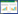文章信息
- 尹舒贤, 赵月华, 刘超, 吕占军, 王秀芳.
- YIN Shu-xian, ZHAO Yue-hua, LIU Chao, LV Zhan-jun, WANG Xiu-fang.
- 人源Alu RNA工程菌的构建和表达
- Construction and Expression of Engineering Bacteria Producing Humanized Alu RNA
- 中国生物工程杂志, 2017, 37(7): 88-96
- China Biotechnology, 2017, 37(7): 88-96
- http://dx.doi.org/DOI:10.13523/j.cb.20170715
-
文章历史
- 收稿日期: 2017-01-16
- 修回日期: 2017-03-24
2. 河北医科大学 河北省实验动物重点实验室 遗传研究室 石家庄 050017
2. Department of Genetics, Hebei Medical University, Hebei Key Lab of Laboratory Animal, Shijiazhuang 050017, China
RNA作为基因表达调节的重要因素越来越引起人们的重视[1]。已经证明RNA能上调或下调基因表达,参与基因表达调节的RNA有microRNA[2-3]、小激活RNA(small activating RNA,saRNA)[4-5]、非编码RNA(non-coding RNA,ncRNA)[6-7]等。细胞中的RNA可以由细胞中的DNA (基因组DNA或转染的质粒等)转录产生,也可以由外源导入[5, 8]。获得外源导入RNA(外源RNA)的方法有RNA合成仪合成[9]、体外转录[10]和从细胞中提取[11]。Alu是人基因组中最重要的非编码序列,占人基因组的10%[12-13],Alu序列影响基因表达[14-15]。Alu家族是灵长类基因组特有的含量丰富的短散在重复序列(short interspersed elements,SINEs),在人基因组中的拷贝数已经达到了100万,在所有已知的基因内含子中几乎都发现了Alu序列。这一基因家族曾被认为是垃圾序列,但随着研究的深入,Alu在基因调控表达网络中的功能逐渐被发现,Alu与人类的疾病密切相关,Alu元件的插入、删除和重组导致了许多先天性遗传疾病和癌症,并且可能影响人类衰老,在人类多样性中也发挥重要作用。目前,对于Alu序列的功能了解得还不透彻,推测主要参与基因调控、基因重排、CpG甲基化、hnRNA选择性剪切、结合转录因子和激素等。本文用Alu RNA为例建立用基因工程菌制备人源Alu RNA(Alu RNA)的方法,该方法也适于制备其他人源RNA。制备的人源RNA可用于细胞学实验。
1 材料与方法 1.1 主要试剂和耗材pET-28α质粒为本研究室保存;尼龙转移膜(Positively CHGD. Nylon transfer membrane,英国GE Healthcare公司);卵白素-过氧化物酶(avidin-peroxidase,武汉博士德生物);生物素标记dUTP (biotin-16-dUTP,瑞士罗氏生物科技公司);BMBL21-DE3(简称DE3)、BMBL21-DE3-pLysS(pLysS)和Trans BL 21(TransBL)感受态细胞(北京博迈德生物);辣根酶化学发光液(美国Life Technologies公司);HindⅢ和NheⅠ等限制酶(大连TaKaRa公司);T4 DNA连接酶(T4 DNA Ligase,Thermo Scientific公司);DNaseⅠ(RNase free,Thermo Scientific公司);RNase A(Solarbio公司);RNase抑制剂(RNase inhibitor, Thermo Scientific公司);鱼精DNA(Solarbio公司);质粒小提试剂盒(北京Tiangen生物公司);异丙基硫代半乳糖苷(IPTG,Solarbio公司);封闭专用脱脂奶粉(北京普利莱基因技术公司)。
