文章信息
- 基于GEO数据库的胃肠上皮化生相关基因与通路的生物信息学分析
- Bioinformatics Analysis of Gastric Intestinal Metaplasia-related Genes and Pathways Based on GEO Database
- 肿瘤防治研究, 2018, 45(10): 768-774
- Cancer Research on Prevention and Treatment, 2018, 45(10): 768-774
- http://www.zlfzyj.com/CN/10.3971/j.issn.1000-8578.2018.17.1651
- 收稿日期: 2017-12-15
- 修回日期: 2018-03-27
2. 200437 上海,上海中医药大学附属岳阳中西医结合医院针灸科;
3. 200437 上海,上海中医药大学附属岳阳中西医结合医院脊柱科
2. Department of Acupuncture, Yueyang Hospital of Integrated Traditional Chinese and Western Medicine Affiliated to Shanghai University of Traditional Chinese Medicine, Shanghai 200437, China;
3. Department of Spine, Yueyang Hospital of Integrated Traditional Chinese and Western Medicine Affiliated to Shanghai University of Traditional Chinese Medicine, Shanghai 200437, China
胃癌是一种全球高发的消化系统肿瘤,严重威胁人类健康。我国每年新发胃癌62.9万例,占新发肿瘤人口的15.8%,仅次于肺癌的17.1%,居总疾病新发率的第二位;每年因胃癌死亡人数为49.8万,占恶性肿瘤死亡人数的17.7%,仅次于肺癌的21.7%,居总疾病死亡率的第二位[1]。目前胃癌的发生机制尚不明确,一般认为其发生发展是多基因、多因素、多阶段的动态演变过程,而肠型胃癌的发生多认为会经历“正常胃黏膜—慢性萎缩性胃炎—肠上皮化生—异型增生—胃癌(肠型)”这一模式[2]。虽然目前对于肠化生的具体分型还没有统一标准,但普遍认为肠化生是胃癌的癌前病变[3-4]。流行病学研究亦证实,胃黏膜肠化生是胃癌发生的高危险因素:You等发现[5],在中国,胃癌高发省份与低发省份相比,肠化生的发病率在明显升高;Shimoyama等[6]在评估日本胃癌风险指数时得出结论,肠化生是肠型胃癌发生的条件;多项长期随访研究也指出,肠化生是胃癌的高发因素[7-8]。
本研究拟从分子水平探讨胃组织从正常胃黏膜演变为肠上皮细胞的发生机制,从而为胃癌的二级预防提供依据。本研究拟从公共基因芯片数据库(GEO)中获取有关正常胃黏膜与肠化标本的芯片,分析两者中存在的差异表达基因,探讨正常胃黏膜肠上皮化生过程中的分子生物学基础,并在胃癌组织中验证这些差异表达基因(Differentially expressed genes, DEGs)是否持续差异表达,以及是否与胃癌的预后相关,进一步判别这些差异基因是否真正在胃癌发生发展过程中起作用。由于非肠型胃癌不一定都经历“正常胃黏膜—慢性萎缩性胃炎—肠上皮化生—异型增生—胃癌(肠型)”这一过程,因此,本研究更侧重于肠型胃癌的发生过程中可能存在的基因差异及机制研究。
1 资料与方法 1.1 基因芯片的选取以及差异基因的筛选本研究从美国国立生物技术信息中心(NCBI)的GEO数据库(https://www.ncbi.nlm.nih.gov/gds)样本中以“intestinal metaplasia”为关键词进行检索,并筛选包含该样本的系列中含有正常对照的系列,检索时间截至2017年12月。并通过GEO2R分析,设定筛选条件P < 0.05,logFC绝对值 > 1.5,分别得到不同芯片的差异基因,并通过FunRich 3.0(http://www.funrich.org/)得到不同芯片之间均表达差异的基因为关键基因。
1.2 差异基因生物学功能及通路分析将得到的差异基因去重,并删除无对应名称的基因片段后,利用生物学信息注释数据库DAVID(https://david.ncifcrf.gov/)对所有差异基因进行GO(gene ontology)生物学过程富集分析和KEGG(KEGG pathway analysis)通路富集分析,以期找出最显著富集的生物学注释,P < 0.01认为是显著富集。
