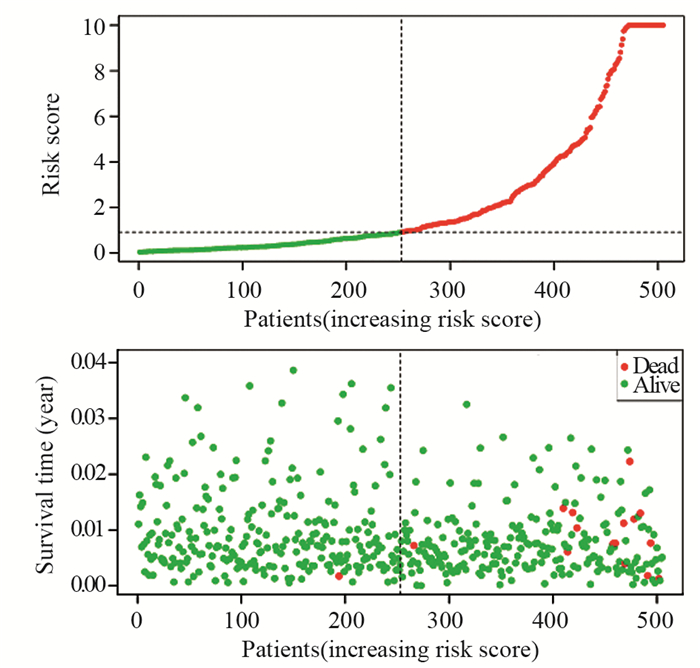
|
| The green dots in the figure represented the surviving PTC patients, and the red dots represented the dead PTC patients. The dotted line represented the median value of risk score. The left side of the dotted line represented the low risk score group, and the right side of the dotted line represented the high risk score group. With the increase of risk score in PTC patients, the number of red dots increased gradually, and the number of dead PTC patients increased. It showed that the high risk group had poorer survival and higher risk of death. 图 6 高低风险评分组患者的生存点图 Figure 6 Survival points diagram of patients in high and low risk groups |
甲状腺癌是最常见的内分泌恶性肿瘤。大多数甲状腺癌起源于甲状腺滤泡细胞。甲状腺癌可分为乳头状癌(75%~80%)、滤泡状癌(10%~15%)、甲状腺未分化癌(0.2%~2%)和由滤泡旁C细胞来源的髓样甲状腺癌(5%~10%)[17]。许多研究表明,miRNA与甲状腺癌的发生和进展有关。Yue等[18]研究表明,miR-7可能通过p21活化激酶1(PAK1)调节甲状腺癌细胞的生长、迁移和侵袭;Wang等[19]研究表明,碘可以通过下调miR-422a、上调MAPK1的表达,促进甲状腺癌的发生;Dong等[20]研究发现miR-141可以通过调控胰岛素受体底物2(IRS2)调控甲状腺癌细胞生长和转移,提示miR-141可以作为治疗甲状腺癌患者的潜在分子靶点;此外,miR-497也被认为是一种通过抑制脑源性神经营养因子发挥作用的甲状腺癌肿瘤抑制因子[21]。因此,miRNA可以作为甲状腺癌诊断、治疗或者预后的标志物。
本研究中,我们通过TCGA数据库构建了基于8个miRNA表达的组合预测甲状腺癌患者预后的风险模型。在这8个miRNA中,有7个miRNA被报道与甲状腺癌或者其他肿瘤相关。MiR-146b被证明在乳头状甲状腺癌中表达上调,并且与肿瘤的侵袭有关[22-23];Fu等[24]利用生物信息学的方法挖掘TCGA数据库发现hsa-mir-196a-2在甲状腺癌中表达显著上调,其与肿瘤进展、淋巴结转移和局部浸润有关,并且可以作为甲状腺癌的独立预后危险因素;miR-509的过表达可以抑制三阴性乳腺癌Hs578T细胞增殖、诱导细胞凋亡和抑制细胞侵袭[25];Tang等[26]利用生物信息学分析发现,miR-6860和miR-509-3与甲状腺癌患者的总体生存率相关,可作为甲状腺癌的预后因子;miR-513c被证明与神经母细胞瘤[27]、肝细胞癌[28]的细胞迁移、入侵和增殖等有关;has-mir-4709被证明与结肠癌[29]、胰腺导管腺癌[30]患者的生存相关。
综上所述,我们构建了基于8个miRNA预测甲状腺癌患者生存的风险模型,该模型具较好的敏感度和特异性,模型中高风险评分组患者的死亡风险较大。研究指出[31],建立多基因预后模型比患者的临床病理指标更能提供精确的预后指导,对个体化治疗方案的选择具有重要的参考价值。我们所构建的8个miRNA模型可有效预测甲状腺癌患者的预后,但仍需要进一步使用大规模的多中心的循证医学证据加以验证。
作者贡献
焦赵爽:研究设计、实施、分析数据及论文撰写
张怀念:数据整理、分析数据及修改论文
| [1] |
董芬, 张彪, 单广良. 中国甲状腺癌的流行现状和影响因素[J]. 中国癌症杂志, 2016, 26(1): 47-52. [Dong F, Zhang B, Shang GL. Distribution and risk factors of thyroid cancer in China[J]. Zhongguo Ai Zheng Za Zhi, 2016, 26(1): 47-52.] |
| [2] |
Raue F, Frank-Raue K. Thyroid Cancer: Risk-Stratified Management and Individualized Therapy[J]. Clinical Cancer Res, 2016, 22(20): 5012-5021. DOI:10.1158/1078-0432.CCR-16-0484 |
| [3] |
Glaser SM, Mandish SF, Gill BS, et al. Anaplastic thyroid cancer: Prognostic factors, patterns of care, and overall survival[J]. Head Neck, 2016, 38(Suppl 1): E2083-E2090. |
| [4] |
林琳, 刘建华, 吴丽娜. 甲状腺癌RNA分子研究进展[J]. 肿瘤防治研究, 2019, 46(11): 1031-1035. [Lin L, Liu Jh, Wu LN. Research Progress of RNA Molecules in Thyroid Cancer[J]. Zhong Liu Fang Zhi Yan Jiu, 2019, 46(11): 1031-1035. DOI:10.3971/j.issn.1000-8578.2019.19.0431] |
| [5] |
Li N, Liu X, Han L, et al. Expression of miRNA-146b-5p in patients with thyroid cancer in combination with Hashimoto's disease and its clinical significance[J]. Oncol Lett, 2019, 17(6): 4871-4876. |
| [6] |