1.2 实验方法 1.2.1 pET-Alu质粒构建本研究室在以往的研究中已经成功构建了C1-Alu×1as(将1拷贝的Alu元件反向插入pEGFP-C1质粒的EGFP基因下游构建而成)、C1-Alu×2as、C1-Alu×4as、C1-Alu×8as、C1-Alu×14as质粒[16]。用HindⅢ和NheⅠ分别酶切这些C1衍生质粒,1%琼脂糖电泳,切胶,分离插入片段。pET-28α用HindⅢ和NheⅠ酶切,电泳,切胶分离质粒片段。将插入片段和pET质粒片段用T4 DNA连接酶连接,将构建的质粒转化DH5α大肠杆菌感受态,按常规方法涂平板、提质粒、酶切和测序鉴定,即构建成pET-Alu×1、pET-Alu×2、pET-Alu×4、pET-Alu×8、pET-Alu×14质粒。
1.2.2 BL-21转化及IPTG诱导本文中使用了3种BL-21大肠杆菌(DE3、pLysS和TransBL),它们的感受态细胞用pET或pET-Alu质粒热激法转化,涂平板,提质粒,酶切鉴定,选正确质粒转化菌,摇菌用LB-卡那霉素(终浓度30μg/ml)培养至600nm OD值为1.0,加入IPTG(终浓度0.2mg/ml),37℃继续摇菌4h。改变IPTG浓度、摇菌时间、培养温度等条件在文中注明。
1.2.3 Alu RNA制备用热酚法制备Alu RNA [11]。具体方法简述如下:10ml菌液离心后,在沉淀中加入2% SDS-0.15mol NaCI 2ml,再加水饱和酚1ml,60℃ 30min,冷却后加氯仿0.5ml,离心取上清液,加3倍体积无水乙醇,4℃沉淀30min,12 000r/min离心,75%乙醇洗沉淀2次,加上清液1/10体积的DNaseⅠ(终浓度0.5U/ml)-RNase抑制剂(终浓度0.5U/ml)消化残存的DNA,获得无DNA污染的Alu RNA。为了检测所获得的RNA是否含有DNA污染,在DNaseⅠ消化的基础上用RNase A(终浓度100μg/ml)消化。提取的Alu RNA用Northern杂交进行检测。
1.2.4 Northern检测Alu RNA1mmol/L的dATP、dCTP、dGTP等量混合, 1mmol/L dTTP的加入量为dATP的2/3,1mmol/L Biotin-16-dUTP的加入量为dATP的1/3,配制出0.25mmol/L Biotin-dNTP。用C1-Alu×8质粒作为模板,Alu140F作为上游引物(5′GTG GTG GCG GGT GCC TGT AG),AluR作为下游引物(5′TGA GAC GGA GTC TCG CTG TG),扩增Alu序列的140bp片段,PCR反应液中含有终浓度为0.1mmol/L的Biotin-dNTP,PCR反应液中的其他成分为常规组成。PCR循环条件:94℃ 30s,50℃ 30s,72℃ 1min,30个循环。即制备出Biotin标记的Alu探针。
