| 土壤中以抗生素为单一碳源的抗性细菌 |
分别选取茶园、森林、养殖场、医院周边作为土样采集点.茶园位于杭州市龙井村, 为丘陵地, 植被为茶树, 长年施用有机肥;森林位于龙井八景丘陵上, 植被为多年生林木, 树种主要有柳树等;养殖场位于浙江大学华家池校区;医院位于浙江大学紫金港校区校医院.在各点周边, 随机取20个表层(0~20 cm)土样, 采集后拣去植物残体混合, 分成2部分, 一部分鲜土直接过2 mm筛用于抗性细菌的分离;另一部分土样风干后, 研磨分别过2 mm和0.25 mm筛, 用于土壤理化性状的测定.供试土壤的基本性质采用常规分析方法进行测定[9], 基本性质见表 1.
| 表1 4种供试土样基本理化性质 Table 1 Basic physical and chemical properties of the four tested soils |
 |
| 点击放大 |
根据DANTAS, 等[8]研究中的配方配制基础培养液, 然后用NaOH或稀HCl调节pH至6.2~6.5, 放入高压灭菌锅中灭菌后冷却, 加入已过0.22 μm微孔滤膜一定量质量浓度的青霉素或四环素溶液即得溶液SCS抗生素培养基;取pH 6.2~6.5的基础培养基, 按15 g/L加入琼脂, 然后放入高压灭菌锅灭菌15 min, 冷却至40~45 ℃时加入已过0.22 μm微孔滤膜一定质量浓度的青霉素或四环素母液, 配成不同质量浓度的固体SCS抗生素培养基.
1.3 以抗生素为单一碳源菌株的筛选称取1 g鲜土加入盛有高压灭菌后的50 mL磷酸盐缓冲溶液(PBS:1.44 g/L Na2HPO4·2H2O, 0.24 g/L KH2PO4, 8.00 g/L NaCl和0.20 g/L KCl的混合溶液)的三角瓶中, 用手摇动三角瓶1 min以混匀.然后吸取1 mL上述土壤溶液加入到装有9 mL抗生素SCS(含抗生素为1 000 mg/L)培养液的试管中, 培养1周后, 重复操作1次, 稀释4次后, 用移液枪吸取稀释液200 μL接种到含抗生素1 000 mg/L的固体SCS-抗生素培养基中, 用涂布棒涂匀, 倒置放入28 ℃培养箱中培养(以上实验过程中所用的枪头、试管, 固体和液体培养基等均经过高压灭菌), 直至固体抗性培养基表面长出菌落, 再通过平板划线得到纯培养物.
1.4 菌株生长曲线绘制取分离菌平板, 无菌操作挑取1环菌落, 接入含有1 000 mg/L青霉素抗生素SCS液体培养基中, 静置培养18 h作种子培养液.每个菌种准备8个盛有相同体积液体培养基的三角瓶(培养基分为含有抗生素100和1 000 mg/L的SCS-抗生素液体培养基), 定量接入种子液, 28 ℃振荡培养, 分别于1:00、4:00、8:00、12:00、16:00、20:00、24:00取出, 立即放入冰箱贮存, 待培养结束进行光密度值(D)测定, 以未接种的培养基作对照,600 nm波长分光光度计上调零点.
1.5 SCS培养液中抗生素质量浓度的测定分别将分离的菌株接种到含抗生素质量浓度为100 mg/L的SCS培养液中进行培养, 分别在培养第1天、第10天、第20天和第30天时用无菌枪头吸出培养液10 mL进行抗生素分析.
青霉素测定方法:青霉素的测定采用高效液相色谱串联质谱法测定.将吸出的SCS-青霉素培养液准确稀释1 000倍.吸取稀释后样品2 mL于10-mL塑料离心管中, 精确加入4 mL乙腈, 1 000 r/min涡旋振荡2 min, 2 800 r/min离心4 min, 精密移取5 mL上清液至另一10-mL塑料离心管中, 加入2.5 mL正己烷, 2 000 r/min涡旋振荡1 min, 静置, 待分层后弃去上层溶液, 下层溶液于45 ℃水浴中氮气吹干, 用1 mL水-乙腈(90:10)溶解残渣, 0.45 μm微孔滤膜滤过, 滤液再用0.2 μm微孔滤膜滤过, 作为供试液, 供LC-MS/MS检测[10].