1.3 关键基因在胃癌中的表达及对胃癌预后的影响通过开源、可视化网站cBioPortal(http://www.cbioportal.org/)分析含有478例样本的胃腺癌与正常标本的TCGA数据,进一步分析这些关键基因是否在胃癌中也表达异常,并进一步通过KM plotter(http://kmplot.com/analysis/)在线网站,分析其mRNA表达水平与胃癌预后的关系。
2 结果 2.1 差异基因筛选共得到与胃黏膜组织发生肠化相关的基因芯片3个,其编号分别是GSE2669、GSE60662和GSE78523,经筛选后三个芯片分别得到的差异基因为19、1 095和217个,去重后共得到1 188个差异表达基因。其中,ALDOB、CLCA1、CLDN7、DMBT1、KRT20、MTTP、OLFM4、REG3A和TFF3基因在三个芯片中均差异表达,将其称为关键基因。这9个基因在肠化组织中均为高表达基因,见图 1。
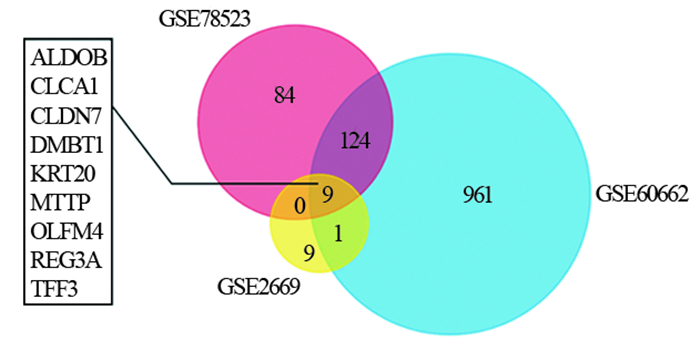
|
| GSE78523, GSE60662, GSE2669: the three microarray datasets were identified in Gene Expression Omnibus (GEO) database. The number in the circles represented the amount of genes in one, both or all the three microarray datasets. ALDOB, CLCA1, CLDN7, DMBT1, KRT20, MTTP, OLFM4, REG3A and TFF3: the nine key genes existed in all three databases 图 1 肠化生组织中异常表达的差异基因 Figure 1 Differentially expressed genes (DEGs) in gastric intestinal metaplasia tissues and normal gastric tissues |
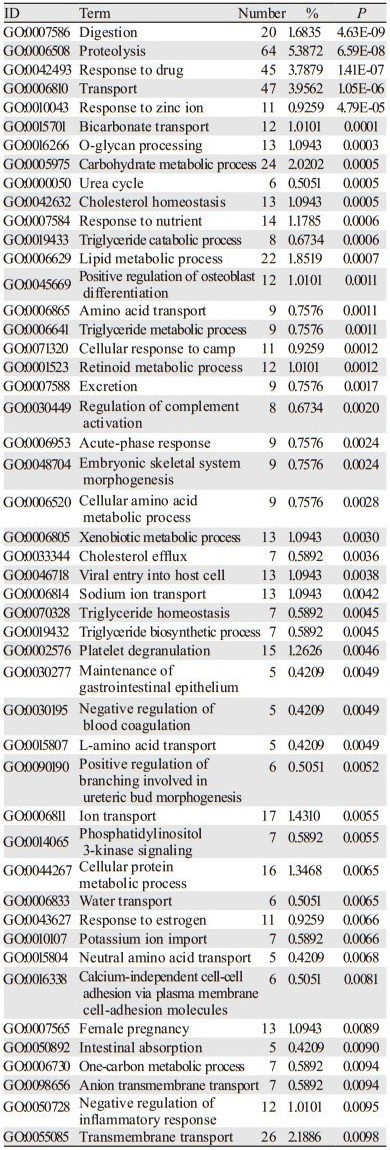
运用在线生物学信息注释数据库DAVID,对上述得到的1 188个差异基因的生物学功能及信号通路分析进行展现和富集分析,得到174个GO生物学过程,其中在消化、蛋白质水解、对药物的反应、物质转运的调节等48个生物学过程中显著富集,见表 1。差异基因富集的KEGG通路有41个,主要体现在包括营养物质的消化吸收、氨基酸合成、消化液分泌、PPAR信号通路等19个KEGG通路,见表 2。
|
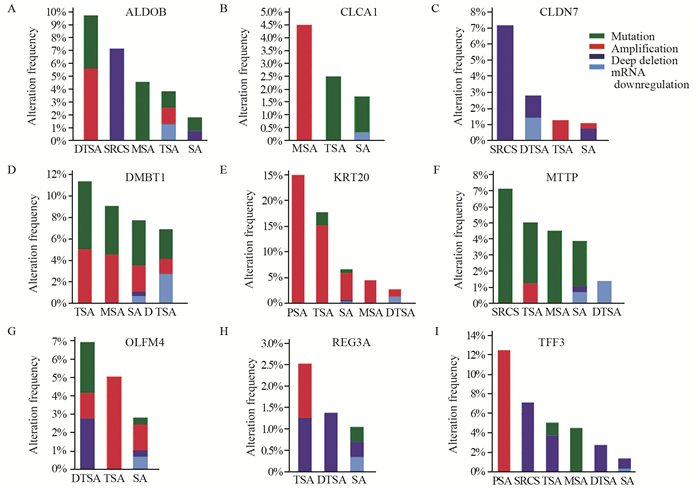
通过cBioPortal分析发现,9个关键基因在包括胃腺癌、弥漫性胃腺癌、印戒细胞癌、胃黏液腺癌、胃管状腺癌、胃乳头状腺癌6种不同的胃癌类型中均存在一定的突变、缺失、扩增或者mRNA水平下调等变化,见图 2。而通过KM plotter生存分析发现,这9个关键基因mRNA表达水平的高低均与胃癌的预后密切相关,见图 3。
|
| DTSA: diffuse type stomach adenocarcinoma; SRCS: signet ring carcinoma of the stomach; MSA: mucinous stomach adenocarcinoma; TSA: tubular stomach adenocarcinoma; SA: stomach adenocarcinoma; PSA: papillary stomach adenocarcinoma; A: The gene ALDOB existed amplification and mutation in DTSA, deep deletion in SRCS, mutation in MSA, mutation, amplification and mRNA down-regulation in TSA, and mutation and deep deletion in SA; B: The gene CLCA1 existed amplification in MSA, mutation in TSA, and mutation and mRNA down-regulation in SA; C: The gene CLDN7 existed deep deletion in SRCS, deep deletion and mRNA down-regulation in DTSA, amplification in TSA, and amplification and deep deletion in SA; D: The gene DMBT1 existed mutation and amplification in TSA, MSA, SA and DTSA, mRNA down-regulation in SA and DTSA, and