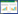文章信息
- 张伟, 刘夺, 李炳志, 元英进.
- ZHANG Wei, LIU Duo, LI Bing-zhi, YUAN Ying-jin.
- 产对香豆酸酿酒酵母菌株的构建及优化
- Construction and Optimization of p-coumaric Acid Producing Saccharomyces cerevisiae
- 中国生物工程杂志, 2017, 37(9): 89-97
- China Biotechnology, 2017, 37(9): 89-97
- http://dx.doi.org/DOI:10.13523/j.cb.20170912
-
文章历史
- 收稿日期: 2017-03-08
- 修回日期: 2017-04-06
2. 天津化学化工协同创新中心合成生物学平台 天津 300072
2. SynBio Research Platform, Collaborative Innovation Center of Chemical Science and Engineering(Tianjin), School of Chemical Engineering and Technology, Tianjin University, Tianjin 300072, China
对香豆酸(p-coumaric acid),即对羟基肉桂酸,广泛存在于自然界植物中,尤其白花蛇舌草、海金沙草、杜仲叶等中草药中[1]。对香豆酸在预防心血管、抗癌、抗氧化、抗炎等方面均发挥着重要的作用[2-4],同时也在糖尿病和高血脂等病症上有着潜在治疗活性[5]。此外,对香豆酸作为一种芳香类氨基酸,也是很多植物次级代谢产物的前体物质,如柚皮素、白藜芦醇、花青素、儿茶素等黄酮类、芪类高附加值的天然产物[6-9]。目前,对香豆酸的主要来源为植物提取、化学合成和生物合成。植物提取面临生长周期长、收益率低及受环境影响大等问题,而化学合成则面临高能耗、低产率及污染环境等问题,生物合成因其具有低成本、高产率和绿色环保等优点,被认为是21世纪最有前途的生产方法。
目前对香豆酸的生物合成法一种是通过苯丙氨酸(phenylalanine,Phe)在苯丙氨酸脱氨酶(phenylalnine ammonialyase)、4-肉桂酸羟化酶(cinnamate 4-hydroxylase)的作用下转化为对香豆酸,但由于4-肉桂酸羟化酶是一种细胞色素P450氧化酶,在微生物中表达有一定的难度,导致转化率较低,因此4-肉桂酸羟化酶被认为是对香豆酸生产的限速酶[10-11];相比于前者,以酪氨酸为底物进行反应,仅需要经过酪氨酸解氨酶(tyrosine ammonia lyase)的作用便可将酪氨酸转化为对香豆酸,反应路径更短、转化效率更高,因此研究者通常采取这条路径进行对香豆酸的合成[12-14]。Xue等[15]将黏红酵母源(Rhodotorula glutinis)的TAL基因导入到大肠杆菌中实现了从酪氨酸到对香豆酸的转化。Li等[16]通过在酿酒酵母中过表达半叶趾虎橙黄扶桑源(Hemiphyllodactylus aurantiacus)和约氏黄杆菌源(Flavobacterium johnsoniae)的TAL基因,并额外添加5mmol/L的酪氨酸分别得到了7.04mg/L和13.71mg/L的对香豆酸。
在底盘细胞的选择上,大肠杆菌由于其自身的内毒素导致其在医药和食品的工业生产上有一定的局限性,而酿酒酵母则被认为是安全的模式微生物工程菌株。酿酒酵母的遗传背景清楚且易于进行基因改造,在天然产物的生产上被广泛应用[17]。本研究以酿酒酵母BY4741为底盘菌株,引入圆红冬孢酵母源(Rhodosporidium toruloides)的TAL基因[18],并通过质粒拷贝数的不同对此基因的表达强度进行调节,在此基础上利用工程学思想对酿酒酵母内源上游路径进行改造[19-20]使其高产对香豆酸,通过delta位点整合[21-22]来过表达酪氨酸的反馈抑制调节的相关基因ARO4及ARO7相应的特定位点突变体ARO4*和ARO7*(带*代表突变后的基因)来解除对关键酶的抑制作用[23];同时对旁路副产物路径的基因进行敲除(图 1),从而避免芳香醇类物质的合成进而使合成芳香氨基酸代谢通量增大,以提高对香豆酸的产量;后期探索了前体酪氨酸的外源添加对对香豆酸产量的影响,为在酿酒酵母中生物合成对香豆酸和后续的苯丙烷类物质和黄酮类物质提供参考依据。
|
| 图 1 酿酒酵母菌中对香豆酸的生物合成路径及改造策略 Figure 1 Biosynthesis pathway and modified strategies of p-coumaric acid in S. cerevisiae |
T4 DNA连接酶、Q5超保真DNA聚合酶和限制性内切核酸酶Bsa I-HF、Not I-HF购自NEB(New England Biolab)公司;质粒小提试剂盒、普通DNA产物纯化试剂盒、琼脂糖凝胶DNA回收试剂盒购自天根公司;对香豆酸标准品(CAS号:501-98-4) 购自上海源叶生物科技有限公司;PEG3350、鲑鱼精DNA、各类氨基酸粉末购自鼎国昌盛公司;去氨基酵母氮源(YNB)购自于Genview公司;醋酸锂购自北方天医化学试剂厂;G418抗生素购自于Genview公司;甲醇购自与Fisher公司,甲酸(Formic acid)购自于DikmaPure公司;大肠杆菌感受态细胞DH5α购自Biomed公司。
