| 荞麦属植物脱落酸不敏感基因序列比较与亲缘关系研究 |
荞麦是蓼科(Polygnaceae)荞麦属(Fagopyrum Mill.)植物[1-2],是我国粮食作物中重要的小宗杂粮,其蛋白质含量高,富含黄酮类化合物,特别是芦丁含量较高,具有显著的降血脂、降血糖和降血压等营养保健功能[2-3]。陈庆富[1]指出,荞麦属至少包括23个种类,根据形态学鉴定可分为大粒组和小粒组2个类群,类群间亲缘关系较远。现有研究普遍认为荞麦起源于中国的西南地区[2]。YAMANE等[4]的研究指出,四倍体金荞麦主要分布于西藏地区,二倍体金荞麦则主要分布在西藏、云南和四川等地,其中分布于云南和四川地区的二倍体金荞麦可分别被聚为一类。近年来,学者们对荞麦的起源与进化关系进行了较为深入的研究,但由于荞麦属植物种间关系比较复杂,至今仍未有一致定论[5-10]。随着分子生物学技术的不断发展,在传统形态学鉴定、蛋白亚基和同工酶技术等基础上,结合DNA分子鉴定技术可为研究荞麦进化关系提供更为可靠的依据。
荞麦主要生长于高寒地区,环境较为恶劣,常遇干旱与冻害等自然灾害,因此,在长期的进化过程中,荞麦在抗旱、抗寒,以及耐贫瘠等方面表现出较强的耐逆特性[11]。脱落酸(abscisic acid, ABA)是重要的植物应激激素,在调控作物抗逆性方面具有重要作用[12-13]。生物或非生物逆境会诱导植物体内迅速合成ABA,进而增强植物逆境耐受能力[14]。ABA信号途径的分子调控网络十分复杂,其中,脱落酸不敏感(ABA-insensitive, ABI)家族蛋白是参与调控ABA信号途径的重要转录因子[15-16]。ABI蛋白中ABI1和ABI2在ABA信号转导途径中起着负调控作用[17],二者功能缺失都会表现出对ABA的高度敏感。ABI3、ABI4和ABI5则通过调控ABA信号转导途径参与调控种子成熟、休眠、萌发和幼苗生长过程[18-22]。郝红梅[23]研究发现,植物ABI序列较为保守,可用于植物进化与亲缘关系研究。迄今为止,尚未见有关荞麦ABI基因研究的相关报道。因此,本研究从转录组测序构建的UniGene数据库中筛选获得荞麦ABI UniGene信息,通过设计特异性引物对荞麦属不同野生种进行目标基因的聚合酶链式反应(polymerase chain reaction, PCR)扩增,利用序列差异分析与聚类分析,探讨荞麦ABI基因在进化过程中的序列变异规律与不同荞麦野生种的起源关系。
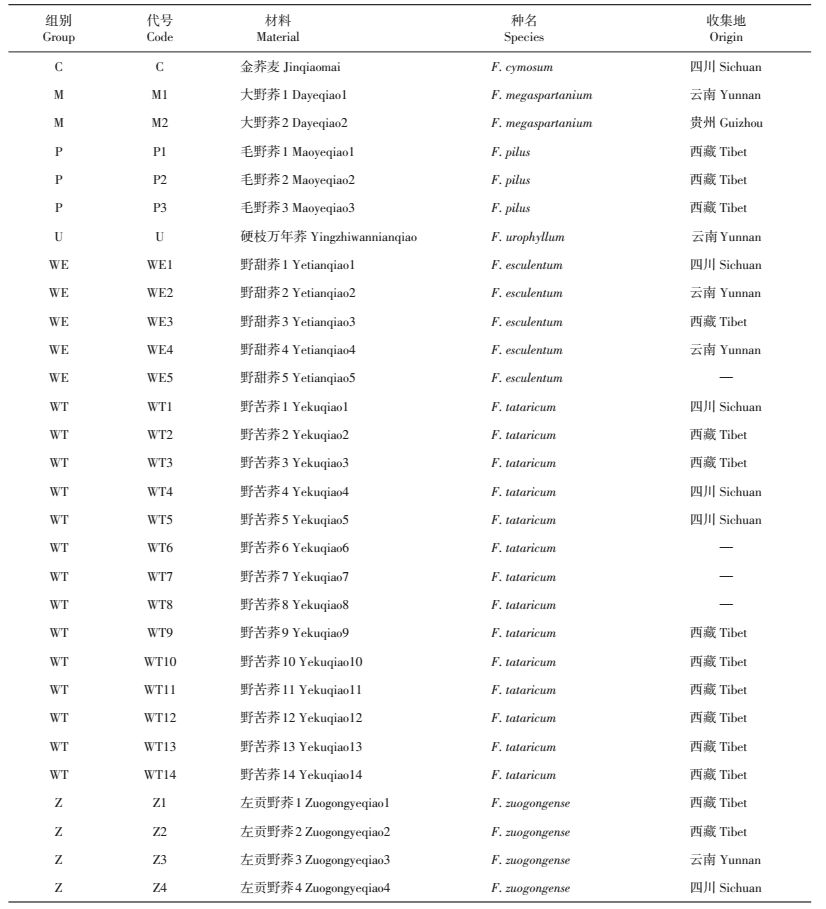
1 材料与方法 1.1 试验材料试验选用7个荞麦野生种,包括野甜荞(F.esculentum)、野苦荞(F.tataricum)、左贡野荞(F.zuogongense)、金荞麦(F.cymosum)、硬枝万年荞(F.urophyllum)、毛野荞(F.pilus)和大野荞(F.megaspartanium),共30份材料(由贵州师范大学荞麦产业技术研究中心提供,表 1)。
| 表1 试验材料 Table 1 Plant materials used in this study |
 |
| 点击放大 |
在苗期对植物材料进行取样,采用CTAB法提取幼嫩叶片的DNA,具体操作步骤参考陈晴晴等[24]的方法。通过转录组测序构建的数据库(登录号:SRS1350375)进行荞麦ABI UniGene筛选。利用Primer 5.0软件对筛选获得的ABI基因进行设计并合成特异性引物[生工生物工程(上海)股份有限公司]。目标基因的PCR扩增与琼脂糖凝胶电泳参考陈晴晴等[24]的方法。将检测合格的PCR扩增产物送至生工生物工程(上海)股份有限公司进行测序。
1.3 测序结果分析与进化树的构建利用NCBI BLAST(http://blast.ncbi.nlm.nih.gov/Blast.cgi)进行序列的在线比对。将序列对齐分值大于130、同一性大于70%的基因序列定义为高度同源。使用DNAsp5软件进行序列差异分析,利用MEGA 5.05软件进行ClustalW多序列比对、遗传距离和聚类分析。
2 结果与分析 2.1 荞麦ABI基因的PCR扩增对转录组基因测序(登录号:SRS1350375)获得的荞麦UniGene数据库进行基因序列比对,筛选得到荞麦ABI的UniGene序列。提取30个荞麦野生种质幼嫩叶片中的DNA,设计ABI UniGene的特征引物,进行基因序列扩增。琼脂糖凝胶电泳检测结果显示,在400~500 bp间出现目标条带(图 1)。
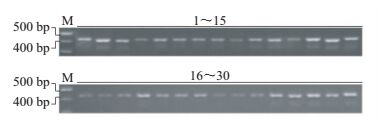 |
