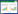文章信息
- 包桂荣, 孙锋
- BAO Guirong, SUN Feng
- 初始阻力运动对男性骨骼肌mRNA表达谱的影响及其作用机制
- Effect of initial resistance exercise on mRNA profiles of skeletal muscles of males and the associated mechanisms
- 中国医科大学学报, 2020, 49(9): 835-840
- Journal of China Medical University, 2020, 49(9): 835-840
-
文章历史
- 收稿日期:2019-05-30
- 网络出版时间:2020-09-10 11:43
阻力运动即抗阻力运动,是一种通过肌肉反复收缩对抗负荷或负重刺激所进行的运动模式。目前普遍认为足够时间与强度的慢性阻力运动可增加肌力并促进骨骼肌肥大[1-2]。目前研究[3]表明,体内外多种因素的共同调控影响肌组织的结构和功能,从而导致肌组织的损伤或重塑。
随着生物信息学的发展,高通量测序技术及分析方法已广泛用于人体健康与疾病发生、发展规律及相关机制的研究。在运动系统领域,已有研究[4-6]探讨了长期慢性或急性单次阻力运动对肌肉组织形态学及代谢的影响。然而,初次阻力运动者进行非常规阻力运动的早期,肌肉细胞的mRNA表达谱如何变化及其相关机制和潜在功能尚不明确[2]。
本研究针对未进行过阻力运动的不吸烟健康成年男性,利用高通量的基因组学分析方法分析基因数据集GSE45426,探讨在初始进行阻力运动的不同阶段骨骼肌mRNA表达谱的改变及其涉及的生物功能与分子机制。
1 材料与方法 1.1 数据收集和分组从美国国家生物技术信息中心(National Center for Biotechnology Information,NCBI)的人类基因表达汇编(gene expression omnibu,GEO)数据库(http://www.ncbi.nlm.nih.gov/geo)下载骨骼肌基因表达数据集GSE45426。研究纳入的16例受试者均是未进行过阻力训练的健康不吸烟成年男性,平均年龄(22±2)岁,体质量指数(23.0±2.0)kg/m2。受试者随机分成运动组和对照组,每组8例。运动过程及标本检测:受试者均采取仰卧位,膝关节屈曲90°,测定最大等长强度。(1)运动组,进行2个周期的向心性阻力运动,每个周期均在等速测力计上以坐姿进行运动,双腿分别先后从膝关节屈曲90°~180°并且以180°/s的角速度进行5回合30次最大等速膝关节伸展,每回合间隔休息1 min。每周期运动后24 h采集优势腿肌肉活检组织,2个周期间隔48 h,2次活检采集部位相距2.5 cm。(2)对照组,不做运动,但在相同时间点和对称的部位采集肌肉活检标本。2组饮食完全相同,具体见文献[2]。
1.2 基因数据的统计与功能分析受试者骨骼肌活组织提取总RNA,应用Human Genome U133 Plus 2.0进行基因表达微阵列分析,数据集的series matrix数据用于后续分析(以方差最大的探针为基本单位)。应用heat mapper软件制作热图。
应用GEO2R对运动组、对照组配对样本(运动/对照基线时分别对比运动/对照1周期或运动/对照2周期时)分别进行差异表达基因(differential expression genes,DEG)分析[7]。log倍数变化(logFC)≥1.5且调整P < 0.05为DEG。应用Volcano Plot绘制火山图。
应用bioconductor/cluster Profiler(Ver.3.6.0)函数包对DEG涉及的生物过程(biological process,BP)、细胞组分(cellular component,CC)和分子功能(molecular function,MF)进行基因本体分析(gene ontology,GO)[8]。P < 0.05且FDR q < 0.05为差异有统计学意义。
应用Gene Set Enrichment analysis 3.0版软件,以运动基线对比运动2个周期为表型,先后以MSigDB在线基因集数据库的hallmark/ KEGG/ BP gene sets作为参考基因集,对基因表达谱进行基因集富集分析(gene set enrichment analysis,GSEA)。随机组合次数设置为1 000次。以P < 0.05且FDR q < 0.25为差异有统计学意义。
对有统计学意义的基因集进行领头亚群基因分析。使用GPL570的注释文件对领头亚群基因分析所选定的探针进行注释,利用genecards网站(www.genecards.org)汇总各个相关基因功能等信息。
2 结果 2.1 2组DEG分析本研究中运动组运动2个周期和1个周期分别与基线(入组时)配对,共纳入54 675个基因进行了配对样本DEG分析。结果显示,对照组各时期的骨骼肌基因表达谱无显著差异。与基线时比较,运动组运动1个周期后基因表达无显著差异;运动2个周期后有231个DEG,其中228个基因表达上调,3个基因表达下调,见图 1。
|
| A, heat map; B, volcano plot. 图 1 与基线时比较,运动组运动2个周期后骨骼肌中的DEG分析结果 Fig.1 DEG in the skeletal muscle cells of the exercise subjects after two sessions of exercise compared to those detected at baseline |