1.2 菌株、质粒和培养基本文涉及的所有质粒和菌株信息详见表 1和表 2。酿酒酵母菌株BY4741[24](MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0),酵母表达载体pRS416为本实验室保存,pRS425K为本实验室构建。
| Plasmids | Description | Source |
| pRS416 | Single copy plasmid with URA3 and Ampr marker | Purchased |
| pRS425K | Replace AmpKX with KanMX in pRS425 | This lab |
| Module 1 | pRS425K: TCYC1-PTDH3-TTEF1 | This study |
| Module 2 | pRS425K: TCYC1-PTDH3-TAL-TTEF1 | This study |
| Module 3 | pRS416: TCYC1-PTDH3-TAL-TTEF1 | This study |
| Module 4 | pRS425K: delta1-URA3-TGPD | This study |
| Module 5 | pRS425K: TGPD-PPGI1-TFBA1 | This study |
| Module 6 | pRS425K: TGPD-PPGI1-ARO4*-TFBA1 | This study |
| Module 7 | pRS425K: TFBA1-PTPI1-TPGK1 | This study |
| Module 8 | pRS425K: TFBA1-PTPI1-ARO7*-TPGK1 | This study |
| Module 9 | pRS425K: TPGK1-delta2 | This study |
| Module 10 | pEASY-Blunt-L-arm1-KANMX4-R-arm1 | This study |
| Module 11 | pEASY-Blunt-L-arm2-KANMX4-R-arm2 | This study |
| Module12 | pRS413: PGAL1-Cre-TCYC1 | This study |
| Strains | Description | Source |
| BY4741 | MATa, his3 Δ 1, leu2 Δ 0, met15 Δ 0, ura3 Δ 0 | Purchased |
| SyBE_Sc02120000 | BY4741, Module 3 | This study |
| SyBE_Sc02120001 | BY4741, Module 2 | This study |
| SyBE_Sc02120002 | BY4741, Delta∷URA3_ TGPD-PPGI1-ARO4*-TFBA1-PTPI1-ARO7*-TPGK1 | This study |
| SyBE_Sc02120003 | SyBE_Sc02120002, Module 2 | This study |
| SyBE_Sc02120004 | SyBE_Sc02120002, ΔARO10 | This study |
| SyBE_Sc02120005 | SyBE_Sc02120004, Module 2 | This study |
| SyBE_Sc02120006 | SyBE_Sc02120002, ΔPDC5 | This study |
| SyBE_Sc02120007 | SyBE_Sc02120006, Module 2 | This study |
| SyBE_Sc02120008 | SyBE_Sc02120002, ΔARO10, ΔPDC5 | This study |
| SyBE_Sc02120009 | SyBE_Sc02120008, Module 2 | This study |
LB培养基(10g/L氯化钠、10g/L胰蛋白胨、5g/L酵母提取物,固体培养基添加20g/L琼脂粉)用于大肠杆菌培养;YPD培养基(20g/L葡萄糖、20g/L胰蛋白胨、10g/L酵母提取物,固体培养基添加20g/L琼脂粉)用于酿酒酵母培养;SC培养基(20g/L葡萄糖、6.7g/L去氨基酵母氮源,2g/L缺亮氨酸、组氨酸、尿嘧啶的混合氨基酸粉末;固体培养基添加20g/L琼脂粉)用于酵母转化子的筛选、培养和发酵;G418固体培养基(20g/L葡萄糖、6.7g/L去氨基酵母氮源,2g/L缺亮氨酸、组氨酸、尿嘧啶的混合氨基酸粉末,200μg/ml G418抗生素,20g/L琼脂粉)用于基因的敲除;YPG培养基(20g/L半乳糖、20g/L胰蛋白胨,10g/L酵母提取物)用于诱导Cre酶的表达。
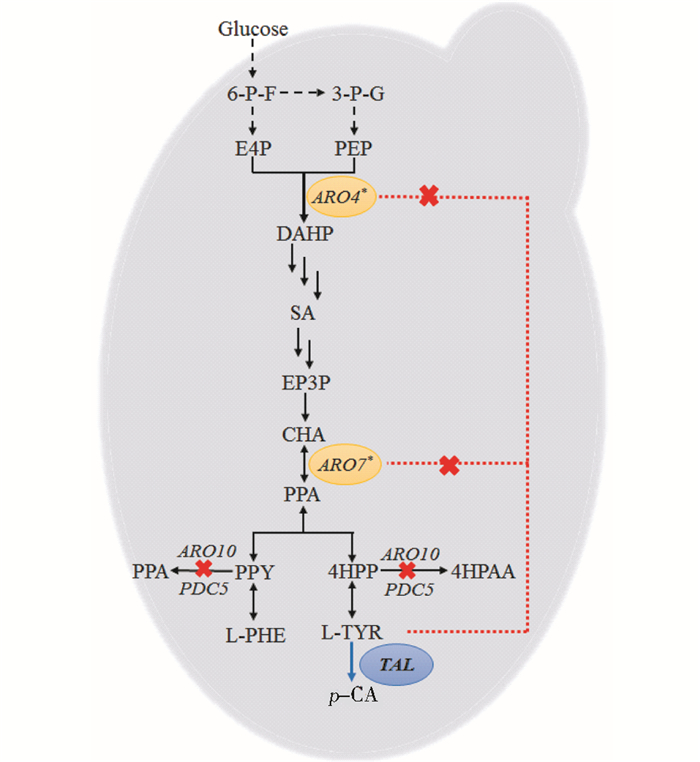