| M:DNA分子质量标志物;(1~15)、(16~30):30个荞麦野生种。 M: DNA marker; (1-15), (16-30): Thirty wild germplasms from Fagopyrum. 图1 利用特异引物获得的PCR扩增产物 Fig. 1 PCR amplification products obtained using specific primers |
将上述扩增得到的所有目标条带送生工生物工程(上海)股份有限公司进行测序,然后用NCBIBLAST对其中的WT1目标片段序列进行比对,发现其与蓖麻(Ricinus communis)ABI5-like2基因(登录号:XM_015729138.1)、葡萄(Vitis vinifera)ABI5-like2基因(登录号:XM_002265711.3)和枣(Ziziphusjujube)ABI5-like2基因(登录号:XM_016020783.1)等序列对齐分值大于130,相似性高于70%。说明扩增所获得的目标片段为荞麦ABI基因。对30份荞麦野生材料ABI基因片段序列中的429个排列位点进行多重比较分析,发现不变位点为342个,多态位点(S)为87个,占20.28%,其中包含84个简约信息位点和3个单型可变位点。说明荞麦属植物种间ABI基因序列差别较小。对14份野苦荞ABI基因的431个排列位点进行多重比较分析,发现不变位点423个,多态位点8个,占1.86%,其中包含6个简约信息位点和2个单型可变位点;对5份野甜荞ABI基因的442个排列位点进行多重比较分析,发现不变位点434个,多态位点8个,占1.81%,其中包含1个简约信息位点和7个单型可变位点。说明荞麦属植物种内ABI基因序列高度保守。
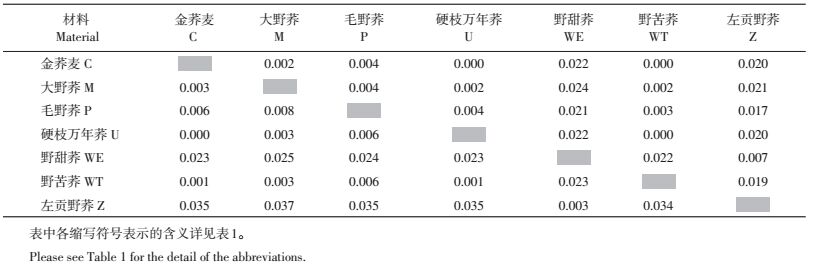
2.3 基于ABI基因序列的亲缘关系对30份荞麦属野生材料的ABI基因片段序列进行遗传距离分析,发现种间平均遗传距离为0.015,其中野甜荞与野苦荞的种间平均遗传距离为0.023,与大野荞为0.025,与毛野荞为0.024,与左贡野荞为0.003。从表 2可知,左贡野荞与大野荞种间遗传距离最大(0.037),硬枝万年荞与金荞麦种间遗传距离最小(0.000)。
| 表2 基于ABI基因片段序列的种间平均遗传距离(下三角)和种间平均净遗传距离(上三角) Table 2 Interspecific genetic distance (below diagonal) and interspecific net genetic distance (above diagonal) based on ABI gene sequence |
 |
| 点击放大 |
利用植物ABI基因序列种内高度保守的特性,对7个野生种共30份野生材料进行聚类分析。结果(图 2)发现:硬枝万年荞、大野荞、野苦荞、金荞麦和毛野荞被聚为一类,节点的自举置信度为98%;野甜荞与左贡野荞被聚为一类,节点的自举置信度为99%;收集于四川的3个野苦荞WT1、WT4和WT5被单独聚为一类;野甜荞与收集于西藏的左贡野荞Z1和Z2被聚为一类;收集于云南和四川的左贡野荞Z3、Z4与Z1、Z2及其他野生荞麦的亲缘关系较远,被单独聚为一类。
 |
| 图上各符号表示的含义详见表 1。 Please see Table 1 for the detail of the code. 图2 基于荞麦ABI基因序列的聚类树状图 Fig. 2 Phylogenetic dendrogram of buckwheat collections based on specific sites of ABI gene fragment sequence |
荞麦生长适应性强,具有较强的抗寒、抗旱、抗辐射和耐贫瘠能力,是一种重要的救灾粮食作物[2,11]。ABI转录因子在调控ABA信号转导介导的逆境胁迫应答方面具有重要作用[25-27]。郝红梅[23]的研究表明,白菜ABI1序列与拟南芥ABI1序列高度保守。本研究发现,7个野生种共30份野生种质的ABI基因片段的多态位点占20.28%,其中,14份野苦荞间多态位点占1.86%,5份野甜荞间多态位点占1.81%:说明荞麦属植物种内ABI基因序列高度保守。因此,荞麦ABI基因序列差异可作为研究荞麦属植物亲缘关系的依据。此外,本研究还发现:收集于四川和云南的左贡野荞Z3和Z4单独聚为一类,与西藏的左贡野荞Z1和Z2存在较明显的差异;同时,收集于四川的野苦荞WT1、WT4和WT5也被单独聚在一起,与其余地区的野苦荞存在较明显差异。说明在进化过程中生长环境对荞麦ABI基因序列变异具有影响。