deep deletion in SA; E: The gene KRT20 existed amplification in PSA, mutation and amplification in TSA, mutation, amplification, deep deletion and mRNA down-regulation in SA, amplification in MSA, and amplification and mRNA down-regulation in DTSA; F: The gene MTTP existed mutation in SRCS, mutation and amplification in TSA, mutation in MSA, mutation, deep deletion and mRNA down-regulation in SA, and mRNA down-regulation in DTSA; G: The gene OLFM4, mutation existed amplification and deep deletion in DTSA, amplification in TSA, mutation, amplification, and deep deletion and mRNA down-regulation in SA; H: The gene REG3A, amplification and deep deletion in TSA, deep deletion in DTSA, mutation, and deep deletion and mRNA down-regulation in SA; I: The gene TFF3, amplification in PSA, deep deletion in SRCS, mutation and deep deletion in TSA, mutation in MSA, deep deletion in DTSA, and deep deletion and mRNA down-regulation in SA 图 2 TCGA数据库在关键基因胃癌组织中的差异变化 Figure 2 Expressions difference of key genes in gastric cancer tissues based on TCGA dataset |

|
| 图 3 关键基因对胃癌患者预后的影响 Figure 3 Correlation between key genes and prognosis of gastric cancer patients |
本研究通过数据分析发现,GEO数据库中共有正常-肠化表达谱基因芯片3组,差异分析显示各组芯片中发生与未发生肠化胃黏膜组织差异表达的基因颇多(共1 188个),但在3组芯片均差异表达的关键基因仅9个,分别为ALDOB、CLCA1、CLDN7、DMBT1、KRT20、MTTP、OLFM4、REG3A和TFF3。CLCA1主要在分泌上皮细胞中表达,虽然目前尚未见其在胃癌中的作用机制的相关报道,但有学者发现,其能影响卵巢癌细胞株OV-90的细胞聚集[9],过表达CLCA1能够明显抑制结肠癌的生长和转移[10],而CLCA1在结肠癌患者中持续低表达[11],其低表达预示着患者预后不良。这与CLCA1在胃癌患者的KM plotter分析结果一致,但CLCA1在肠化中是高表达基因,而在癌症中却变为低表达基因,这可能是因为肠化是肠型胃癌可能的进展过程,但KM plotter分析是对所有类型胃癌患者进行分析,可能混杂有非肠型胃癌,因此该KM plotter分析结果可能与现有文献不符,从而导致这种差异。
而在其余8个关键基因中,ALODB在肝癌中表达下调,过表达ALODB能明显抑制肝癌细胞株的侵袭转移能力,其低表达意味着肝癌患者预后不良[12];Li等[13]发现ALODB中在结肠癌中的作用正好相反,沉默其表达能够明显抑制结肠癌细胞的增殖、侵袭和迁移能力,其表达量升高意味着结肠癌患者预后不良;He等[14]发现ALODB表达下调意味着胃癌患者预后不良。还有研究发现,CLDN7与胰腺癌[15]、结肠癌[16]、肺癌[17]等多种肿瘤的转移和侵袭相关,实验研究[16]显示CLDN7敲除可明显减弱HT29和SW948细胞的增殖、迁移和侵袭能力;而在胃癌相关研究中发现,CLDN7在早期胃癌[18]、肠型胃癌[19]中异常表达,细胞实验进一步证实其能促进AGS胃癌细胞的增殖、侵袭和转移[20],且Jun等[21]对134例临床样本的检测发现,CLDN7的高表达与胃癌患者预后不良密切相关。DMBT1最早在胶质细胞瘤中发现,后在多种肿瘤中发现其表达异常,并与患者预后相关,逐渐成为一种候选的抑癌基因[22-23]。