四环素测定方法:采用高效液相色谱法测定培养液中的四环素.将吸出的SCS-四环素培养液精确稀释1 000倍.用C18吸附柱固相萃取法提取水样.分别用10 mL甲醇和10 mL EDTA (2 g/L), 以3.0 mL/min速度对C18吸附柱进行活化处理.吸取稀释后培养液8 mL用稀硫酸调至pH < 3, 以10.00 mL/min的速度流过CL8吸附柱,用5 mL纯水以5.0 mL/min速度清洗小柱, 以除去EDTA, 然后用氮气吹吸附柱20 min以除去吸附柱上的水分,用8 mL含0.01 moL/L草酸的甲醇以1.0 mL/min速度淋洗吸附柱(其中2 mL先浸泡吸附柱), 收集淋洗液[11].
1.6 菌株观察和染色菌株的肉眼和光学显微镜观察参照《伯杰细菌鉴定手册》[12].革兰染色参考《微生物学实验》[13].
1.7 分子生物学鉴定所分离的抗生素耐药菌按照传统方法进行DNA的提取, 采用细菌通用引物27F(5′-AGAGTTTGAT CCTGGCTCAG-3′), 1492R(5′-TACCTTGTTAC GACTT-3′)分别对菌株总DNA中的细菌16S rDNA片段进行扩增. 20 μL反应体系如下:10倍PCR缓冲液2 μL, dNTP混合液(各2.5 mmol/L) 1.6 μL, MgCl2(25 mmol/L)1.2 μL, 引物27F(10 μmol/L)1 μL, 引物1492R(10 μmol/L)1 μL, DNA模板1 μL, Taq酶(5 U/μL)0.1 μL, ddH2O 12.1 μL.PCR反应条件:94 ℃预变性5 min; 94 ℃变性30 s, 55 ℃退火45 s, 72 ℃延伸45 s, 35个循环; 最后72 ℃延伸7 min.扩增产物直接送上海生工生物工程公司进行测序.
1.8 数据分析采用Microsoft Excel 2007软件对数据进行处理和绘图, 采用MEGA5.0, 用邻接法构建系统发育树.
2 结果与分析 2.1 以青霉素或四环素为单一碳源的生长抗性细菌筛选从4个不同利用类型的土壤中共筛选到以青霉素为碳源生长的菌株2株(p4, p5), 均分离自森林土壤;以四环素为碳源生长的菌株3株(t1, t5, t9), 其中t1分离自茶园土壤, t5和t9分离自森林土壤.各菌株革兰染色和形态观察结果(表 2)表明,5种菌株均为革兰阴性菌, 菌落大小在0.1~2 mm, 菌落均为圆形且表面凸起.目前,在全球范围内, 革兰阴性致病菌感染引起的致病率和病死率占主导地位[14].可见, 浙江土壤中存在能以抗性素为单一碳源生长的抗性细菌, 虽然相对数量较小, 但是根据研究报道这些能以抗性素为碳源的细菌具有耐药的多样性, 而且许多与致病性相关[15].有趣的是, 虽然养殖场和医院用地土壤由于抗生素的应用抗性基因增加, 本次实验中却未分离到相关微生物, 而森林土壤未受到干扰却分离到了以抗生素为碳源生长的抗性细菌.相关研究表明大多数产抗生素菌株都携带对其抗生素的耐药性基因[16], 自然界中的风、水流、野鸟等都是抗生素抗性基因传播的重要驱动力[17], 因此即使是未受人为影响的森林土, 其土著微生物都有可能获得耐药性基因而变成耐药菌.本研究结果说明土壤即使未受干扰的林地存在能以抗生素为单一碳源的菌株, 是耐药菌的储存库, 应引起足够的重视.