刘延晴, 林岩松. 碘难治性分化型甲状腺癌诊治策略及预后[J]. 中国实用外科杂志, 2019, 39(3): 216-220. [Liu YQ, Lin YS. Emerging management and its impact on radioiodine refractory differentiated thyroid cancer[J]. Zhongguo Shi Yong Wai Ke Za Zhi, 2019, 39(3): 216-220.] |
| [7] |
杨进宝, 李小毅, 商中华. 甲状腺乳头状癌各亚型临床病理特点的研究进展[J]. 癌症进展, 2015, 13(1): 55-60. [Yang JB, Li XY, Shang ZH. Advances in clinicopathological characteristics of various subtypes of papillary thyroid cancer[J]. Ai Zheng Jin Zhan, 2015, 13(1): 55-60.] |
| [8] |
Abdullah MI, Junit SM, Ng KL, et al. Papillary Thyroid Cancer: Genetic Alterations and Molecular Biomarker Investigations[J]. Int J Med Sci, 2019, 16(3): 450-460. DOI:10.7150/ijms.29935 |
| [9] |
Raposo L, Morais S, Oliveira MJ, et al. Trends in thyroid cancer incidence and mortality in Portugal[J]. Eur J Cancer Prev, 2017, 26(2): 135-143. DOI:10.1097/CEJ.0000000000000229 |
| [10] |
Bray F, Ferlay J, Laversanne M, et al. Cancer Incidence in Five Continents: Inclusion criteria, highlights from Volume X and the global status of cancer registration[J]. Int J Cancer, 2015, 137(9): 2060-2071. DOI:10.1002/ijc.29670 |
| [11] |
Grogan RH, Kaplan SP, Cao H, et al. A study of recurrence and death from papillary thyroid cancer with 27 years of median follow-up[J]. Surgery, 2013, 154(6): 1436-1446. DOI:10.1016/j.surg.2013.07.008 |
| [12] |
马珊, 尹乐平. 甲状腺乳头状癌手术方式的选择[J]. 实用医院临床杂志, 2017, 14(3): 124-127. [Ma S, Yin LP. Choice of surgical method of thyroid papillary carcinoma[J]. Yi Yong Yi Yuan Lin Chuang Za Zhi, 2017, 14(3): 124-127. DOI:10.3969/j.issn.1672-6170.2017.03.044] |
| [13] |
Trzybulska D, Bobjer J, Giwercman A, et al. Serum microRNAs in male subfertility-biomarkers and a potential pathogenetic link to metabolic syndrome[J]. J Assist Reprod Genet, 2017, 34(10): 1277-1282. DOI:10.1007/s10815-017-0989-0 |
| [14] |
Hill M, Tran N. MicroRNAs Regulating MicroRNAs in Cancer[J]. Trends Cancer, 2018, 4(7): 465-468. DOI:10.1016/j.trecan.2018.05.002 |
| [15] |
Yu L, Xiang L, Feng J, et al. miRNA-21 and miRNA-223 expression signature as a predictor for lymph node metastasis, distant metastasis and survival in kidney renal clear cell carcinoma[J]. J Cancer, 2018, 9(20): 3651-3659. DOI:10.7150/jca.27117 |
| [16] |
Liu Q, Diao R, Feng G, et al. Risk score based on three mRNA expression predicts the survival of bladder cancer[J]. Oncotarget, 2017, 8(37): 61583-61591. DOI:10.18632/oncotarget.18642 |
| [17] |
Butz H, Patocs A. MicroRNAs in endocrine tumors[J]. EJIFCC, 2019, 30(2): 146-164. |
| [18] |
Yue K, Wang X, Wu Y, et al. microRNA-7 regulates cell growth, migration and invasion via direct targeting of PAK1 in thyroid cancer[J]. Mol Med Rep, 2016, 14(3): 2127-2134. DOI:10.3892/mmr.2016.5477 |
| [19] |