2.2 运动组运动2个周期后DEG的GO分析
结果显示,与入组时比较,运动组运动2个周期后表达上调的DEG主要参与了胞外间质组织形成、炎症反应、胶原纤维组织形成、细胞黏附、胶原代谢、骨骼系统发育和Toll样受体4(Toll-like receptor 4,TLR4)信号通路等生物过程。表达上调的DEG主要存在于细胞外区域、间隙、间质、外泌体和胶原蛋白三聚体、蛋白质性细胞外间质、基底膜、内质网腔、血小板α颗粒腔、血液微粒和细胞表面。表达上调的DEG调控的分子功能主要包括细胞外间质组成成分调节、胶原结合、整合素结合、肝素结合等。表达下调的DEG在GO分析中无显著阳性结果,见表 1。
| Analysis category | Code | Count | P | FDR |
| GO-BP | ||||
| Extracellular matrix organization | 0030198 | 28 | 1.08E-23 | 1.74E-20 |
| Collagen fibril organization | 0030199 | 13 | 1.32E-15 | 2.15E-12 |
| Cell adhesion | 0007155 | 26 | 2.16E-12 | 3.48E-09 |
| Collagen catabolic process | 0030574 | 10 | 1.01E-08 | 1.63E-05 |
| Inflammatory response | 0006954 | 19 | 2.76E-08 | 4.45E-05 |
| Skeletal system development | 0001501 | 10 | 7.24E-06 | 0.011 7 |
| Toll-like receptor 4 signaling pathway | 0034142 | 5 | 2.30E-05 | 0.037 1 |
| GO-CC | ||||
| Extracellular space | 0005615 | 59 | 3.07E-24 | 3.91E-21 |
| Extracellular matrix | 0031012 | 30 | 1.70E-21 | 2.16E-18 |
| Extracellular exosome | 0070062 | 79 | 2.86E-21 | 3.64E-18 |
| Extracellular region | 0005576 | 57 | 5.82E-19 | 7.40E-16 |
| Collagen trimer | 0005581 | 15 | 1.48E-13 | 1.88E-10 |
| Proteinaceous extracellular matrix | 0005578 | 20 | 7.65E-12 | 9.73E-09 |
| Basement membrane | 0005604 | 11 | 3.17E-09 | 4.03E-06 |
| Endoplasmic reticulum lumen | 0005788 | 12 | 1.92E-06 | 0.002 4 |
| Platelet alpha granule lumen | 0031093 | 7 | 1.20E-05 | 0.015 2 |
| Blood microparticle | 0072562 | 10 | 1.30E-05 | 0.016 5 |
| Cell surface | 0009986 | 18 | 1.32E-05 | 0.016 8 |
| GO-MF | ||||
| Extracellular matrix structural constituent | 0005201 | 13 | 9.79E-13 | 1.30E-09 |
| Collagen binding | 0005518 | 11 | 1.72E-10 | 2.29E-07 |
| Integrin binding | 0005178 | 11 | 4.62E-08 | 6.15E-05 |
| Heparin binding | 0008201 | 10 | 1.79E-05 | 0.023 8 |
2.3 运动组运动2个周期后GSEA与领头亚群基因分析
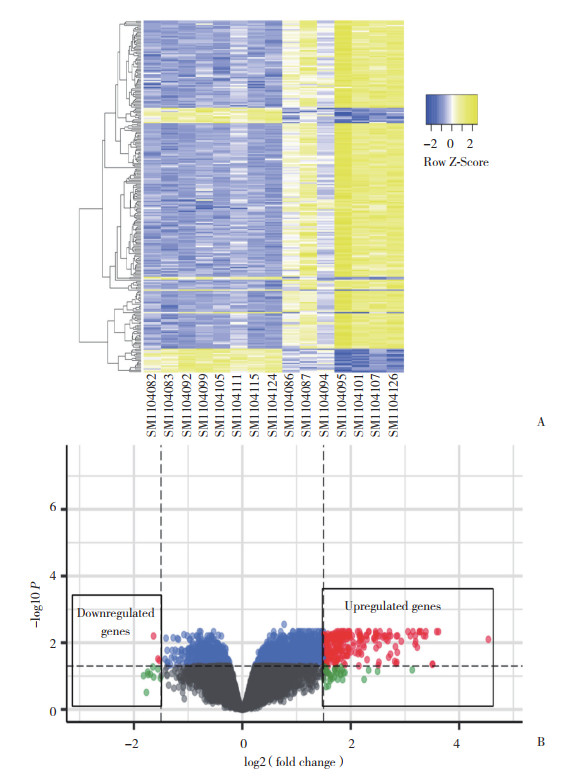
结果显示,与入组时比较,运动组运动2个周期后标本中富集了炎症反应、白细胞介素-6、干扰素-γ等炎症相关通路,以及PI3K、KRAS等细胞增生通路和上皮-间质转化、血管新生、凋亡、缺氧、P53等相关通路,见图 2。在炎症相关通路的领头亚群基因分析中发现,炎症反应、白细胞介素-6、γ干扰素3条通路中均出现CXCL9、CXCL11、CXCL10、IL4R和IRF1基因,其中IRF1在3条炎症通路和补体基因集中均可表达,见图 3。
|
| A, pathway of inflammatory response; B, pathway of IL6-JAK-STAT3 signaling; C, pathway of interferon-γ response; D, pathway of complement. 图 2 运动组运动2个周期后骨骼肌中基因表达谱的GSEA结果 Fig.2 The results of gene set enrichment analysis of the gene expression profile of skeletal muscle cells of the exercise subjects after two sessions of exercise |