Alu RNA经甲醛变性胶电泳后转尼龙膜[17],用次甲基蓝染色,预杂交液(含1%脱脂专用奶粉,10mg/ml鱼精DNA)封闭,换为杂交液(预杂交液中加入Biotin标记的Alu探针),于HL-2000 HybriLinker杂交炉中,42℃旋转杂交过夜。冲洗后,加入1:200稀释的卵白素-过氧化物酶,摇床反应1h,冲洗后加入辣根酶化学发光液,伯乐成像分析系统(ChemiDocTM Touch,Bio-RAD公司)照相。用Gel-Pro-analyzer软件分析积分光密度(IOD)值。
2 结果与分析 2.1 表达载体构建及酶切鉴定Hind Ⅲ/Nhe Ⅰ酶切C1-Alu×1as、C1-Alu×2as、C1-Alu×4as、C1-Alu×8as、C1-Alu×14as质粒,1%琼脂糖电泳,切胶分离小片段,插入Hind Ⅲ/Nhe Ⅰ酶切的pET质粒,获得pET-Alu×1正(简称pET-Alu×1)、pET-Alu×2、pET-Alu×4、pET-Alu×8、pET-Alu×14质粒。插入位置示意图见图 1a。Hind Ⅲ/Nhe Ⅰ酶切电泳EB染色图见图 1b,酶切电泳结果符合预计结果。
|
| 图 1 质粒构建和酶切鉴定示意图 Figure 1 The diagrammatic sketch of plasmid construction and enzyme digestion (a) Schematic diagram of plasmid construction (b) Agarose gel electrophoresis images of plasmids being digested lane 1: pET-Alu×1; lane 2: pET-Alu×2; lane 3: pET-Alu×4; lane 4: pET-Alu×8; lane 5: pET-Alu×14; lane 6: Marker |
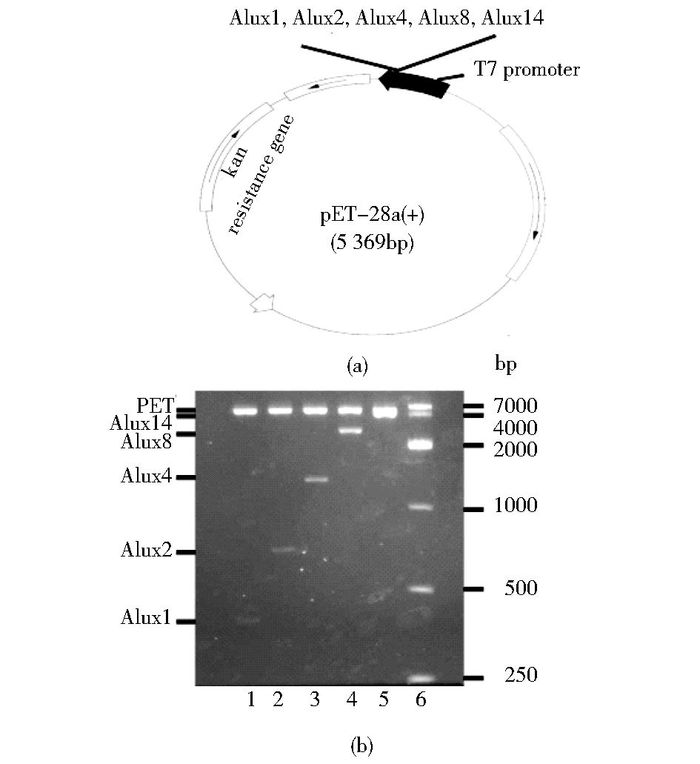
pET-Alu×8 DE3菌、pET DE3菌、DE3菌,共3种菌摇菌培养,调600nm OD=1.0。不加IPTG或加IPTG,培养1h、2h、3h、4h、5h、7h、9h,分别取出1份测定600nm OD值。发现3种菌不加IPTG诱导,OD值在2.6左右进入平台期;加IPTG诱导,pET-Alu×8 DE3菌、pET DE3菌在1.3左右进入平台期,说明IPTG抑制该2种菌的生长,加IPTG对DE3菌生长影响不大(图 2)。
|
| 图 2 IPTG诱导对菌体生长的影响 Figure 2 Effects of IPTG induction on growth of bacteria |
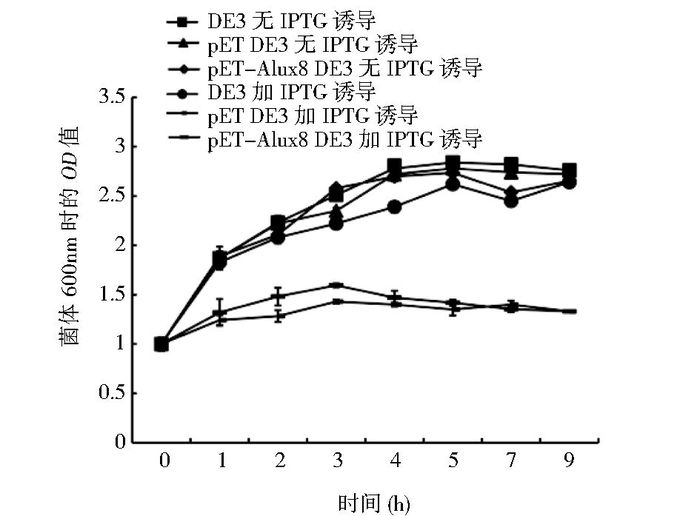
pET-Alu×8 DE3菌在37℃培养至600nm OD值为1.0时,加IPTG 0.2mg/ml诱导2h、4h、6h、8h、10h、12h、14h、16h,提取RNA进行Northern检测,发现2~4h RNA表达达到高峰,8~14h维持在相对恒定的低水平,16h时RNA表达明显下降(图 3a),图 3b是次甲基蓝染色显示的上样RNA量。用Gel-Pro-analyzer分析软件计算图 3a中每一个泳道的条带积分光密度(IOD),所得结果如图 3c所示。

|
| 图 3 IPTG诱导时间对Alu RNA产量的影响 Figure 3 Effect of IPTG induction time on the yield of Alu RNA (a) The results of northern blotting after different time induced by IPTG (b) Dyeing results of methylene blue (c) IOD values of each lane of the northern blotting results (means of three independent experiments) |