Garay等[24]发现DMBT1不仅在人胃癌癌前病变标本中异常表达,而且与野生型小鼠相比,DMBT1基因敲除可明显促进感染HP后的胃黏膜炎性反应及肠上皮化生,说明DMBT1在胃癌的发生发展过程中起到重要作用。KRT20是细胞角蛋白家族的一员,是细胞骨架主要成分之一,多局限在胃肠上皮细胞中。早在2002年就有胃癌相关研究发现,KRT20在肠化和正常胃黏膜组织[25]中存在巨大的表达差异。乔建梁等[26]发现,KRT20可以作为胃癌术后辅助化疗的监测指标,Sun等[27]通过152例临床病例的研究发现,KRT20的高表达,意味着为胃癌腹膜转移的风险增大。MTTP是一种新发现的脂质转运蛋白,主要存在于肝细胞和小肠细胞微粒体腔内,在脂蛋白的正常组装、分泌中发挥重要作用。虽然其在肿瘤中的作用未见相关报道,但通过KM plotter生存分析发现,其表达下调意味着胃癌患者预后不良。OLFM4是一种新发现的糖蛋白,在多种肿瘤组织中均高表达。在消化系统肿瘤尤其是胃癌中亦不例外[28]。研究发现OLFM4可参与胃癌早期阶段的形成,且与晚期胃癌的预后有关[29],可以通过作用于PI3K/PKB下游等多种途径[30]促进胃癌细胞增殖;此外,作为一种分泌性糖蛋白,OLFM4已经逐渐成为胃癌发生发展的一个标志物[31]。REG3A是REG蛋白家族的一员,主要与胰岛细胞及肝细胞的再生、激活胰腺星形细胞相关[32-33],但Chen等[34]研究发现REG3A在胃癌中高表达,并预示着预后较差,通过JAK2/STAT3信号通路促进细胞增殖、侵袭和转移。TFF3是目前广泛研究的一个分子,其在正常胃黏膜中表达量很少,但在胃癌发生后,表达量显著上升,并且可通过EMT[35]、VEGF[36]、NF-κB[37]等多种机制影响胃癌的发生、发展,是胃癌非侵入性筛查潜在的理想标志物,并可用于胃肠道肿瘤化疗药效与预后的评价[38]。可见除CLCA1仅与肿瘤生成、转移关系密切有关而未在胃癌中得到证实外,其余8个关键基因与胃癌的产生与发展均具有一定相关性,这与本文的研究结果一致。
而对差异基因和关键基因进行进一步的GO富集和KEGG通路分析发现,发生肠化组织与正常组织相比,生理功能的差异性主要集中于与消化吸收功能密切相关的消化液合成分泌、蛋白质氨基酸代谢、物质转运调节功能方面,这与胃为人体主要的消化器官密切相关。也提示正常-肠化-胃癌的组织结构改变与胃的消化功能改变相关、与患者的饮食习惯亦或有密切联系。现代研究发现,饮酒[39]、高盐饮食及辛辣刺激饮食[40]与肠化生的发生密切相关,这进一步佐证了上述推论。
此外,通过TCGA数据库检索发现,上述9个关键基因在肠化-胃癌组织中亦具有差异表达,这在一定程度上表明,上述9个关键基因在正常-肠化-胃癌的生理病理演变过程中发挥着持续作用。而通过KM plotter分析进一步探究发现,这9个关键基因与胃癌患者预后相关,再次佐证了这9个关键基因在胃癌的发生、发展中有着重要作用。
由于目前关于肠化与正常的基因芯片数据较少,9个关键基因的数目主要受限于GSE2669的数据,其完成时间较早,受限于当时的技术,故导致最终差异基因较少。此外,GSE60662和GSE78523两个数据集共有128个差异基因,这些基因涉及多个不同的生理生化过程,亦值得后续研究和探讨。
本研究基于GEO肠化生基因表达谱及TCGA数据库对正常、肠化、胃癌组织进行了基因、通路分析,揭示了与正常-肠化-胃癌演变可能存在密切相关性的9个关键基因,并通过生存分析进一步探究了关键基因与胃癌预后的关系。这是从生物信息学角度对胃癌演变规律的一次探索,为后期抑制正常胃黏膜向肠化生的演变,以及逆转肠化生过程提供依据,并为进一步的机制研究指明了方向,对肠型胃癌临床及基础研究具有一定借鉴意义。
| [1] | Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015[J]. CA Cancer J Clin, 2016, 66(2): 115–32. DOI:10.3322/caac.21338 |
| [2] | Correa P, Piazuelo MB. The gastric precancerous cascade[J]. J Dig Dis, 2012, 13(1): 2–9. DOI:10.1111/cdd.2012.13.issue-1 |
| [3] | Yoon H, Kim N. Diagnosis and management of high risk group for gastric cancer[J]. Gut Liver, 2015, 9(1): 5–17. DOI:10.5009/gnl14118 |
| [4] | Malik TH, Sayahan MY, Al Ahmed HA, et al. Gastric Intestinal Metaplasia: An Intermediate Precancerous Lesion in the Cascade of Gastric Carcinogenesis[J]. J Coll Physicians Surg Pak, 2017, 27(3): 166–72. |