Wang J, Yang H, Si Y, et al. Iodine Promotes Tumorigenesis of Thyroid Cancer by Suppressing Mir-422a and Up-Regulating MAPK1[J]. Cell Physiol Biochem, 2017, 43(4): 1325-1336. DOI:10.1159/000481844 |
| [20] |
Dong S, Meng X, Xue S, et al. microRNA-141 inhibits thyroid cancer cell growth and metastasis by targeting insulin receptor substrate 2[J]. Am J Transl Res, 2016, 8(3): 1471-1481. |
| [21] |
Wang P, Meng X, Huang Y, et al. MicroRNA-497 inhibits thyroid cancer tumor growth and invasion by suppressing BDNF[J]. Oncotarget, 2017, 8(2): 2825-2834. DOI:10.18632/oncotarget.13747 |
| [22] |
Chou CK, Liu RT, Kang HY. MicroRNA-146b: A Novel Biomarker and Therapeutic Target for Human Papillary Thyroid Cancer[J]. Int J Mol Sci, 2017, 18(3): E636. DOI:10.3390/ijms18030636 |
| [23] |
Qiu Z, Li H, Wang J, et al. miR-146a and miR-146b in the diagnosis and prognosis of papillary thyroid carcinoma[J]. Oncol Rep, 2017, 38(5): 2735-2740. DOI:10.3892/or.2017.5994 |
| [24] |
Fu YT, Zhang DQ, Zhou L, et al. Has-MiR-196a-2 is up-regulated and acts as an independent unfavorable prognostic factor in thyroid carcinoma[J]. Eur Rev Med Pharmacol Sci, 2018, 22(9): 2707-2714. |
| [25] |
Zhang G, Liu Z, Han Y, et al. Overexpression of miR-509 Increases Apoptosis and Inhibits Invasion via Suppression of Tumor Necrosis Factor-alpha in Triple-Negative Breast Cancer Hs578T Cells[J]. Oncol Res, 2016, 24(4): 233-238. DOI:10.3727/096504016X14648701447977 |
| [26] |
Tang J, Kong D, Cui Q, et al. Bioinformatic analysis and identification of potential prognostic microRNAs and mRNAs in thyroid cancer[J]. Peer J, 2018, 6: e4674. DOI:10.7717/peerj.4674 |
| [27] |
Xia HL, Lv Y, Xu CW, et al. MiR-513c suppresses neuroblastoma cell migration, invasion, and proliferation through direct targeting glutaminase (GLS)[J]. Cancer Biomark, 2017, 20(4): 589-596. DOI:10.3233/CBM-170577 |
| [28] |
Zhang K, Zhao Z, Yu J, et al. LncRNA FLVCR1-AS1 acts as miR-513c sponge to modulate cancer cell proliferation, migration, and invasion in hepatocellular carcinoma[J]. J Cell Biochem, 2018, 119(7): 6045-6056. DOI:10.1002/jcb.26802 |
| [29] |
Wei HT, Guo EN, Liao XW, et al. Genomescale analysis to identify potential prognostic microRNA biomarkers for predicting overall survival in patients with colon adenocarcinoma[J]. Oncol Rep, 2018, 40(4): 1947-1958. |
| [30] |
Liao X, Wang X, Huang K, et al. Genome-scale analysis to identify prognostic microRNA biomarkers in patients with early stage pancreatic ductal adenocarcinoma after pancreaticoduodenectomy[J]. Cancer Manag Res, 2018, 10: 2537-2551. DOI:10.2147/CMAR.S168351 |
| [31] |
贾晓晨, 贾勇圣, 孟文静, 等. 基于TCGA数据库建立的八基因预后模型在乳腺癌中的应用[J]. 天津医药, 2018, 46(8): 856-861. [Jia XC, Jia YS, Meng WJ, et al. Identification of prognostic eight-gene signature model in breast cancer using integrated TCGA database[J]. Tianjin Yi Yao, 2018, 46(8): 856-861.] |
 2020, Vol. 47
2020, Vol. 47