pET-Alu×8 DE3菌液,不加IPTG诱导,没有Alu RNA产生(图 4泳道1),0.1~0.4mg/ml IPTG诱导时,Alu RNA产量没有区别,偏离该浓度时,RNA产量略下降(图 4)。

|
| 图 4 IPTG浓度对Alu RNA产量的影响 Figure 4 Effects of IPTG concentrations on yield of Alu RNA (a) Northern detection results after 4 hours induction by different concentrations of IPTG (b) Dyeing results of methylene blue |
pET-Alu×1 DE3菌、pET-Alu×2 DE3菌、pET-Alu×4、pET-Alu×8 DE3菌、pET-Alu×14 DE3菌,OD 1.0时加IPTG诱导4h,提取RNA Northern检测,随Alu拷贝数的增加,Alu RNA产量上升(图 5a),图 5b是次甲基蓝染色显示的上样RNA量。图 5c是根据图 5a绘制的杂交信号强度图。
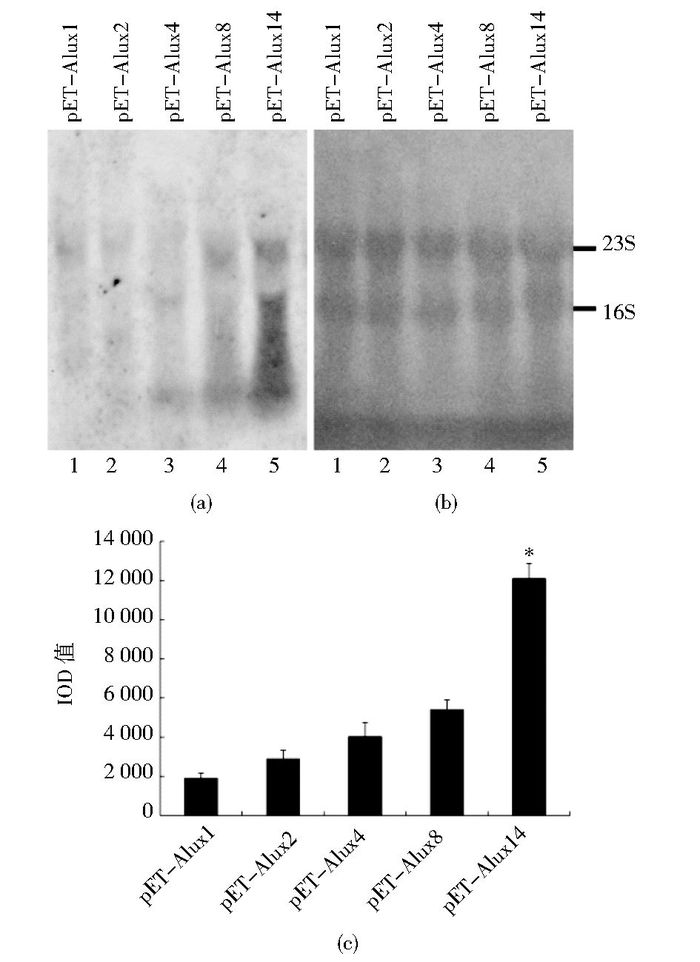
|
| 图 5 Alu拷贝数对Alu RNA产生的影响 Figure 5 Effect of copy number of Alus on Alu RNA production (a) The results of northern detection 1, 2, 4, 8 or 14 copies of Alu elements were inserted into the pET plasmid to construct pET-Alu plasmids that were transformed into DE3 bacteria, the northern blot results showed that Alu RNA production amount increased with the increase of Alu copy number (b) Dyeing result of methylene blue (c) IOD values of each lane of the northern blotting results (means of three independent experiments *: The Alu RNA yield of pET-Alu 14 plasmid was significantly higher than that of other plasmids |
pET-Alu×8 DE3菌在37℃培养至600nm OD值为1.0时,加入终浓度为0.2mg/ml的IPTG于34℃、37℃和40℃培养4h,提取RNA,Northern杂交结果和次甲基蓝染色结果见图 6a和图 6b;图 6c为图 6a的软件分析IOD结果。显示,37℃培养Alu RNA产量最大(泳道1),其次为34℃培养(泳道2),40℃培养几乎没有Alu RNA产生(泳道3)。

|