| 表2 革兰染色和菌落形态观察 Table 2 Gram staining and the morphology observation of the five strains |
 |
| 点击放大 |
为了解菌株的生长特性与培养液中抗生素的关系, 分别将p4、p5接种到青霉素质量浓度为100和1 000 mg/L的SCS培养基中, 将t1、t5、t9接种到四环素质量浓度为100和1 000 mg/L的SCS培养基中.生长曲线结果(图 1)表明,5个菌株在监测的60 h内都呈现了完整的生长过程(迟缓区、对数区、稳定期、衰亡期).p4, p5菌株在100 mg/L青霉素SCS中40 h时吸光值最大, 而在1 000 mg/L抗生素时生长速度明显加快, 在20 h时D(600 nm)值达到最大, 并且菌株稳定期更长.t1, t5, t9在2种质量浓度抗生素培养液中的生长曲线规律与p4, p5类似, 3种菌株在较低质量浓度抗生素培养液中迟缓期和对数期时间较短, 在较高质量浓度抗生素吸光值增加较快, 衰亡期持续时间较长.5种抗生素在不同质量浓度培养液中的生长曲线表明,抗生素是菌株生长的限制因素, 抗生素质量浓度增加可刺激菌株生长.
 |
| 图1 菌株在不同抗生素质量浓度培养液中的生长曲线 Fig. 1 Growth curves of strains in different antibiotic concentration culture |
为进一步求证所分离菌株为以抗生素为单一碳源菌株, 将平板上保存的菌株接种到抗生素单一碳源培养液中, 研究抗生素质量浓度变化, 考虑到抗生素的自身降解同时做空白实验(即不接种菌株).由图 2可知, 青霉素和四环素会发生自身降解, 培养30 d时, 青霉素和四环素的自身降解率分别为22.6%和16.5%.向培养液中接种菌株后, 培养液中抗生素的质量浓度显著低于同期对照.其中接种p4培养第30天时, 溶液中的青霉素质量浓度比同期空白降低19.5%, 通过统计分析差异呈极显著水平(P < 0.01).同样, 接种t5和t9后四环素质量浓度同样比对照显著降低, 培养30 d时, 溶液中四环素质量浓度比对照分别降低29.1%和24.9%.结果说明分离的菌株利用培养液中抗生素的碳源实现了自身生长.
 |
| 图2 接种菌株后培养液中抗生素质量浓度变化 Fig. 2 Antibiotic concentration changes after strains inoculated in medium |
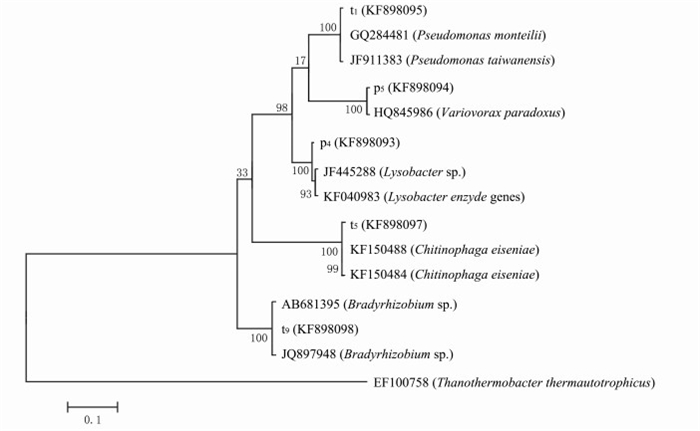
从抗性平板上分离得到的5株抗性细菌分别进行总DNA提取、16S rRNA基因扩增, 克隆测序后提交GenBank, 获得序列号为KF898093、KF898094、KF898095、KF898096、KF898097.应用MEGA 5.0软件采用Neighbor-Joining法Bootstrap为1000构建系统发育树, 确定其进化地位(图 3).