1.3 酿酒酵母表达载体、整合质粒载体和敲除质粒载体的构建基因表达载体、过表达基因模块和敲除模块的构建如图 2所示。以BY4741基因组DNA为模板,通过特异性引物PCR扩增CYC1、TEF1、GPD、FBA1、PGK1终止子,TDH3、PGI1、TPI1启动子,L-arm1(ARO10基因起始密码子上游398bp),R-arm1(ARO10基因终止密码子下游409bp),L-arm2(PDC5基因起始密码子上游441bp),R-arm2(PDC5基因终止密码子下游450bp)和URA3 ORF框;通过引物设计氨基酸突变点,以BY4741基因组为模板PCR扩增ARO4*和ARO7*两个具有定点氨基酸位点突变的基因;通过特异性引物,以含有loxP和G418基因的质粒为模板扩增基因敲除的标签。通过OE-PCR(over-lap PCR)扩增得到TCYC1-PTDH3-TTEF1片段并以Not I酶切连接到pRS425K,命名为Module1;将TAL基因以Bsa I酶切到Module1中,命名为Module2;将T}YC1C-PTDH3-TAL-TTEF1片段Not I酶切连入pRS416中,命名为Module3;通过OE-PCR扩增得到delta1-URA3-TGPD片段并以Not I酶切连接到pRS425K,命名为Module4;相同的,通过OE-PCR扩增得到TGPD-PPGI1-TFBA1片段并以Not I酶切连接到pRS425K,命名为Module5;通过OE-PCR扩增得到TFBA1-PTPI1-TPGK1片段并以Not I酶切连接到pRS425K,命名为Module7;将ARO4*基因以Bsa I酶切到Module5中,命名为Module6;将ARO7*基因以Bsa I酶切到Module7中,命名为Module8;通过OE-PCR扩增得到TPGK1-delta2片段并以Not I酶切连接到pRS425K,命名为Module9;通过OE-PCR扩增得到L-arm1-KANMX4-R-arm1片段并以Not I酶切连接到pEASY-Blunt,命名为Module10;通过OE-PCR扩增得到L-arm2-KANMX4-R-arm2片段并以Not I酶切连接到pEASY-Blunt,命名为Module11。
|
| 图 2 外源基因表达载体、基因过表达盒及基因敲除盒的构建 Figure 2 Construction of heterogenous gene expression plasmid, gene overexpression and gene deletion cassette (a)Construction of heterogenous gene plasmid (b)Construction of heterogenous gene overexpression cassette (c)Construction of heterogenous gene deletion cassette |
酪氨酸解氨酶基因TAL来源于圆红冬孢酵母(Rhodosporidium toruloides),基因经过Jcat按照酿酒酵母密码子偏好性进行优化并规避常用酶切位点,并在两端添加Bsa I酶切位点后交由金斯瑞公司合成。合成回来的基因质粒用Bsa I酶切,获得基因片段,同时将相应质粒也用Bsa I酶切,纯化回收后经T4连接酶连接,得到相应的基因表达载体。
1.4 人工酵母菌株的发酵将得到的人工酿酒酵母菌株接种于5ml种子培养基中,在30℃、250r/min培养11~13h,以初始菌体浓度OD600=0.2分别接种于50ml二级种子培养基中,培养8~12h,以初始菌体浓度OD600=0.2接种于50ml发酵培养基中,于30℃、250r/min条件下培养,以此条件继续培养至96h。
1.5 对香豆酸的提取和检测取500μl菌液于干净的2ml EP管中,加入500μl乙酸乙酯,置于振荡器中涡旋10min,12 000r/min离心5min,取上层乙酸乙酯有机相于干净的2ml EP管中,再向余下的菌液中加入500μl乙酸乙酯重复上述操作一次。将两次乙酸乙酯有机相抽提液(共1ml)氮吹浓缩,加入500μl色谱纯甲醇溶解,并采用0.2μm有机滤膜过滤,利用高效液相色谱(high performance liquid chromatography, HPLC)检测对香豆酸浓度。色谱条件:色谱柱:BDS HYPERSIL C18色谱柱(150mm×4.6mm,5μm);检测器:Waters2489UV/Vis;泵:Waters e2695;流动相:甲醇和0.1%甲酸-水;柱温:30℃;流速:0.2ml/min;进样体积:5μl。对香豆酸检测波长为310nm。
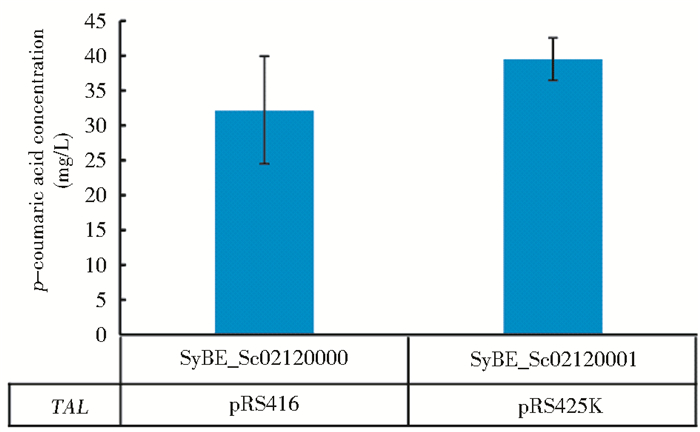
2 结果与分析 2.1 生产对香豆酸酿酒酵母菌株的构建及表达水平的调节酪氨酸解氨酶基因TAL经文献报道的来源有很多种[25],本研究选取的是圆红冬孢酵母源[18](Rhodosporidium toruloides),在对其进行酿酒酵母最适密码子优化之后,连入pRS416的单拷贝质粒(拷贝数为1个)中,即Module3,并通过醋酸锂转化法[26]转化至酿酒酵母菌株BY4741中,摇瓶发酵后经HPLC检测产物,在保留时间为7.50min时检测到对香豆酸(与标准品保留时间一致),产量约为32.21mg/L。异源合成产物的关键基因的拷贝数影响产物的产量[27],为了提高TAL基因在胞内的表达量,本研究构建了将TAL连接至多拷贝质粒pRS425K(拷贝数为20~50个)的模块,即Module2,转化至BY4741中后摇瓶发酵。实验结果表明(图 3),多拷贝与单拷贝质粒相比在对香豆酸产量上提高了约23%,说明通过增加基因拷贝数来提高酪氨酸解氨酶在胞内的表达量,进而提高对香豆酸的产量的策略具有可行性。
|
| 图 3 TAL基因不同的表达水平对产物产量的影响 Figure 3 Effects of TAL in different expression levels on production of p-coumaric acid |