|
| A, the Venn diagram to illustrate the leading-edge genes in the 3 gene-sets including INFLAMMATORY-RES, IL6_STAT3 and INTERFERON-GAMMA.5, the number of leading-edge genes shared by the 3 gene sets; 8/14/7/71, the number of the leading-edge genes shared by 2 of the 3 gene sets; 51/20/71, the number of the leading-edge genes not shared by the gene sets. B, the Venn diagram to illustrate the leading-edge genes in the 4 gene-sets including IL6_JAK_STAT3, INFLAMMATORY-RES, COMPLEMENT and INTERFERON-GAMMA. 1, the number of leading-edge genes shared by the 4 gene sets, and the name of the gene is IRF1;2/1, the number of the leading-edge genes shared by 3 of the 4 gene sets; 12/14/7/0/1/10, the number of the leading-edge genes shared by 2 of the 4 gene sets; 41/10/60/58, the number of the leading-edge genes not shared by the gene sets. 图 3 运动组运动2个周期后骨骼肌中基因表达谱的GSEA结果 Fig.3 The results of gene set enrichment analysis of the gene expression profile of skeletal muscle cells of the exercise subjects after two sessions of exercise |
3 讨论
本研究结果显示,从未进行过阻力运动的健康不吸烟成年男性进行2个周期阻力运动训练后,骨骼肌中的基因表达谱与入组时相比具有显著差异。C-C基序趋化因子配体2(C-C motif chemokine ligand 2,CCL2)、补体成分3(complement component 3,C3)等基因表达升高。与入组时比较,表达上调的DEG大多位于细胞外区域或组织中,通过组成或结合细胞外间质成分,参与胞外间质组织调节与炎症反应等相关生物过程;运动2个周期的骨骼肌中富集了炎症、增生、转化与凋亡等相关通路;而运动1个周期的骨骼肌中DEG不显著。
目前认为骨骼肌可在阻力运动的刺激下发生结构与代谢等方面的改变,而且仅有 < 2%的基因表达改变同时出现在急性单次和长期慢性阻力运动后的骨骼肌中[5]。本研究发现初次进行阻力运动的24 h对骨骼肌的基因表达谱无显著影响,而间隔48 h后的再次阻力运动训练即可显著改变运动者骨骼肌的基因表达,导致大量基因表达上调。因而证明,骨骼肌的基因表达在阻力运动的早期即存在动态演变。已有研究[9-10]从骨骼肌代谢与分子水平证明阻力运动后的骨骼肌损伤与修复是连续的过程,本研究从基因水平进一步证实,阻力运动早期的骨骼肌除了富集炎症反应相关通路外,还富集缺氧、血管新生、上皮-间质转化、细胞凋亡和细胞增生等信号通路。
本研究还发现炎症反应相关通路的关键基因包括CXCL10、CXCL9、CXCL11、IL4R和IRF1。CXCL9、CXCL10和CXCL11均属于CXC细胞因子家族,均通过结合细胞因子受体CXCR3发挥趋化作用[11]。CXCL10又称为γ干扰素诱导蛋白10,通过活化或记忆T细胞而发挥多重免疫功能[12-13]。人类细胞学与小鼠实验发现骨骼肌损伤后CXCL10表达增加,但在骨骼肌再生的过程中不是必不可少的[14]。CXCL9是γ干扰素诱导的单核因子,为γ干扰素诱导的T细胞趋化因子[15]。在肌损伤的CXCL10敲除小鼠中CXCL9和CXCL11表达增加[14]。IL4R为Ⅰ型细胞因子受体,通过与白细胞介素-4结合,在B细胞中促进IgE产生,在T细胞中促进2型辅助T细胞分化[16-17]。目前发现IL4R异常与多种恶性肿瘤的发生发展密切相关[18]。IRF1最初发现可以激活细胞因子γ干扰素的表达,后来证实可与干扰素刺激反应原件结合激活多种靶基因的表达,在机体免疫反应、凋亡调控、DNA损伤与肿瘤抑制等过程中发挥重要作用[19-22]。本研究结果显示IRF1是唯一在多条炎症相关通路中均发挥重要作用的基因。因此,CXCL10、CXCL9、CXCL11、IL4R和IRF1可能是非常规阻力运动后骨骼肌发生炎症反应而导致损伤或再修复的关键基因。
综上所述,本研究利用生物信息学方法,基于在线数据GSE45426分析证实未进行过阻力运动的健康成年男性进行阻力运动的早期,骨骼肌在缺氧、凋亡、炎症、血管新生与细胞增生等多重通路上同时发生基因表达谱的变化,而且这种变化多发生于骨骼肌细胞外区域或结构中。本研究对于探究阻力运动导致的骨骼肌损伤与肥大的调控机制以及进一步寻找调控靶点与措施具有重要意义。