| [5] | You WC, Zhang L, Gail MH, et al. Precancerous lesions in two counties of China with contrasting gastric cancer risk[J]. Int J Epidemiol, 1998, 27(6): 945–8. DOI:10.1093/ije/27.6.945 |
| [6] | Shimoyama T, Fukuda S, Tanaka M, et al. Evaluation of the applicability of the gastric carcinoma risk index for intestinal type cancer in Japanese patients infected with Helicobacter pylori[J]. Virchows Arch, 2000, 436(6): 585–7. DOI:10.1007/s004289900179 |
| [7] | Pittayanon R, Rerknimitr R, Klaikaew N, et al. The risk of gastric cancer in patients with gastric intestinal metaplasia in 5-year follow-up[J]. Aliment Pharmacol Ther, 2017, 46(1): 40–5. DOI:10.1111/apt.14082 |
| [8] | Lee TY, Wang RC, Lee YC, et al. The Incidence of Gastric Adenocarcinoma Among Patients With Gastric Intestinal Metaplasia: A Long-term Cohort Study[J]. J Clin Gastroenterol, 2016, 50(7): 532–7. DOI:10.1097/MCG.0000000000000406 |
| [9] | Musrap N, Tuccitto A, Karagiannis GS, et al. Comparative Proteomics of Ovarian Cancer Aggregate Formation Reveals an Increased Expression of Calcium-activated Chloride Channel Regulator 1 (CLCA1)[J]. J Biol Chem, 2015, 290(28): 17218–27. DOI:10.1074/jbc.M115.639773 |
| [10] | Li X, Hu W, Zhou J, et al. CLCA1 suppresses colorectal cancer aggressiveness via inhibition of the Wnt/beta-catenin signaling pathway[J]. Cell Commun Signal, 2017, 15(1): 38. DOI:10.1186/s12964-017-0192-z |
| [11] | Yang B, Cao L, Liu J, et al. Low expression of chloride channel accessory 1 predicts a poor prognosis in colorectal cancer[J]. Cancer, 2015, 121(10): 1570–80. DOI:10.1002/cncr.v121.10 |
| [12] | Tao QF, Yuan SX, Yang F, et al. Aldolase B inhibits metastasis through Ten-Eleven Translocation 1 and serves as a prognostic biomarker in hepatocellular carcinoma[J]. Mol Cancer, 2015, 14: 170. DOI:10.1186/s12943-015-0437-7 |
| [13] | Li Q, Li Y, Xu J, et al. Aldolase B Overexpression is Associated with Poor Prognosis and Promotes Tumor Progression by Epithelial-Mesenchymal Transition in Colorectal Adenocarcinoma[J]. Cell Physiol Biochem, 2017, 42(1): 397–406. DOI:10.1159/000477484 |