目前的研究表明,在酿酒酵母的酪氨酸生物合成路径中涉及两步有关反馈抑制的反应,第一个反应是编码DAHP(3-deoxy-D-arabino-heptulosonate-7-phosphate)合成酶的ARO4基因,为酪氨酸的反馈抑制基因。将ARO4的229位的赖氨酸突变为亮氨酸后,此基因便不再受酪氨酸的反馈抑制作用[28];由ARO7编码的分支酸变位酶(chorismate mutase)则被认为是第二个受酪氨酸反馈抑制的酶,但将141位的丝氨酸变为甘氨酸之后,酪氨酸对其的反馈抑制也被解除[29]。Li等[16]通过过表达ARO4K229L、ARO7G141S以及白藜芦醇生物合成路径上的基因获得的白藜芦醇产量为对照菌株的36倍。本研究通过OE-PCR将受酪氨酸反馈抑制酶的相应基因进行定点氨基酸突变(将成功克隆的片段进行测序验证),在酿酒酵母中以delta位点整合[30-31]至基因组的方式实现基因的稳定存在。用Not I切下Module4、Module6、Module8、Module9,将4个片段共转化至酿酒酵母菌株BY4741,提取酵母基因组进行PCR验证,出现条带大小正确的目标条带,从而确定阳性克隆,对整合完成后正确的转化子导入Module2对产物通过高效液相色谱法进行筛选验证。由图 4可知,采用delta位点进行基因整合时,不同菌株之间对香豆酸的产量有较为明显的差异。其中菌株SyBE_Sc02120002-5的对香豆酸产量最高为41.93mg/L,菌株SyBE_Sc02120002-9的产量最低为13.06mg/L,差异达到3倍,其余菌株的对香豆酸产量呈正态分布。Delta位点整合产生的不同菌株之间的差异,可能是由于整合位置和数目引起的,这在王瑞钊等[32]合成β-胡萝卜素菌株的构建中也有所体现。将对香豆酸产量最高的菌株命名为SyBE_Sc02120002。与未过表达突变基因的菌株相比,对香豆酸产量也有一定的提升。

|
| 图 4 Delta位点整合过表达基因人工酿酒酵母菌株的筛选 Figure 4 Screening of engineered S. cerevisiae of gene overexpression by delta site integration |
丙苯酮酸(PPY)和4-羟基苯丙酮酸(4-HPP)是生物合成酪氨酸途径上重要的前体物质(图 1)。PPY和4-HPP在ARO10和PDC5基因编码的酶的作用下,分别生成旁路物质苯乙醛和4-羟基乙酸[33]。本研究从阻断旁路物质合成的角度出发,实现苯丙酮酸和4-羟基苯丙酮酸的积累,从而为生物合成对香豆酸提供更多的前体。
以SyBE_Sc02120002为底盘菌株,分别构建单基因敲除ARO10、PDC5以及双基因敲除ARO10和PDC5的菌株。将Module10、Module11用Not I切下L-arm1-loxP-KANMX4-loxP-R-arm1和L-arm2-loxP-KANMX4-loxP-R-arm2片段,采用乙酸锂转化法转化至酿酒酵母菌株SyBE_Sc02120002中,将得到的转化子进行划线分纯培养后提取酵母基因组进行4组特异性引物PCR验证,并将PCR得到的正确条带琼脂糖凝胶回收送至金唯智公司测序,得到完全匹配的测序结果则可确定相应基因已被敲除,将正确敲除基因的酵母菌株命名为SyBE_Sc02120004和SyBE_Sc02120006。然后向敲除基因的菌株内通过乙酸锂转化法导入Module12,将正确的克隆接种到YPG培养基中,在半乳糖存在的条件下诱导Cre酶的表达[34],从而使KANMX4标签两端的同向的loxP位点重叠,回收G418标签,再通过三组特异性引物PCR验证KANMX4基因已被敲除。将已收回G418标签的菌株在YPD中传代丢失质粒Module12。在此菌株上继续进行另一个基因的敲除,得到酿酒酵母菌株SyBE_Sc02120008。通过导入Module2来验证基因敲除对对香豆酸产量的影响。
由图 5的发酵结果可知,单独敲除ARO10或PDC5基因时均可以提高对香豆酸的产量,且单独敲除两个基因对产量的影响无明显差异,与对照菌株相比,对香豆酸产量提高了87%。当两个基因均被敲除之后,相较于单敲菌株对香豆酸的产量有所提高,在阻断旁路物质的合成之后,使向合成对香豆酸的代谢通量增大,进而产量得到了提高。菌株SyBE_Sc02120009的对香豆酸产量为87.56mg/L,是对照菌株的2.05倍。Rodriguez等[35]以CEN.PK为底盘菌株在单独敲除ARO10或PDC5时,得到的对香豆酸的产量仅比对照菌株提高了25%,但双敲除菌株的产量为对照菌株的2.29倍左右。由此可见,选取不同的酿酒酵母菌株作为底盘时,在改造的过程产量和终产量上也会有一定的差异,同样的结果在Zhang和Stephanopoulous[36]的大肠杆菌的工作中也有所体现。
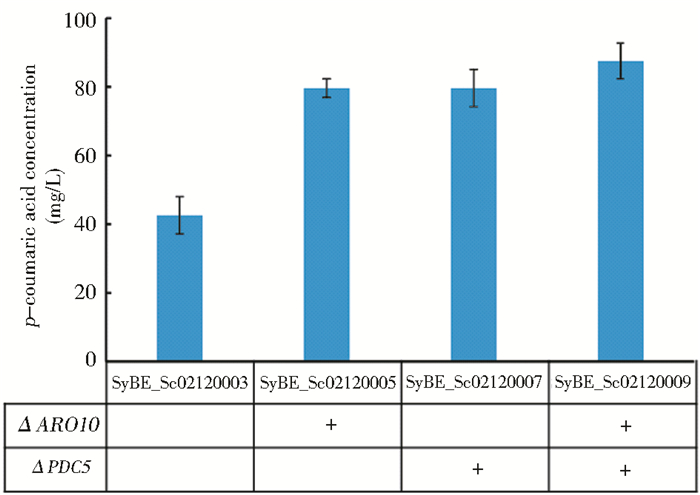
|
| 图 5 敲除基因ARO10和PDC5对对香豆酸产量的影响 Figure 5 Effects of ARO10 and PDC5 knock out on production of p-coumaric acid |
目前利用微生物异源生产苯丙素类、黄酮类化合物时,往往采取外源添加前体物质的策略[37-38]。在对香豆酸的生物合成上,一般采用添加L-酪氨酸来提高产量。Wang等[39]通过外源添加3mmol/L的酪氨酸在大肠杆菌中合成了160.5mg/L的对香豆酸,Santos等[40]也通过向培养基中添加500mg/L的酪氨酸并以不同的大肠杆菌作为宿主细胞分别得到了104mg/L和213mg/L的对香豆酸。在酿酒酵母中也同样采取外源添加的方式,Shin等[41]通过添加12mmol/L酪氨酸得到13.6mg/L对香豆酸。本研究在人工酿酒酵母SyBE_Sc02120009的基础上,设计外源添加前体酪氨酸的实验,来考察不同浓度的前体氨基酸的添加对对香豆酸产量的影响。本研究先验证L-酪氨酸能否穿过酵母细胞壁到达胞内参与反应,实验结果证明可以实现,继而本研究选取了0.25mmol/L、0.5mmol/L、0.75mmol/L、1mmol/L和1.25mmol/L五个浓度梯度,并以未添加酪氨酸的作为对照,发酵结束后测定对香豆酸的产量。实验结果表明,在酪氨酸添加浓度小于1mmol/L时,对香豆酸的产量随着酪氨酸添加的浓度增大而增大,在浓度为1mmol/L时达到峰值,为(174.57±0.30) mg/L,当酪氨酸浓度增加到1.25mmol/L时,对香豆酸的产量反而降低,这表明外源添加酪氨酸虽然能提高对香豆酸的产量,但高浓度的酪氨酸可能由于其抑制细胞生长或其他原因导致对香豆酸产量下降。所以,最终确定酪氨酸的添加量为1mmol/L。