| [1] |
D'ANTONA G, LANFRANCONI F, PELLEGRINO MA, et al. Skeletal muscle hypertrophy and structure and function of skeletal muscle fibres in male body builders[J]. J Physiol (Lond), 2006, 570(Pt 3): 611-627. DOI:10.1113/jphysiol.2005.101642 |
| [2] |
MURTON AJ, BILLETER R, STEPHENS FB, et al. Transient transcriptional events in human skeletal muscle at the outset of concentric resistance exercise training[J]. J Appl Physiol, 2014, 116(1): 113-125. DOI:10.1152/japplphysiol.00426.2013 |
| [3] |
CSAPO R, GUMPENBERGER M, WESSNER B. Skeletal muscle extracellular matrix-what do we know about its composition, regulation, and physiological roles? a narrative review[J]. Front Physiol, 2020, 11: 253. DOI:10.3389/fphys.2020.00253 |
| [4] |
GORDON PM, LIU DM, SARTOR MA, et al. Resistance exercise training influences skeletal muscle immune activation:a microarray analysis[J]. J Appl Physiol, 2012, 112(3): 443-453. DOI:10.1152/japplphysiol.00860.2011 |
| [5] |
PHILLIPS BE, WILLIAMS JP, GUSTAFSSON T, et al. Molecular networks of human muscle adaptation to exercise and age[J]. PLoS Genet, 2013, 9(3): e1003389. DOI:10.1371/journal.pgen.1003389 |
| [6] |
RAUE U, TRAPPE TA, ESTREM ST, et al. Transcriptome signature of resistance exercise adaptations:mixed muscle and fiber type specific profiles in young and old adults[J]. J Appl Physiol, 2012, 112(10): 1625-1636. DOI:10.1152/japplphysiol.00435.2011 |
| [7] |
RITCHIE ME, PHIPSON B, WU D, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies[J]. Nucleic Acids Res, 2015, 43(7): e47. DOI:10.1093/nar/gkv007 |
| [8] |
YU GC, WANG LG, HAN YY, et al. Cluster profiler:an R package for comparing biological themes among gene clusters[J]. OMICS, 2012, 16(5): 284-287. DOI:10.1089/omi.2011.0118 |
| [9] |
PEAKE JM, NEUBAUER O, DELLA GATTA PA, et al. Muscle damage and inflammation during recovery from exercise[J]. J Appl Physiol, 2017, 122(3): 559-570. DOI:10.1152/japplphysiol.00971.2016 |
| [10] |
NEVES RVP, ROSA TS, SOUZA MK, et al. Dynamic, not isometric resistance training improves muscle inflammation, oxidative stress and hypertrophy in rats[J]. Front Physiol, 2019, 10: 4. DOI:10.3389/fphys.2019.00004 |
| [11] |
TENSEN CP, FLIER J, VAN DER RAAIJ-HELMER EM, et al. Human IP-9:a keratinocyte-derived high affinity CXC-chemokine ligand for the IP-10/Mig receptor (CXCR3)[J]. J Invest Dermatol, 1999, 112(5): 716-722. DOI:10.1046/j.1523-1747.1999.00581.x |