 |
| 图3 菌株16S rRNA基因的系统发育树 Fig. 3 Phylogenetic tree of strain 16S rRNA gene sequences |
从图 3可以看出, 5种菌株(p4, p5, t1, t5, t9)处于5个不同的分支.p4菌株与1株从根际土壤分离的菌株(SNNU513, JF445288)相似性达到了99%, 结合形态观察可初步确定p4菌株属于溶杆菌属(Lysobacter)的产酶溶杆菌(Lysobacter enzymogenes).溶杆菌属的微生物为化能异养革兰阴性杆菌, 好氧, 生长pH范围为5~10, 能产水溶性的棕色色素, 对几丁质及其他多糖有降解作用[8].p5菌株与分离自溴苯腈辛酸酯废水处理池1株溴苯腈辛酸酯降解菌株(XB3, HQ845986)相似性为100%, 该菌株为争论贪噬菌(Variovorax paradoxus), 因此p5属于贪噬菌属(Variovorax), 与争论贪噬菌Variovorax paradoxus的亲缘关系较近.Variovorax paradoxus能够分解利用多种有机硫化物、芳香烃、金属离子和其他化合物[18].
t1与分离自印度水样的Pseudomonas monteilii菌株(PCWCW13, GQ284481)和分离自活性污泥产聚羟基脂肪酸的Pseudomonas taiwanensis菌株(EPAn59, JF911383)相似性同为99%, 因此t1菌株大致可确定为假单胞菌属(Pseudomona).李显志,等[19]对假单胞菌属细菌的研究表明, 假单胞菌作为1种机会致病菌, 可以介导多种抗生素获得耐药性.同样, DANTAS, 等[8]研究分离到的75株以抗生素为单一碳源的抗性菌株中有24%属于假单胞菌属(Pseudomona).t5与分离自钾质粗面岩的2个Chitinophaga eiseniae菌株(JN251, KF150488)和(JN246, KF150484)相似性同为99%, 由此可确定t5属于噬几丁质菌属(Chitinophaga).t9与1株慢生根瘤菌属(Bradyrhizobium)的菌株(PeniS4C4, JQ897948)相似性为99%, 可初步确定t9为慢生根瘤菌属(Bradyrhizobium).可见, 分离到的以抗生素为碳源生长的抗性细菌具有群落多样性.
3 结论通过青霉素或四环素单一碳源选择性培养基(SCS),从未受干扰的森林土壤和长期施有机肥的茶园土壤中共分离出5株生长良好、菌落特征不同的菌株, 均为革兰阴性菌.抗生素是抗性菌株生长的限制因素, 抗生素质量浓度增加可以刺激菌株生长.分子生物学鉴定结果表明分离的菌株具有群落结构多样性, 分别属于溶杆菌属(Lysobacter)、贪噬菌属(Variovorax)、假单胞菌属(Pseudomona)、噬几丁质菌属(Chitinophaga)和慢生根瘤菌属(Bradyrhizobium), 这些菌株与一些复杂化合物难降解菌和机会致病菌的系统发育比较接近, 应引起重视.
| [1] | SARMAH A K, MEYER M T, BOXALL A. A global perspective on the use, sales, exposure pathways, occurrence, fate and effects of veterinary antibiotics (VAs) in the environment. Chemosphere, 2006, 65(5): 725-759. DOI:10.1016/j.chemosphere.2006.03.026 |
| [2] | HVISTENDAHL M. China takes aim at rampant antibiotic resistance. Science, 2012, 336(6083): 795-795. DOI:10.1126/science.336.6083.795 |
| [3] | RICHARDSON B J, LAM P K, MARTIN M. Emerging chemicals of concern: pharmaceuticals and personal care products (PPCPs) in Asia, with particular reference to Southern China. Marine Pollution Bulletin, 2005, 50(9): 913-920. DOI:10.1016/j.marpolbul.2005.06.034 |
| [4] | ALCOCK R E, SWEETMAN A, JONES K C. Assessment of organic contaminant fate in waste water treatment plants I: selected compounds and physicochemical properties. Chemosphere, 1999, 38(10): 2247-2262. DOI:10.1016/S0045-6535(98)00444-5 |
| [5] | KNAPP C W, DOLFING J, EHLERT P A, et al. Evidence of increasing antibiotic resistance gene abundances in archived soils since 1940. Environmental Science & Technology, 2010, 44(2): 580-587. |
| [6] | TORSVIK V, GOKSOYR J, DAAE F L. High diversity in DNA of soil bacteria. Applied and Environmental Microbiology, 1990, 56(3): 782-787. |
| [7] | THIELE S. Adsorption of the antibiotic pharmaceutical compound sulfapyridine by a long-term differently fertilized loess chernozem. Journal of Plant Nutrition and Soil Science, 2000, 163(6): 589-594. DOI:10.1002/(ISSN)1522-2624 |