|
| 图 6 外源添加L-酪氨酸对发酵产物对香豆酸产量的影响 Figure 6 Effects of added L-tyrosine concentration on production of p-coumaric acid |
本研究通过引入外源基因TAL实现了异源合成对香豆酸,并对TAL进行不同表达水平的调节;在此基础上,对酿酒酵母内源路径进行改造优化,其中包括对L-酪氨酸的负反馈抑制基因进行氨基酸突变,并采用delta位点整合至酵母基因组,既增加了基因拷贝数同时也避免了多拷贝质粒容易丢失的问题,delta位点整合由于其整合位置和数目的不确定性本研究对构建成功的菌株进行筛选;其次,本研究也对合成对香豆酸的旁路路径进行相关基因的敲除工作,促进代谢流更多地流向对香豆酸,从而实现了在酿酒酵母中高产对香豆酸。另外,本研究对L-酪氨酸的外源添加对产物的影响进行探究,为生物合成对香豆酸及其衍生物打下基础。
| [1] |
冉桂梅, 何彬, 杨凌, 等. 对香豆酸在大鼠体内的药动学研究. 中国药学杂志, 2005, 40(24): 1889-1891. Ran G M, He B, Yang L, et al. Study on pharmacokinetics of p-coumaric acid in rat. Chinese Pharmaceutical Journal, 2005, 40(24): 1889-1891. DOI:10.3321/j.issn:1001-2494.2005.24.016 |
| [2] |
Cheng J C, Dai F, Zhou B, et al. Antioxidant activity of hydroxycinnamic acid derivatives in human low density lipoprotein:Mechanism and structure-activity relationship. Food Chemistry, 2007, 104(1): 132-139. DOI:10.1016/j.foodchem.2006.11.012 |
| [3] |
Abdelwahab M H, Elmahdy M A, Abdellah M F, et al. Influence of p-coumaric acid on doxorubicin-induced oxidative stress in rat's heart. Pharmacological Research, 2003, 48(5): 461-465. DOI:10.1016/S1043-6618(03)00214-7 |
| [4] |
Bodini S F, Manfredini S, Epp M, et al. Quorum sensing inhibition activity of garlic extract and p-coumaric acid. Letters in Applied Microbiology, 2009, 49(5): 551-555. DOI:10.1111/lam.2009.49.issue-5 |
| [5] |
Alam M A, Subhan N, Hossain H, et al. Hydroxycinnamic acid derivatives:a potential class of natural compounds for the management of lipid metabolism and obesity. Nutrition & Metabolism, 2016, 13(1): 1-13. |
| [6] |
Fowler Z L, Koffas M A. Biosynthesis and biotechnological production of flavanones:current state and perspectives. Applied Microbiology and Biotechnology, 2009, 83(5): 799-808. DOI:10.1007/s00253-009-2039-z |
| [7] |
Shin S Y, Jung S M, Kim M D, et al. Production of resveratrol from tyrosine in metabolically engineered Saccharomyces cerevisiae. Enzyme & Microbial Technology, 2012, 51(4): 211-216. |
| [8] |
Yan Y, Chemler J, Huang L, et al. Metabolic engineering of anthocyanin biosynthesis in Escherichia coli. Applied & Environmental Microbiology, 2005, 71(7): 3617-3623. |