| 图 6 pET-Alu×8 DE3菌培养温度对Alu RNA产量的影响 Figure 6 Effects of culture temperature of pET-Alu×8 DE3 bacteria on production amount of Alu RNA (a) The results of Northern blotting pET-Alu×8 DE3 bacteria were cultured at 34℃, 37℃ or 40℃ and then were induced by IPTG for 4 hours, we found Alu RNA production was the highest under the 37℃ cultivation (b) Dyeing result of methylene blue (c) IOD values of each lane of the northern blotting results (means of three independent experiments) *: The Alu RNA yield was the highest under the 37℃ cultivation and was significantly higher than that of other culture temperature |
pET-Alu×8TransBL菌、pET-Alu×8 DE3菌和pET-Alu×8pLysS菌,分别培养到600nm OD值为1.0,加入终浓度为0.2mg/ml的IPTG,于37℃培养4h,提取RNA,Northern杂交结果和次甲基蓝染色结果见图 7a、b;图 7c为图 7a的软件分析IOD结果。显示,pET-Alu×8 DE3菌Alu RNA产量最大(泳道2),其次为pET-Alu×8pLysS菌(泳道3),pET-Alu×8TransBL菌没有Alu RNA产生(泳道1)。

|
| 图 7 pET-Alu×8转化不同类型BL-21宿主菌对Alu RNA产量的影响 Figure 7 Effects of types of BL-21 host bacteria on production amount of Alu RNA (a) The result of northern detection. pET-Alu×8 plasmid was transformed into three kinds of BL-21 cells, including DE3, BMBL21-DE3-pLysS (pLysS) and Trans BL-21 (TransBL), Northern blotting showed that Alu RNA production was the highest when using pET-Alu×8 DE3 (b) Dyeing results of methylene blue (c) IOD values of each lane of the northern blotting results (means of three independent experiments) *: The Alu RNA yield was the highest using DE3 engineering bacteria and was significantly higher than that of other bacteria (P < 0.01) |
pET-Alu×8 DE3菌IPTG诱导,热酚法提取RNA,DNaseⅠ消化去除残存的DNA(图 8a,泳道1),不用DNaseⅠ消化(图 8a,泳道2),DNaseⅠ和RNase A消化,去除残存的DNA和RNA(图 8a,泳道3),然后做Northern杂交检测Alu RNA,结果见图 8。泳道1和泳道2的IOD值没有明显的区别,说明热酚法提取的RNA中质粒DNA所造成的探针杂交信号不起主要作用。泳道3没有杂交信号,说明RNA在杂交信号出现中起主要作用。
