| Transcriptomic analysis reveals polygalacturonase genes in Pachypeltis micranthus |
2. Institute of Forestry Protection, Yunnan Academy of Forestry, Kunming 650204, China;
3. Bureau of Forestry Pest Control and Quarantine of Yunnan Province, Kunming 650051, China
2. 云南省林业科学院森林保护研究所, 昆明650204;
3. 云南省林业有害生物防治检疫局, 昆明 650051
The mirid bug, Pachypeltis micranthus (Hemiptera: Miridae), feeds on the climbing hemp vine, Mikania micrantha, which has been firstly discovered in Dehong, Yunnan in 2008 and known as an important biological agent of this notorious invasive alien weed[1]. Due to the efficient control effect on climbing hemp vine, it has been well characterized on ecological and biological aspects[2-3], while the information about this insect in the other aspects remains to be explored.
Similar to other species of mirid bugs, P. micranthus nymphs and adults utilize their piercing-sucking mouthparts to imbibe the contents from the cell wall components and lyses cells of the plant tissue degraded by the injected saliva[4]. In the saliva, there are various digestive enzymes secreted by the salivary glands. Of these, polygalacturonase (PG) is one of the most important components involved in the induction of visible plant injury by degrading the pectin in the cell walls. PG is a cell wall hydrolytic enzyme that catalyzes the random hydrolysis, and disassembly of the 1, 4-α-D-galactosiduronic linkages of pectin were shown to be the main cause of mirid bugs feeding damags[5-6]. To our knowledge, although multiple forms of PG genes have been identified from Apolygus lucorum, Lygus hesperus and L. lineolaris, little is still known about the molecular features of this enzyme in mirid bugs[4, 7-8]. Due to the importance of PG, knowledge of it is crucial to understanding how P. micranthus feeds and interacts with its host plants.
To compile molecular resources for further studies on P. micranthus, and explore the PG gene family of this species, we here present the results of de novo transcriptome sequencing of P. micranthus using a highthroughput sequencing method with the advantage of next generation sequencing that has made it a new powerful approach to characterize the genomic background of insect species lacking a fully sequenced genome on a large scale[9-10]. Additionally, PG genes were identified from the transcriptomic database.
1 Materials and methods 1.1 RNA isolationThe adults of P. micranthus were collected from the suburbs of Dehong, Yunnan Province, China. Whole bodies of 20 adult females and 20 adult males after 5 days emergence were immediately homogenated into Trizol regent (Invitrogen). During transferring from the collection field to the laboratory, the samples were stored at a cooler containing dry ice for preservation. Then, total RNA was extracted using Trizol regent following the manufacturer’ s protocol and treated with RNase-free DNase (Qiagen). The quantity and quality of the RNA aliquots were evaluated by a bioanalyzer 2100 (Agilent).
1.2 Library construction and sequencingThe mRNA was purified from approximately 10 μg of the total RNA with magnetic oligo-dT beads according to the Illumina manufacturer’ s instructions. After fragmented into short sequences, the first second strand cDNA was synthesized using transcriptase and random primers. Then the second strand cDNA synthesis was performed using DNA polymerase I and RNaseH. Fragments were end repaired, A-tailed and indexed adapters were ligated, which were purified and enriched with polymerase chain reaction (PCR) to create the final cDNA library. The library was sequenced using Illumina HiSeqTM 2000.
1.3 Transcript assembly and annotationThe image data were transformed by base calling into raw reads, which were submitted to DNA Data Bank of Japan (DDBJ) (http://www.ddbj.nig.ac.jp/) sequence read archive (SRA) under the accession number of DRA001048[11]. The raw reads were filtered by removing adapter sequences and low-quality sequences into clean reads. They were assembled into contigs and unigenes using Trinity software[12]. During assembly, the minimum contig length and pair number cutoff of trinity were respectively set as 100 bp and 4, and other default parameters were used to run it. The assembled unigenes were submitted to decide the sequence direction using BlastX algorithm (E-value<10-5) to search against the protein databases including the National Center for Biotechnology Information (NCBI) non-redundant (Nr) database, Swiss-Prot, Gene Ontology (GO), Clusters of Orthologous Groups (COG), and Kyoto Encyclopedia of Genes and Genomes (KEGG). If results of different databases were conflicting, a priority order was followed to above list of the databases. ESTScan was used to define the sequence direction when a unigene has no search response in the above databases. Then, all unigenes were used for subsequent bioinformatics analysis. Briefly, unigenes were searched against Nr database with using BlastX algorithm with E-value cutoff of 10-5. GO annotations were performed in the Blast2GO software suit[13] by mapping the blast results to GO terms using default parameters according to molecular function, biological process and cellular component ontologies. To predict and classify possible functions, unigenes were aligned to the COG database. KEGG annotation was performed to assign the unigenes into pathways. COG and KEGG analysis were carried out in Blast2GO.