| [9] |
Zhao S, Jones J A, Lachance D M, et al. Improvement of catechin production in Escherichia coli through combinatorial metabolic engineering. Metabolic Engineering, 2015, 28: 43-53. DOI:10.1016/j.ymben.2014.12.002 |
| [10] |
Trantas E, Panopoulos N, Ververidis F. Metabolic engineering of the complete pathway leading to heterologous biosynthesis of various flavonoids and stilbenoids in Saccharomyces cerevisiae. Metabolic Engineering, 2009, 11(6): 355-366. DOI:10.1016/j.ymben.2009.07.004 |
| [11] |
Yan Y, Kohli A, Koffas M A. Biosynthesis of natural flavanones in Saccharomyces cerevisiae. Applied & Environmental Microbiology, 2005, 71(9): 5610-5613. |
| [12] |
Hwang E I, Kaneko M, Ohnishi Y, et al. Production of plant-specific flavanones by Escherichia coli containing an artificial gene cluster. Applied & Environmental Microbiology, 2003, 69(5): 2699-2706. |
| [13] |
Vannelli T, Wei Q W, Sweigard J, et al. Production of p-hydroxycinnamic acid from glucose in Saccharomyces cerevisiae and Escherichia coli by expression of heterologous genes from plants and fungi. Metabolic Engineering, 2007, 9(2): 142-151. DOI:10.1016/j.ymben.2006.11.001 |
| [14] |
Santos C N, Koffas M, Stephanopoulos G. Optimization of a heterologous pathway for the production of flavonoids from glucose. Metabolic Engineering, 2011, 13(4): 392-400. DOI:10.1016/j.ymben.2011.02.002 |
| [15] |
Xue Z, Mccluskey M, Cantera K, et al. Improved production of p -hydroxycinnamic acid from tyrosine using a novel thermostable phenylalanine/tyrosine ammonia lyase enzyme. Enzyme & Microbial Technology, 2007, 42(1): 58-64. |
| [16] |
Li M, Kildegaard K R, Chen Y, et al. De novo production of resveratrol from glucose or ethanol by engineered Saccharomyces cerevisiae. Metabolic Engineering, 2015, 32: 1-11. DOI:10.1016/j.ymben.2015.08.007 |
| [17] |
Siddiqui M S, Thodey K, Trenchard I, et al. Advancing secondary metabolite biosynthesis in yeast with synthetic biology tools. Fems Yeast Research, 2012, 12(2): 144. DOI:10.1111/j.1567-1364.2011.00774.x |
| [18] |
Liu D, Li B, Liu H, et al. Profiling influences of gene overexpression on heterologous resveratrol production in Saccharomyces cerevisiae. Frontiers of Chemical Science & Engineering, 2017, 11(1): 117-125. |
| [19] |
Liu D, Jin D U, Zhao G, et al. Applications of synthetic biology in medicine and energy. Ciesc Journal, 2011, 62(9): 2391-2397. |