| [14] | He J, Jin Y, Chen Y, et al. Downregulation of ALDOB is associated with poor prognosis of patients with gastric cancer[J]. Onco Targets Ther, 2016, 9: 6099–109. DOI:10.2147/OTT |
| [15] | Thuma F, Zoller M. EpCAM-associated claudin-7 supports lymphatic spread and drug resistance in rat pancreatic cancer[J]. Int J Cancer, 2013, 133(4): 855–66. DOI:10.1002/ijc.v133.4 |
| [16] | Philip R, Heiler S, Mu W, et al. Claudin-7 promotes the epithelial-mesenchymal transition in human colorectal cancer[J]. Oncotarget, 2015, 6(4): 2046–63. |
| [17] | Akizuki R, Shimobaba S, Matsunaga T, et al. Claudin-5, -7, and -18 suppress proliferation mediated by inhibition of phosphorylation of Akt in human lung squamous cell carcinoma[J]. Biochim Biophys Acta, 2017, 1864(2): 293–302. DOI:10.1016/j.bbamcr.2016.11.018 |
| [18] | Johnson AH, Frierson HF, Zaika A, et al. Expression of tight-junction protein claudin-7 is an early event in gastric tumorigenesis[J]. Am J Pathol, 2005, 167(2): 577–84. DOI:10.1016/S0002-9440(10)62999-9 |
| [19] | Park JY, Park KH, Oh TY, et al. Up-regulated claudin 7 expression in intestinal-type gastric carcinoma[J]. Oncol Rep, 2007, 18(2): 377–82. |
| [20] | Zavala-Zendejas VE, Torres-Martinez AC, Salas-Morales B, et al. Claudin-6, 7, or 9 overexpression in the human gastric adenocarcinoma cell line AGS increases its invasiveness, migration, and proliferation rate[J]. Cancer Invest, 2011, 29(1): 1–11. DOI:10.3109/07357907.2010.512594 |
| [21] | Jun KH, Kim JH, Jung JH, et al. Expression of claudin-7 and loss of claudin-18 correlate with poor prognosis in gastric cancer[J]. Int J Surg, 2014, 12(2): 156–62. DOI:10.1016/j.ijsu.2013.11.022 |
| [22] | Müller H, Renner M, Helmke BM, et al. Elevated DMBT1 levels in neonatal gastrointestinal diseases[J]. Histochem Cell Biol, 2016, 145(2): 227–37. DOI:10.1007/s00418-015-1381-8 |
| [23] | Müller H, Nagel C, Weiss C, et al. Deleted in malignant brain tumors 1 (DMBT1) elicits increased VEGF and decreased IL-6 production in type Ⅱ lung epithelial cells[J]. BMC Pulm Med, 2015, 15: 32. DOI:10.1186/s12890-015-0027-x |