1.4 Identification of PG genesPG amino acid sequences reported from A. lucorum, L. hesperus and L. lineolaris were used to search against protein sequences predicted from the assembled unigenes using BlastP with an E-value cut off of lower than 10-5. The identified PG genes sequences will be available upon request. Multiple alignment of PG amino acid sequences were conducted with ClustalX version 1.83[14]. A neighborjoining (NJ) tree of PGs from mirid bugs using bootstrap values (1 000 replicates) and Macrophomina phaseolina MS6 glycoside hydrolase served as the outgroup was constructed with MEGA version 5[15].
2 Results 2.1 Illumina sequencing, assembly and transcript annotationDeep sequencing generated 62 558 584 raw reads, which yielded 52 204 818 clean reads after filtering. The GC content was 46.19%. This represented more than 5.2 Gbps of genomic sequences. These high-quality clean reads were assembled into 143 006 contigs and 57 739 unigenes. The mean length of unigenes was 765 bp, with an N50 of 1 204 bp. Of these unigenes, 23 549 (40.86%) were shown to have putative homologues in the Nr database (Fig. 1). Although we used an E-value threshold of E<10-5 for this analysis, the majority of matches were below this threshold. Also, most of matches had similarities between 40% and 80%. The greatest number of matches (49.95%) was with sequences from the non-typical model insect species followed by the sequences from Tribolium castaneum, Acyrthosiphon pisum and Nasonia vitripennis. Based on Blast matches with sequences whose function is previously known, 7 048 unigenes were annotated in GO, producing 15 646 terms for biological process, 9 980 for cellular component, and 6 177 for molecular function (Fig. 2). Among the biological process, a high percentage of genes were assigned for cellular process, metabolic process and biological regulation. In cellular component, cell and cell part were dominant. The molecular function terms were associated predominantly with binding and catalytic activity. In respect of COG classification, 8 548 unigenes were assigned to 25 COG categories (Fig. 3). The cluster for general function prediction only represents the largest group (17.58%) followed by transcription (9.65%), translation, ribosomal structure (7.41%) and biogenesis and replication, recombination and repair (7.07%). A total of 16 135 of the assembled unigenes were mapped to 255 KEGG pathways. Of the pathways identified, metabolic pathways contained 2 552 unigenes (15.85%) was significantly larger than other pathways.