| [8] | DANTAS G, SOMMER M O A, OLUWASEGUN R D, et al. Bacteria subsisting on antibiotics. Science, 2008, 320(5872): 100-103. DOI:10.1126/science.1155157 |
| [9] |
鲁如坤. 土壤农业化学分析方法. 北京: 中国农业科技出版社, 2000. LU R K. Analytical Method of Soil Agricultural Chemistry. Beijing: Chinese Agricultural Science and Technology Press, 2000. (in Chinese with English abstract) |
| [10] |
赵维, 杜钢, 李向荣. 高效液相色谱-串联质谱法测定牛奶中9种青霉素类药物的残留量. 浙江大学学报(医学版), 2012(2): 11. ZHAO W, DU G, LI X R. Determination of nine penicillin residues in milk by high-performance liquid chromatography-mass spectrometry. Journal of Zhejiang University (Medical Sciences), 2012(2): 11. (in Chinese with English abstract) |
| [11] |
胡冠九, 王冰, 孙成. 高效液相色谱法测定环境水样中5种四环素类抗生素残留. 环境化学, 2007, 26(1): 106-107. HU G J, WANG B, SUN C. Determination of five tetracycline residues in environmental water samples by high-performance liquid chromatography. Environmental Chemistry, 2007, 26(1): 106-107. (in Chinese with English abstract) |
| [12] |
中国科学院微生物研究所.伯杰细菌鉴定手册:8版.北京:科学出版社, 1984:274-313. Institute of Microbiology Chinese Academy of Science. Berger Bacterial Identification Manual: 8th. Beijing: Science Press, 1984:274-313. (in Chinese) |
| [13] |
肖明, 王雨净. 微生物学实验. 北京: 科学出版社, 2008: 50-57. XIAO M, WANG Y J. Microbiology Experiment. Beijing: Science Press, 2008: 50-57. (in Chinese with English abstract) |
| [14] | ZOWAWI H M, BALKHY H H, WALSH T R, et al. β-lactamase production in key gram-negative pathogen isolates from the Arabian Peninsula. Clinical Microbiology Reviews, 2013, 26(3): 361-380. DOI:10.1128/CMR.00096-12 |
| [15] | ABDELM Y, MONIB M, HAZEM A. Chloramphenicol, a simultaneous carbon and nitrogen source for Streptomyes sp. from Egyptian soil. Nature, 1961, 189(476): 775-776. |
| [16] | HOPWOOD D A. How do antibiotic-producing bacteria ensure their self-resistance before antibiotic biosynthesis incapacitates them?. Molecular Microbiology, 2007, 63(4): 937-940. DOI:10.1111/mmi.2007.63.issue-4 |
| [17] | ALLEN H K, DONATO J, WANG H H, et al. Call of the wild: antibiotic resistance genes in natural environments. Nature Reviews Microbiology, 2010, 8(4): 251-259. DOI:10.1038/nrmicro2312 |
| [18] | SATOLA S W, FARLEY M M, ANDERSON K F, et al. Comparison of detection methods for heteroresistant vancomycin-intermediate Staphylococcus aureus, with the population analysis profile method as the reference method. Journal of Clinical Microbiology, 2011, 49(1): 177-183. DOI:10.1128/JCM.01128-10 |
| [19] |
李显志. 铜绿假单胞菌主动外排泵介导的多重抗生素耐药性. 中国抗生素杂志, 2003, 28(10): 577-596. LI Z X. Efflux-mediated multiple antibiotic resistance in Pseudomonas aeruginosa. Chinese Journal of Antibiotics, 2003, 28(10): 577-596. (in Chinese with English abstract) |
 2016, Vol. 42
2016, Vol. 42