| [24] | Garay J, Piazuelo MB, Lopez-Carrillo L, et al. Increased expression of deleted in malignant brain tumors (DMBT1) gene in precancerous gastric lesions: Findings from human and animal studies[J]. Oncotarget, 2017, 8(29): 47076–89. |
| [25] | Jovanovic I, Tzardi M, Mouzas IA, et al. Changing pattern of cytokeratin 7 and 20 expression from normal epithelium to intestinal metaplasia of the gastric mucosa and gastroesophageal junction[J]. Histol Histopathol, 2002, 17(2): 445–54. |
| [26] | 乔建梁, 孟兴凯, 张俊晶, 等. 胃癌患者术后化疗前后检测外周血细胞角蛋白20mRNA的临床意义[J]. 中华胃肠外科杂志, 2009, 12(1): 32–5. [ Qiao JL, Meng XK, Zhang JJ, et al. Expression of CK20 mRNA in the peripheral blood around postoperative chemotherapy in patients with gastric cancer and its clinical significance[J]. Zhonghua Wei Chang Wai Ke Za Zhi, 2009, 12(1): 32–5. DOI:10.3760/cma.j.issn.1671-0274.2009.01.010 ] |
| [27] | Sun GR, Dong XY, He QS, et al. Expression and clinical significance of CK19 and CK20 expressions in transverse mesocolon biopsies from patients with gastric carcinoma[J]. Cell Biochem Biophys, 2012, 62(2): 361–4. DOI:10.1007/s12013-011-9293-2 |
| [28] | Oue N, Sentani K, Noguchi T, et al. Serum olfactomedin 4 (GW112, hGC-1) in combination with Reg Ⅳ is a highly sensitive biomarker for gastric cancer patients[J]. Int J Cancer, 2009, 125(10): 2383–92. DOI:10.1002/ijc.v125:10 |
| [29] | Jang BG, Lee BL, Kim WH. Olfactomedin-related proteins 4 (OLFM4) expression is involved in early gastric carcinogenesis and of prognostic significance in advanced gastric cancer[J]. Virchows Arch, 2015, 467(3): 285–94. DOI:10.1007/s00428-015-1793-9 |
| [30] | Sommer KW, Schamberger CJ, Schmidt GE, et al. Inhibitor of apoptosis protein (IAP) survivin is upregulated by oncogenic c-H-Ras[J]. Oncogene, 2003, 22(27): 4266–80. DOI:10.1038/sj.onc.1206509 |
| [31] | Guette C, Valo I, Vetillard A, et al. Olfactomedin-4 is a candidate biomarker of solid gastric, colorectal, pancreatic, head and neck, and prostate cancers[J]. Proteomics Clin Appl, 2015, 9(1-2): 58–63. DOI:10.1002/prca.v9.1-2 |