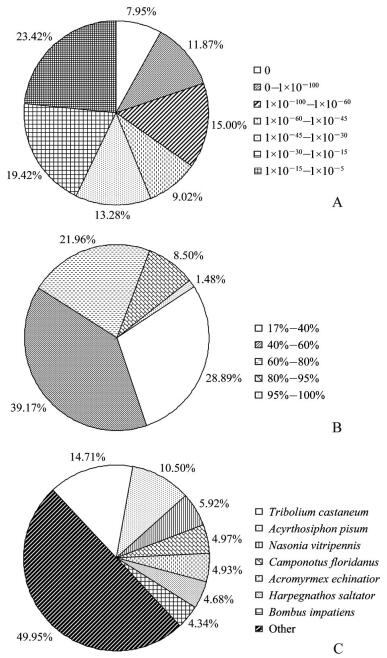 |
| The first BlastX hit of each unigene with a cut-off E-value of 1×10-5 was used for analysis. A: E-value distribution; B: similarity distribution; C: species distribution. Fig. 1 Nr classification of unigenes |
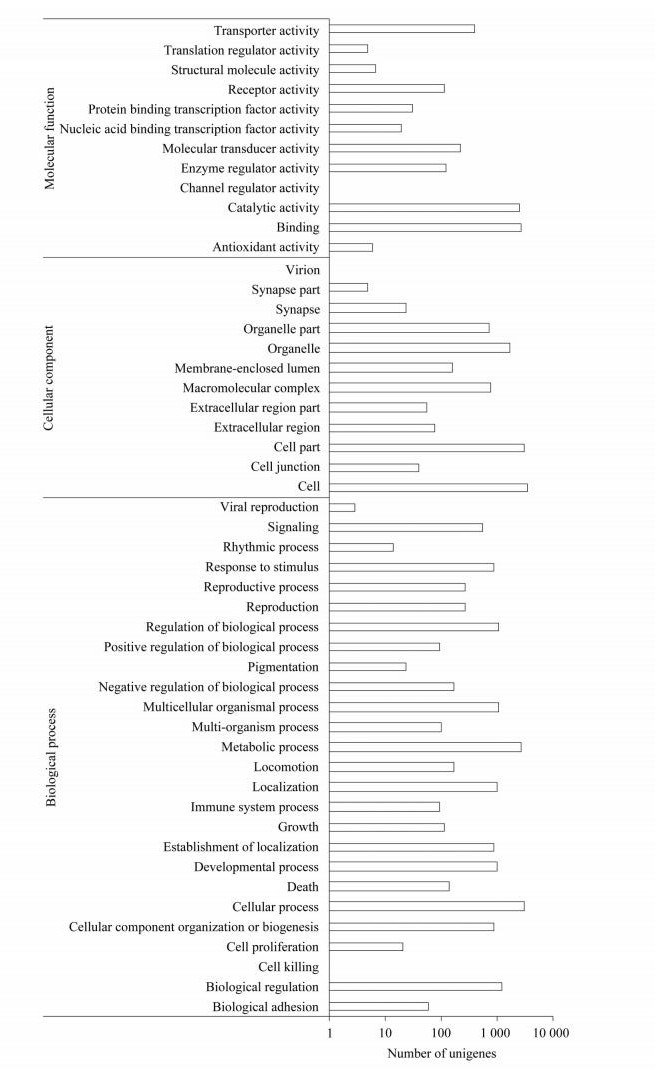 |
| Unigenes were annotated in three main categories: molecular function, cellular component and biological process. Fig. 2 GO annotation of unigenes |
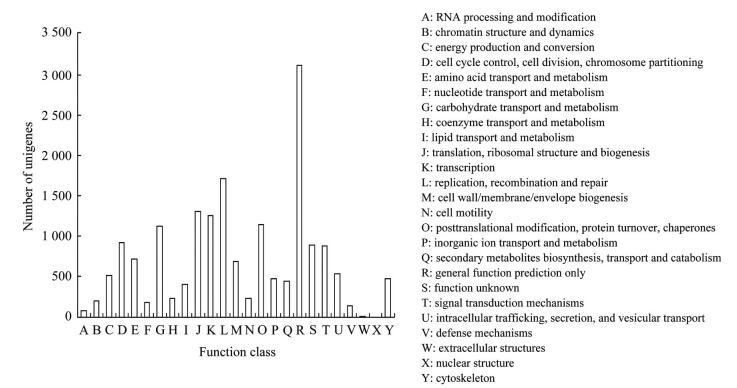 |
| In total, 8 548 unigenes have a COG classification among the 25 categories. Fig. 3 COG classification of unigenes |
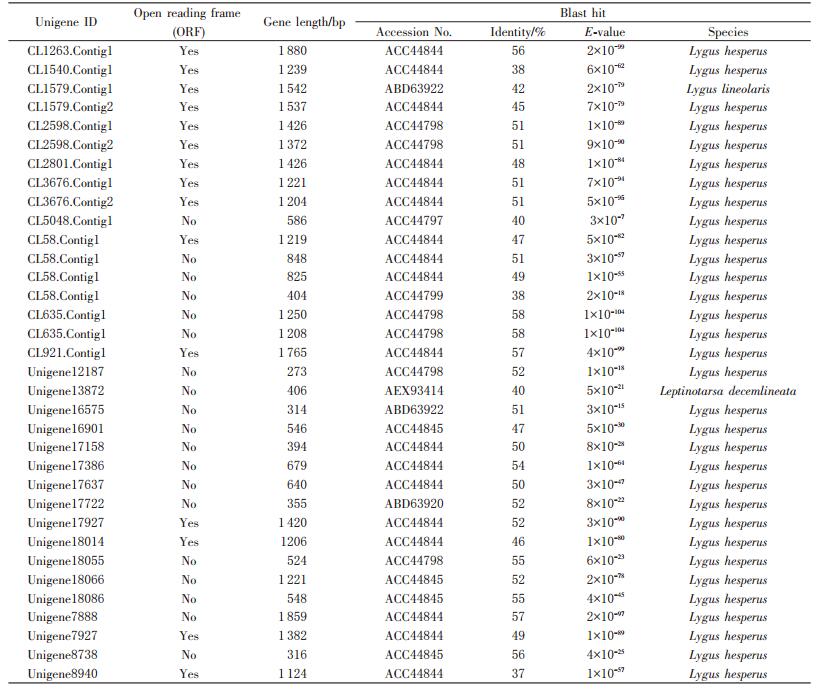
Twenty four putative PG transcripts were identified in the P. micranthus transcriptome (Table 1). Of these, 15 full-length protein sequences were predicted. Sequence similarity analysis indicated that their protein sequences encoded by them all showed 32%-96% identity with each other (Fig. 4). The typical features including signal peptides, two disulfide bridges and enzymatically critical amino acid motifs were conserved across these PGs. A PG phylogenetic tree was constructed to facilitate comparison of PGs from P. micranthus and other three mirid bugs (Fig. 5). The phylogenetic tree showed that P. micranthus PGs formed four distinct clades, of which contained four, six, two and three members, respectively. Of interest, PGs from P. micranthus were distinctly separated from other groups formed by A. lucorum, L. hesperus and L. lineolaris PGs.
| Table 1 Putative PG genes identified in P. micranthus transcriptome |
 |
| 点击放大 |
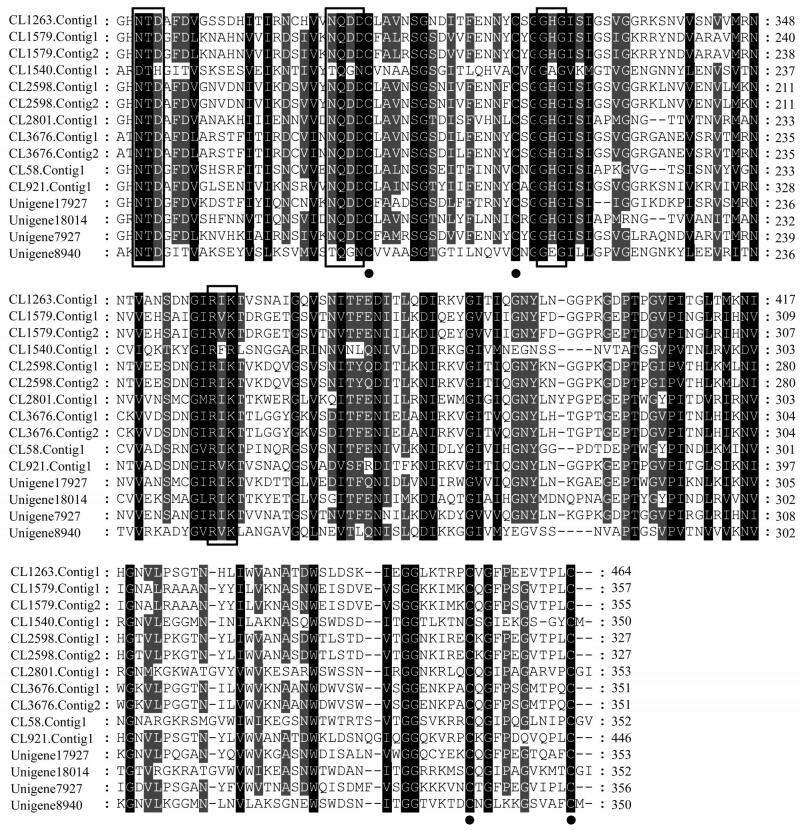 |
| Conserved motifs sequences were boxed, and predicted disulfide bridges were marked by solid circle. Fig. 4 Multiple deduced amino sequence alignments of P. micranthus PG genes |
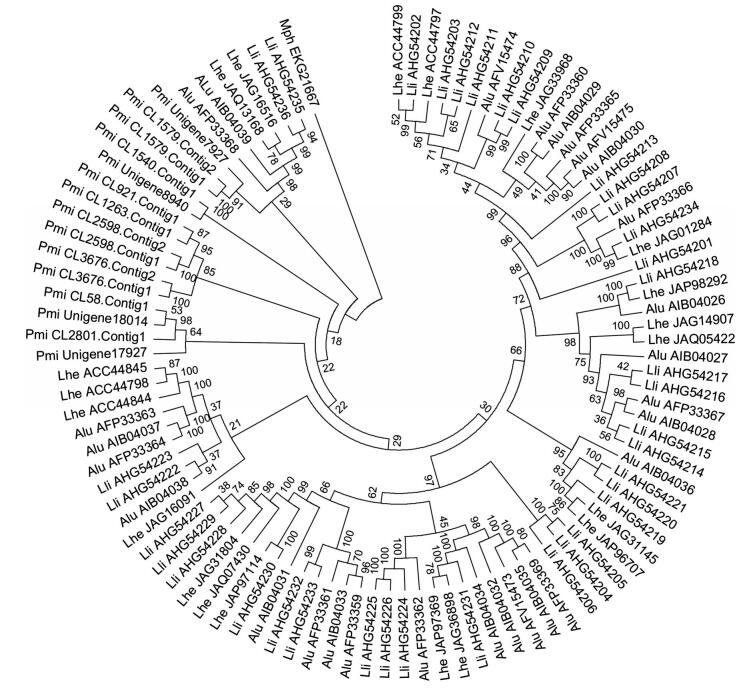 |
| The accession number for PGs were followed by the abbreviations represented the species. A glycoside hydrolase from M. phaseolina MS6 was used as an outgroup. Alu: A. lucorum; Lhe: Lygus hesperus; Lli: L. lineolaris; Mph: M. phaseolina; Pmi: P. micranthus. Fig. 5 Phylogenetic tree of PGs from of P. micranthus and other three mirid bugs |
This is the first report of a large-scale genetic information from P. micranthus. The raw reads were in a mean length of 90 bp, which was consistent with the Illumina sequencing capacity. Regarding the GC content of 46.19%, it was comparable to that value of other insect and eukaryote sequencing projects[16]. After quality filtering, clean reads were assembled into the unigenes with the average length of 765 bp, which is similar to the length of assembled sequences reported by other insects for Illumina technology[17-19]. It suggests that the generated P. micranthus transcriptome data were effectively assembled. Results for the 59.14% unigenes unable to match against the Nr database might be caused by the fact that there was no abundant sequence related to the available close species of P. micranthus. Of these transcripts, novel sequences might be many of them. The P. micranthus transcriptome was compared with other sequenced insect species, and the results clearly showed that the majority unigenes had similarity to the sequences from the non-typical model insect species. It is due to that there is no genome sequenced Hemiptera available and only limited gene sequences belonging to this order have been deposited. With respect of the dominant functional groups according to GO, COG and KEGG analyses, similar observations for metabolic pathways were reported in transcriptomic studies of other insects[20-21].