| [20] |
肖文海, 周嗣杰, 王颖, 等. 如何工程化生物学. 化工进展, 2016, 35(6): 1827-1836. Xiao W H, Zhou S J, Wang Y, et al. How to make biology more "engineering". Chemical industry and Engineering Progress, 2016, 35(6): 1827-1836. |
| [21] |
Fink G, Farabaugh P, Roeder G, et al. Transposable elements (Ty) in yeast. Cold Spring Harbor Symposia on Quantitative Biology, 1981, 45(pt 2): 575-580. |
| [22] |
Dujon B. The yeast genome project:what did we learn. Trends in Genetics, 1996, 12(7): 263-270. DOI:10.1016/0168-9525(96)10027-5 |
| [23] |
Luttik M A, Vuralhan Z, Suir E, et al. Alleviation of feedback inhibition in Saccharomyces cerevisiae aromatic amino acid biosynthesis:quantification of metabolic impact. Metabolic Engineering, 2008, 10(4): 141-153. |
| [24] |
Brachmann C B, Davies A, Cost G J, et al. Designer deletion strains derived from Saccharomyces cerevisiae S288C:A useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast, 1998, 14(2): 115-132. DOI:10.1002/(ISSN)1097-0061 |
| [25] |
Jendresen C B, Stahlhut S G, Li M, et al. Novel highly active and specific tyrosine ammonia-lyases from diverse origins enable enhanced production of aromatic compounds in bacteria and yeast. Applied & Environmental Microbiology, 2015, 81(13): 4458-4476. |
| [26] |
Gietz R D, Schiestl R H. High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nature Protocols, 2007, 2(1): 38-41. DOI:10.1038/nprot.2007.15 |
| [27] |
Wang R Z, Gu X L, Yao M D, et al. Engineering of β-carotene hydroxylase and ketolase for astaxanthin overproduction in Saccharomyces cerevisiae. Frontiers of Chemical Science & Engineering, 2017, 11(1): 89-99. |
| [28] |
Hartmann M, Schneider T R, Pfeil A, et al. Evolution of feedback-inhibited beta/alpha barrel isoenzymes by gene duplication and a single mutation. Proceedings of the National Academy of Sciences of the United States of America, 2003, 100(3): 862-867. DOI:10.1073/pnas.0337566100 |
| [29] |
Krappmann S, Lipscomb W N, Braus G H. Coevolution of transcriptional and allosteric regulation at the chorismate metabolic branch point of Saccharomyces cerevisiae. Proceedings of the National Academy of Sciences, 2000, 97(25): 13585-13590. DOI:10.1073/pnas.240469697 |
| [30] |
F W F L, Silva N A D. Sequential δ-Integration for the regulated insertion of cloned genes in Saccharomyces cerevisiae. Biotechnology Progress, 1997, 13(4): 368-373. DOI:10.1021/bp970055d |
| [31] |