| [32] | Liu X, Wang J, Wang H, et al. REG3A accelerates pancreatic cancer cell growth under IL-6-associated inflammatory condition: Involvement of a REG3A-JAK2/STAT3 positive feedback loop[J]. Cancer Lett, 2015, 362(1): 45–60. DOI:10.1016/j.canlet.2015.03.014 |
| [33] | Wang J, Zhou H, Han Y, et al. SOCS3 methylation in synergy with Reg3A overexpression promotes cell growth in pancreatic cancer[J]. J Mol Med(Berl), 2014, 92(12): 1257–69. |
| [34] | Chen ZF, Huang ZM, Xue HB, et al. REG3A promotes the proliferation, migration, and invasion of gastric cancer cells[J]. Onco Targets Ther, 2017, 10: 2017–23. DOI:10.2147/OTT |
| [35] | Zheng Q, Gao J, Li H, et al. Trefoil factor 3 peptide regulates migration via a Twist-dependent pathway in gastric cell[J]. Biochem Biophys Res Commun, 2013, 438(1): 6–12. DOI:10.1016/j.bbrc.2013.06.115 |
| [36] | Guleng B, Han J, Yang JQ, et al. TFF3 mediated induction of VEGF via hypoxia in human gastric cancer SGC-7901 cells[J]. Mol Biol Rep, 2012, 39(4): 4127–34. DOI:10.1007/s11033-011-1195-2 |
| [37] | Loncar MB, Al-azzeh ED, Sommer PS, et al. Tumour necrosis factor alpha and nuclear factor kappaB inhibit transcription of human TFF3 encoding a gastrointestinal healing peptide[J]. Gut, 2003, 52(9): 1297–303. DOI:10.1136/gut.52.9.1297 |
| [38] | 王琳, 黄建坤. TFF3作为肿瘤生物标志物的研究[J]. 肿瘤防治研究, 2015, 42(11): 1156–60. [ Wang L, Huang JK. Review on researches of TFF3 as tumor biomarker[J]. Zhong Liu Fang Zhi Yan Jiu, 2015, 42(11): 1156–60. DOI:10.3971/j.issn.1000-8578.2015.11.022 ] |
| [39] | Chacaltana Mendoza A, Soriano álvarez C, Frisancho Velarde O. Associated risk factors in patients with gastric intestinal metaplasia with mild gastroduodenal disease. Is it always related to Helicobacter pylori infection?[J]. Rev Gastroenterol Peru, 2012, 32(1): 50–7. |
| [40] | 柯丽, 张迪, 陈瑜, 等. 中国西北地区胃粘膜肠上皮化生危险因素调查[J]. 现代生物医学进展, 2016, 16(34): 6639–43. [ Ke L, Zhang D, Chen Y, et al. Risk Factors Of Gastric Intestinal Metaplasia in Northwest of China[J]. Xian Dai Sheng Wu Yi Xue Jin Zhan, 2016, 16(34): 6639–43. ] |
 2018, Vol. 45
2018, Vol. 45