Based on the constructed transcriptomic database, 24 PG transcripts have been revealed. Twenty eight putative PG genes were identified in the L. lineolaris transcriptome, and 45 and 14 PG genes were respectively characterized from L. lineolaris and A. lucorum by sequencing cDNA library of the salivary glands[4, 8, 22]. Although the transcriptome of adult whole body other than salivary glands of P. micranthus were used to reveal the PG genes, the gene number identified is among the range reported from the above described studies. The analysis of the reveled PG genes with ORFs indicated that low degree of sequence identity was found among them. However, the strictly conserved amino acid resides involved in hydrolysis across all fungal and bacterial PGs were observed in all of them[23], indicating they are potential with the biological function of plant cell wall degrading. There are four disulfide bridges found from coleopterans and fungi, while only two disulfide bridges in the identified PGs from P. micranthus and other mirid bugs[4, 8, 22]. This might be due to that PGs from mirid bugs secreted into the saliva for digestion of plant cell wall are uniquely evolved by their originated environment that is different from beetle and fungal PGs found in the guts and fungal environments[23]. According to the phylogenetic relationships of PGs from four mirid bugs, P. micranthus PGs were separately grouped into four clades. But PGs from the others three species all fall into different clusters with each other, which is similar to the results from previous studies[4, 8]. It can be found that all PGs from different species are of ancient origin with expansion trends, implying that gene duplication is contributed to the evolution of PG genes.
In summary, the above generated unigenes and annotation information provide a fundamental resource at the genomic level for future molecular investigation in P. micranthus. The discovery of PG genes from the transcriptomic database was performed in an effort to better understand how P. micranthus feeds and interacts with its host plant.
Acknowledgment: We are grateful to Prof. LIU Guoqing working at Institute of Entomology, Nankai University, China for the identification of the study species.| [1] |
泽桑梓, 季梅, 闫争亮, 等.薇甘菊颈盲蝽性信息素的初步验证及后肠挥发物的鉴定.
动物学研究,2011,32 :147–153.
ZE S Z, JI M, YAN Z L, et al. Analysis and identification of hindgut volatiles for Pachypeltis sp. and field lure. Zoological Research, 2011,32 :147–153. (in Chinese with English abstract) |
| [2] |
泽桑梓, 苏尔广, 闫争亮, 等.季梅薇甘菊颈盲蝽对薇甘菊的控制作用.
西部林业科学,2013,42 :46–52.
ZE S Z, SU E G, YAN Z L, et al. Effects of Pachypeltis sp. against Mikania micrantha. Journal of West China Forest Science, 2013,42 :46–52. (in Chinese with English abstract) DOI: 10.3969/j.issn.1672-8246.2013.01.009. |
| [3] |
季梅, 泽桑梓, 赵宁, 等.颈盲蝽取食对薇甘菊叶片防御性酶活性的影响.
浙江农业学报,2014,26 :748–751.
JI M, ZE S Z, ZHAO N, et al. Effects of Pachypeltis sp. feeding on defense enzyme activities in Mikania micrantha leaves. Acta Agricultural Zhejiangensis, 2014,26 :748–751. (in Chinese with English abstract) DOI: 10.3969/j.issn.1004-1524.2014.03.35. |
| [4] |
SHOWMAKER K C, BEDNáOVá A, GRESHAM C, et al. Insight into the salivary gland transcriptome of Lygus lineolaris (Palisot de Beauvois).
PLoS ONE, 2016,11 :e0147197. DOI: 10.1371/journal.pone.0147197. |
| [5] |
HADFIELD K A, BENNETT A B. Polygalacturonases: many genes in search of a function.
Plant Physiology, 1998,117 :337–343. DOI: 10.1104/pp.117.2.337. |
| [6] |
SHACKEL K A, DE LA PAZCELORIO-MANCERA M, AHMADI H, et al. Micro-injection of Lygus salivary gland proteins to simulate feeding damage in alfalfa and cotton flowers.
Archives of Insect Biochemistry and Physiology, 2005,58 :69–83. DOI: 10.1002/(ISSN)1520-6327. |
| [7] |
HULL J J, GEIB S M, FABRICK J A, et al. Sequencing and de novo assembly of the western tarnished plant bug (Lygus hesperus) transcriptome.
PLoS ONE, 2013,8 :e55105. DOI: 10.1371/journal.pone.0055105. |
| [8] |
ZHANG L, XU P, XIAO H, et al. Molecular characterization and expression profiles of polygalacturonase genes in Apolygus lucorum (Hemiptera: Miridae).