荞麦属包括至少23个种,其种间亲缘关系较为复杂[1]。陈庆富[1,28]通过形态学鉴定发现左贡野荞与甜荞较为相似。梁成刚等[29]对荞麦属植物13S球蛋白基因序列进行分析发现,甜荞及野甜荞与左贡野荞亲缘关系较近。根据ABI基因片段序列差异,在本研究中收集于西藏的左贡野荞Z1、Z2和野甜荞聚为一类,说明其亲缘关系较近。前人通过形态学观察和同工酶研究等方式推测甜荞与苦荞可能由金荞进化而来[7-9]。陈庆富[1,28]研究发现金荞中存在3种类型且彼此间生殖隔离,因此,将其中2类二倍体种分别命名为大野荞与毛野荞,并将两者与金荞一起归为金荞复合物一类。张以忠等[6]利用过氧化物酶同工酶鉴定,将甜荞与大野荞、苦荞与毛野荞分别聚为一类。LI等[5]则通过十二烷基硫酸钠-聚丙烯酰胺凝胶电泳(SDS-PAGE)鉴定将甜荞、左贡野荞与大野荞聚为一类,将苦荞、金荞、毛野荞和巨荞聚为一类。任翠娟等[30]通过随机扩增多态性DNA标记技术(RAPD)鉴定,发现大野荞与甜荞、毛野荞与苦荞亲缘关系较近。由此,陈庆富[1]推测甜荞的祖先种可能为大野荞,而苦荞的祖先种可能为毛野荞。然而,甜荞是典型的异花授粉植物,苦荞为典型的自花授粉植物,其各自的进化历程十分复杂[9]。郑亚迪[31]基于ITS.matK和psbA-trnH单基因与叶绿体matK+psbA-trnH和ITS+matK+psbA-trnH合并数据集分析,发现四川海螺沟野荞与其他荞麦类群均存在较远的亲缘关系,推测其可能为最原始的荞麦种。本研究利用ABI基因序列差异鉴定发现,大野荞与野苦荞、金荞麦聚为一类,然后再与毛野荞聚为一类,而大野荞与野甜荞遗传距离较远,这与梁成刚等[29]的研究结果一致。然而,目前尚不能确定荞麦不同种间的具体进化历程。
硬枝万年荞的遗传多样性非常丰富,其进化关系较为复杂。史建强等[32]利用简单重复序列(SSR)分子标记鉴定,将硬枝万年荞与疏穗小野荞、细柄野荞、小野荞和齿翅野荞聚为一类。梁成刚等[29]利用13S球蛋白基因序列差异鉴定,将细柄野荞、硬枝万年荞、苦荞和部分野苦荞聚为一类。郑亚迪[31]利用叶绿体matK和psbA-trnH序列联合分析将硬枝万年荞与苦荞聚为一类,然后再与金荞和汶川苦荞聚为一类,并通过综合分析认为硬枝万年荞是较汶川野荞和金荞更原始的野生荞麦种。本研究发现,硬枝万年荞的ABI基因片段序列与大野荞及部分野苦荞聚为一类,说明其差异较小,亲缘关系较近。
| [1] |
陈庆富. 荞麦属植物科学. 北京: 科学出版社, 2012: 5-55. CHEN Q F. Plant Sciences on Genus Fagopyrum. Beijing: Science Press, 2012: 5-55. (in Chinese with English abstract) |
| [2] | RAINA A P, GUPTA V. Evaluation of buckwheat (Fagopyrum species) germplasm for rutin content in seeds. Indian Journal of Plant Physiology, 2015, 20(2): 167-171. DOI:10.1007/s40502-015-0147-6 |
| [3] | AHMED A, KHALID N, AHMAD A, et al. Phytochemicals and biofunctional properties of buckwheat: A review. Journal of Agricultural Science, 2013, 152(3): 349-369. |
| [4] | YAMANE K, OHNISHI O. Phylogenetic relationships among natural populations of perennial buckwheat, Fagopyrum cymosum Meisn., revealed by allozyme variation. Genetic Resources and Crop Evolution, 2001, 48(1): 69-77. DOI:10.1023/A:1011265212293 |
| [5] | LI J H, CHEN Q F, ZELLER F J. Variation in seed protein subunits among species of the genus Fagopyrum Mill. Plant Systematics and Evolution, 2008, 274(3/4): 193-202. |
| [6] |
张以忠, 李艳娟, 邓琳琼, 等. 荞麦过氧化物酶同工酶研究. 种子, 2011, 30(2): 52-54. ZHANG Y Z, LI Y J, DENG L Q, et al. Study on peroxidase isozyme of buckwheat. Seed, 2011, 30(2): 52-54. (in Chinese with English abstract) |