Sakai A, Shimizu Y, Hishinuma F. Integration of heterologous genes into the chromosome of Saccharomyces cerevisiae using a delta sequence of yeast retrotransposon Ty. Applied Microbiology and Biotechnology, 1990, 33(3): 302-306. DOI:10.1007/BF00164526 |
| [32] |
王瑞钊, 潘才惠, 王颖, 等. 高产β-胡萝卜素酿酒酵母菌株的设计与构建. 中国生物工程杂志, 2016, 36(7): 83-91. Wang R Z, Pan C H, Wang Y, et al. Design and construction of highβ-carotene producing Saccharomyces cerevisiae. China Biotechnology, 2016, 36(7): 83-91. |
| [33] |
Koopman F, Beekwilder J, Crimi B, et al. De novo production of the flavonoid naringenin in engineered Saccharomyces cerevisiae. Microbial Cell Factories, 2012, 11(1): 155. DOI:10.1186/1475-2859-11-155 |
| [34] |
Hegemann J H, Güldener U, Köhler G J. Gene disruption in the budding yeast Saccharomyces cerevisiae. Methods in Molecular Biology, 2006, 313: 129-144. |
| [35] |
Rodriguez A, Kildegaard K R, Li M, et al. Establishment of a yeast platform strain for production of p-coumaric acid through metabolic engineering of aromatic amino acid biosynthesis. Metabolic Engineering, 2015, 31: 181-188. DOI:10.1016/j.ymben.2015.08.003 |
| [36] |
Zhang H, Stephanopoulos G. Co-culture engineering for microbial biosynthesis of 3-amino-benzoic acid in Escherichia coli. Biotechnology Journal, 2016, 11(7): 149-150. |
| [37] |
Shin S Y, Han N S, Park Y C, et al. Production of resveratrol from p-coumaric acid in recombinant Saccharomyces cerevisiae expressing 4-coumarate:coenzyme A ligase and stilbene synthase genes. Enzyme & Microbial Technology, 2011, 48(1): 48-53. |
| [38] |
Sun P, Liang J L, Kang L Z, et al. Increased resveratrol production in wines using engineered wine strains Saccharomyces cerevisiae EC1118 and relaxed antibiotic or auxotrophic selection. Biotechnol Prog, 2014, 31(3): 650-655. |
| [39] |
Wang S, Zhang S, Xiao A, et al. Metabolic engineering of Escherichia coli for the biosynthesis of various phenylpropanoid derivatives. Metabolic Engineering, 2015, 29(9): 153-159. |
| [40] |
Santos C N, Koffas M, Stephanopoulos G. Optimization of a heterologous pathway for the production of flavonoids from glucose. Metabolic Engineering, 2011, 13(4): 392-400. DOI:10.1016/j.ymben.2011.02.002 |
| [41] |
Shin S Y, Jung S M, Kim M D, et al. Production of resveratrol from tyrosine in metabolically engineered Saccharomyces cerevisiae. Enzyme & Microbial Technology, 2012, 51(4): 211-216. |
 2017, Vol. 37
2017, Vol. 37