PLoS ONE, 2015,10 :e0126391. DOI: 10.1371/journal.pone.0126391. |
| [9] |
KARATOLOS N, PAUCHET Y, WILKINSON P, et al. Pyrosequencing the transcriptome of the greenhouse whitefly, Trialeurodes vaporariorum reveals multiple transcripts encoding insecticide targets and detoxifying enzymes.
BMC Genomics, 2011,12 :56. DOI: 10.1186/1471-2164-12-56. |
| [10] |
ZHU J Y, YANG P, ZHANG Z, et al. Transcriptomic immune response of Tenebrio molitor pupae to parasitization by Scleroderma guani.
PLoS ONE, 2013,8 :e54411. DOI: 10.1371/journal.pone.0054411. |
| [11] |
ZHU J Y, SANG Z Z, CAO L J, et al. Development of microsatellite markers for the plant bug, Pachypeltis micranthus (Hemiptera: Miridae).
Applied Entomology and Zoology, 2016,51 :327–331. DOI: 10.1007/s13355-015-0392-1. |
| [12] |
GRABHERR M G, HAAS B J, YASSOUR M, et al. Full length transcriptome assembly from RNA-Seq data without a reference genome.
Nature Biotechnology, 2011,29 :644–652. DOI: 10.1038/nbt.1883. |
| [13] |
GÖTZ S, GARCíA-GóMEZ J M, TEROL J, et al. High-throughput functional annotation and data mining with the Blast2GO suite.
Nucleic Acids Research, 2008,36 :3420–3435. DOI: 10.1093/nar/gkn176. |
| [14] |
THOMPSON J D, GIBSON T J, PLEWNIAK F, et al. The Clustal_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools.
Nucleic Acids Research, 1997,25 :4876–4882. DOI: 10.1093/nar/25.24.4876. |
| [15] |
TAMURA K, PETERSON D, PETERSON N, et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods.
Molecular Biology and Evolution, 2011,28 :2731–2739. DOI: 10.1093/molbev/msr121. |
| [16] |
PRICE D P, NAGARAJAN V, CHURBANOV A, et al. The fat body transcriptomes of the yellow fever mosquito Aedes aegypti, pre-and post-blood meal.
PLoS ONE, 2011,6 :e22573. DOI: 10.1371/journal.pone.0022573. |
| [17] |
XUE J, BAO Y Y, LI B L, et al. Transcriptome analysis of the brown planthopper Nilaparvata lugens.
PLoS ONE, 2010,5 :e14233. DOI: 10.1371/journal.pone.0014233. |
| [18] |
ZHU J Y, ZHAO N, YANG B. Global transcriptome profiling of the pine shoot beetle, Tomicus yunnanensis (Coleoptera: Scolytinae).
PLoS ONE, 2012,7 :e32291. DOI: 10.1371/journal.pone.0032291. |
| [19] |
ZIMMER C T, MAIWALD F, SCHORN C, et al. A de novo transcriptome of European pollen beetle populations and its analysis, with special reference to insecticide action and resistance.
Insect Molecular and Biology, 2014,23 :511–526. DOI: 10.1111/imb.2014.23.issue-4. |
| [20] |
BAI X, MAMIDALA P, RAJARAPU S P, et al. Transcriptomics of the bed bug (Cimex lectularius).
PLoS ONE, 2011,6 :e16336. DOI: 10.1371/journal.pone.0016336. |
| [21] |
STAFFORD-BANKS C A, ROTENBERG D, JOHNSON B R, et al. Analysis of the salivary gland transcriptome of Frankliniella occidentalis.
PLoS ONE, 2014,9 :e94447. DOI: 10.1371/journal.pone.0094447. |
| [22] |
ZHU Y C, YAO J, LUTTRELL R. Identification of genes potentially responsible for extra-oral digestion and overcoming plant defense from salivary glands of the tarnished plant bug (Hemiptera: Miridae) using cDNA sequencing.
Journal of Insect Science, 2016,16 :60. DOI: 10.1093/jisesa/iew041. |
| [23] |
ALLEN M L, MERTENS J A. Molecular cloning and expression of three polygalacturonase cDNAs from the tarnished plant bug, Lygus lineolaris.
Journal of Insect Science, 2008,8 :27. |
 2016, Vol. 43
2016, Vol. 43