| [7] |
赵钢, 唐宇. 荞麦过氧化物同工酶研究. 荞麦动态, 1990(2): 10-15. ZHAO G, TANG Y. Study on peroxidase isoenzyme of buckwheat. Buckwheat Dynamic, 1990(2): 10-15. (in Chinese with English abstract) |
| [8] | HEDBERG O. Pollen morphology in the genus Polygonum L. s. lat. and its taxonomical significance. Svensk Botanisk Tidskrift, 1946, 40(4): 371-404. |
| [9] |
任翠娟, 陈庆富. 荞麦(Fagopyrum)的形态解剖学及系统学研究. 贵州教育学院学报(自然科学), 2008, 19(3): 23-27. REN C J, CHEN Q F. The current status and prospect of the study on morphological anatomy and phylogeny of Fagopyrum. Journal of Guizhou Educational Institute (Natural Science), 2008, 19(3): 23-27. (in Chinese with English abstract) |
| [10] | SMITH C. Encyclopedia of global archaeology//WEISSKOPF A, FULLER D Q. Buckwheat: Origins and Development. New York, USA: Springer, 2014:1025-1028. |
| [11] | JACQUEMART A L, CAWOY V, KINET J M, et al. Is buckwheat (Fagopyrum esculentum Moench) still a valuable crop today. The European Journal of Plant Science and Biotechnology, 2012, 6(Special Issue 2): 1-10. |
| [12] | RAGHAVENDRA A S, GONUGUNTA V K, CHRISTMANN A, et al. ABA perception and signalling. Trends in Plant Science, 2010, 15(7): 395-401. DOI:10.1016/j.tplants.2010.04.006 |
| [13] | GOMEZ-CADENAS A, VIVES V, ZANDALINAS S I, et al. Abscisic acid: A versatile phytohormone in plant signaling and beyond. Current Protein and Peptide Science, 2015, 16(5): 413-434. DOI:10.2174/1389203716666150330130102 |
| [14] | HAYASHI K I, KINOSHITA T. Plant signaling: Abscisic acid receptor hole-in-one. Nature Chemical Biology, 2014, 10(6): 414-415. DOI:10.1038/nchembio.1529 |
| [15] | KOORNNEEF M, REULING G, KARSSEN C M. The isolation and characterization of abscisic acid-insensitive mutants of Arabidopsis thaliana. Physiologia Plantarum, 1984, 61(3): 377-383. DOI:10.1111/ppl.1984.61.issue-3 |
| [16] | FINKELSTEIN R R. Abscisic acid-insensitive mutations provide evidence for stage-specific signal pathways regulating expression of an Arabidopsis late embryo genesis-abundant (lea) gene. Molecular and General Genetics, 1993, 238(3): 401-408. DOI:10.1007/BF00291999 |