| [12] |
KOPER OM, KAMINSKA J, SAWICKI K, et al. cxcL9, cxcL10, cxcL11, and their receptor (CXCR3) in neuroinflammation and neurodegeneration[J]. Adv Clin Exp Med, 2018, 27(6): 849-856. DOI:10.17219/acem/68846 |
| [13] |
DUFOUR JH, DZIEJMAN M, LIU MT, et al. IFN-gamma-inducible protein 10(IP-10;CXCL10) -deficient mice reveal a role for IP-10 in effector T cell generation and trafficking[J]. J Immunol, 2002, 168(7): 3195-3204. DOI:10.4049/jimmunol.168.7.3195 |
| [14] |
DEYHLE MR, HAFEN PS, PARMLEY J, et al. CXCL10 increases in human skeletal muscle following damage but is not necessary for muscle regeneration[J]. Physiol Rep, 2018, 6(8): e13689. DOI:10.14814/phy2.13689 |
| [15] |
LEE HH, FARBER JM. Localization of the gene for the human MIG cytokine on chromosome 4q21 adjacent to INP10 reveals a chemokine "mini-cluster"[J]. Cytogenet Cell Genet, 1996, 74(4): 255-258. DOI:10.1159/000134428 |
| [16] |
王建荣, 冯瑞华, 多力坤·木扎帕尔, 等. 白细胞介素-4受体基因多态性及IgE水平与哮喘预测指数相关性研究[J]. 中国当代儿科杂志, 2015, 17(12): 1306-1310. |
| [17] |
ZENG H, CHI H. MTOR signaling in the differentiation and function of regulatory and effector T cells[J]. Curr Opin Immunol, 2017, 46: 103-111. DOI:10.1016/j.coi.2017.04.005 |
| [18] |
LIN Y, YUAN Q, QIAN F, et al. Polymorphism rs4787951 in IL-4R contributes to the increased risk of renal cell carcinoma in a Chinese population[J]. Gene, 2019, 685: 242-247. DOI:10.1016/j.gene.2018.11.070 |
| [19] |
ESCALANTE CR, YIE J, THANOS D, et al. Structure of IRF-1 with bound DNA reveals determinants of interferon regulation[J]. Nature, 1998, 391(6662): 103-106. DOI:10.1038/34224 |
| [20] |
JANFESHAN S, YAGHOBI R, EIDI A, et al. Expression profile of interferon regulatory factor 1 in chronic hepatitis B virus-infected liver transplant patients[J]. Exp Clin Transplant, 2017, 15(6): 669-675. DOI:10.6002/ect.2015.0302 |
| [21] |
WAN P, ZHANG J, DU Q, et al. The clinical significance and biological function of interferon regulatory factor 1 in cholangiocarcinoma[J]. Biomed Pharmacother, 2018, 97: 771-777. DOI:10.1016/j.biopha.2017.10.096 |
| [22] |
YAN R, VAN MEURS M, POPA ER, et al. Endothelial interferon regulatory factor 1 regulates lipopolysaccharide-induced VCAM-1 expression independent of NF κB[J]. J Innate Immun, 2017, 9(6): 546-560. DOI:10.1159/000477211 |
 2020, Vol. 49
2020, Vol. 49