| [17] | HIRAYAMA T, SHINOZAKI K. Perception and transduction of abscisic acid signals: Keys to the function of the versatile plant hormone ABA. Trends in Plant Science, 2007, 12(8): 343-351. DOI:10.1016/j.tplants.2007.06.013 |
| [18] | LIU H, STONE S L. Regulation of ABI5 turnover by reversible posttranslational modifications. Plant Signaling and Behavior, 2014, 9(1): e27577. DOI:10.4161/psb.27577 |
| [19] | YU F, WU Y, XIE Q. Precise protein post-translational modifications modulate ABI5 activity. Trends in Plant Science, 2015, 20(9): 569-575. DOI:10.1016/j.tplants.2015.05.004 |
| [20] | REEVES W M, LYNCH T J, MOBIN R, et al. Direct targets of the transcription factors ABA-insensitive (ABI) 4 and ABI5 reveal synergistic action by ABI4 and several bZIP ABA response factors. Plant Molecular Biology, 2011, 75(4/5): 347-363. |
| [21] | FINKELSTEIN R, LYNCH T, REEVES W, et al. Accumulation of the transcription factor ABA-insensitive (ABI) 4 is tightly regulated post-transcriptionally. Journal of Experimental Botany, 2011, 62(11): 3971-3979. DOI:10.1093/jxb/err093 |
| [22] | LOPEZ- MOLINA L, MONGRAND S, MCLACHLIN D T, et al. ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. The Plant Journal, 2002, 32(3): 317-328. DOI:10.1046/j.1365-313X.2002.01430.x |
| [23] |
郝红梅. 白菜PP2C型蛋白磷酸酶ABI1同源蛋白(BraABI1b)参与植物ABA反应的初步功能分析. 陕西, 杨凌: 西北农林科技大学, 2015: 1-44. HAO H M. Functional analysis of an ABA insensitive 1 like type-2C protein phosphatase in Brassica rapa. Yanglin, Shaanxi: Northwest Agricultural and Forestry University, 2015: 1-44. (in Chinese with English abstract) |
| [24] |
陈晴晴, 石桃雄, 陈庆富. 荞麦不同种间BW10KD过敏蛋白基因序列比较. 广东农业科学, 2013, 40(9): 133-139. CHEN Q Q, SHI T X, CHEN Q F. Comparison of BW10KD allergen protein gene sequences among different buckwheat species. Guangdong Agricultural Sciences, 2013, 40(9): 133-139. (in Chinese with English abstract) |
| [25] | KASHIWAKURA Y I, KOBAYASHI D, JIKUMARU Y, et al. Highly sprouting tolerant wheat grain exhibits extreme dormancy and cold imbibition resistant accumulation of abscisic acid. Plant and Cell Physiology, 2016, 57(4): 715-732. DOI:10.1093/pcp/pcw051 |
| [26] | BROCARD I M, LYNCH T J, FINKELSTEIN R R. Regulation and role of the Arabidopsis abscisic acid-insensitive 5 gene in abscisic acid, sugar, and stress response. Plant Physiology, 2002, 129(4): 1533-1543. DOI:10.1104/pp.005793 |
| [27] | OOMS J J, LEON-KLOOSTERZIEL K M, BARTELS D, et al. Acquisition of desiccation tolerance and longevity in seeds of Arabidopsis thaliana: A comparative study using abscisic acidinsensitive abi3 mutants. Plant Physiology, 1993, 102(4): 1185-1191. DOI:10.1104/pp.102.4.1185 |
| [28] | CHEN Q F. A study of resources of Fagopyrum (Polygonaceae) native to China. Botanical Journal of the Linnean Society, 1999, 130(1): 53-64. DOI:10.1111/boj.1999.130.issue-1 |
| [29] |
梁成刚, 陈晴晴, 石桃雄, 等. 荞麦属种间13S球蛋白基因序列比较研究. 植物遗传资源学报, 2016, 17(3): 541-546. LIANG C G, CHEN Q Q, SHI T X, et al. Sequence analysis of 13S globulin gene of genus Fagopyrum species. Journal of Plant Genetic Resources, 2016, 17(3): 541-546. (in Chinese with English abstract) |
| [30] |
任翠娟, 陈庆富. 荞麦属(Fagopyrum Mill)植物资源的RAPD研究. 种子, 2009, 28(11): 37-44. REN C J, CHEN Q F. Study on RAPD of genus Fagopyrum resources. Seed, 2009, 28(11): 37-44. (in Chinese with English abstract) DOI:10.3969/j.issn.1005-2690.2009.11.024 |
| [31] |
郑亚迪. 西南地区荞麦属植物分子系统发育研究. 成都: 四川农业大学, 2012: 32-38. ZHENG Y D. Analysis of phylogenetic relationships of Fagopyrum Mill based on ITS, matK and psbA-trnH sequences. Chengdu: Sichuan Agricultural University, 2012:32-38. (in Chinese with English abstract) http://cdmd.cnki.com.cn/Article/CDMD-10626-1013157687.htm |
| [32] |
史建强, 李艳琴, 张宗文, 等. 荞麦及其野生种遗传多样性分析. 植物遗传资源学报, 2015, 16(3): 443-450. SHI J Q, LI Y Q, ZHANG Z W, et al. Genetic diversity of buckwheat and its wild species. Journal of Plant Genetic Resources, 2015, 16(3): 443-450. (in Chinese with English abstract) |
 2017, Vol. 43
2017, Vol. 43