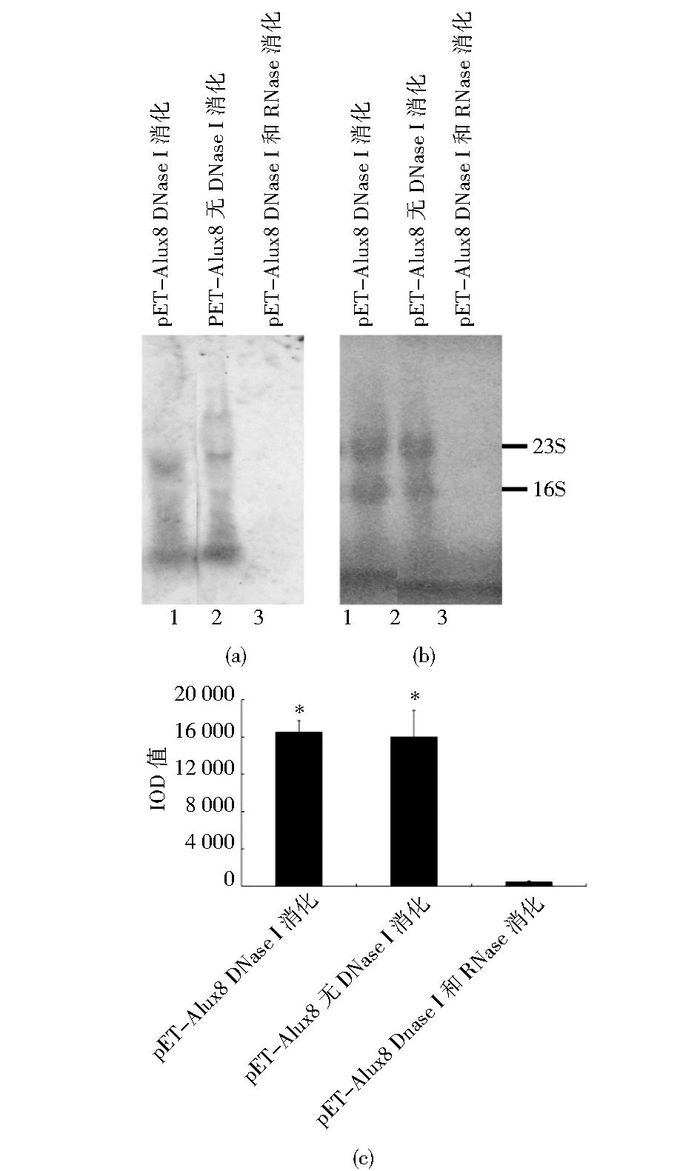
|
| 图 8 DNA酶和RNA酶消化的影响 Figure 8 Effects of digestion of DNA enzyme and RNA enzyme (a) The result of northern detection RNA was extracted from pET-Alu×8 DE3 bacteria induced by IPTG and digested with DNaseⅠ(lane 1), without DNaseⅠ(lane 2), or with DNaseⅠ plus RNase A (lane 3). RNA was detected using Northern blotting (b) Dyeing results of methylene blue (c) IOD values of each lane of the northern blotting results (means of three independent experiments) *: The Alu RNA yield of pET-Alu 8 DNaseⅠdigestion and pET-Alu 8 without DNaseⅠdigestion was significantly higher than that of pET-Alu 8 DNaseⅠplus RNase A digestion (P < 0.01) |
因为pET-Alu×14 DE3菌的Alu RNA产量最高(图 5,泳道5),因此用pET-Alu×14 DE3菌进行实验,对Alu RNA的产量进行估算。将菌体培养至OD值为1.0时加入0.2mg/ml IPTG,37℃培养4h,提取RNA,6次实验每100ml菌液获得的RNA产量为2.94±0.59mg。RNA经Northern检测,同时以正向Alu单链DNA作为参照(称为参照Alu),RNA上样量5μg/孔,Alu参照0.2μg/孔,图 9a显示Northern杂交结果,图 9b是两个泳道的IOD值。Alu RNA孔的信号强度是DNA信号强度的3.95倍。如果忽略RNA和DNA结合能力的差异,那么所制备的Alu RNA中有15.8%属于纯Alu RNA。纯Alu RNA在所提取的Alu RNA中所占比例的计算步骤如下:参照Alu μg数(本文为0.2μg)×(Alu RNA泳道的IOD值/参照Alu的IOD值)=Alu RNA泳道中的纯Alu RNA的μg数;(纯Alu RNA μg/Alu RNA上样量μg)×100%=纯Alu RNA在所提取Alu RNA中所占的比例。每100ml菌液可提取2.94mg RNA,其中Alu RNA的含量占15.8%,那么每100ml菌液纯Alu RNA产量平均为0.46mg。
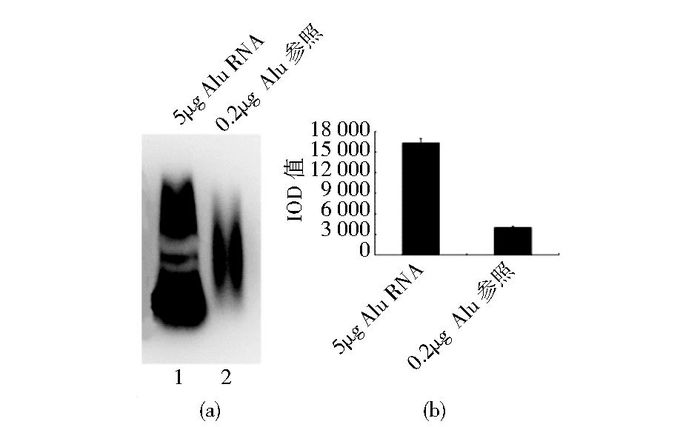
|
| 图 9 对Alu RNA的产量进行估算 Figure 9 The estimation of production amount of Alu RNA (a) The results of northern detection. RNA was extracted from pET-Alu×14 DE3 bacteria induced by IPTG and digested with DNaseⅠ. 5μg RNA digested with DNaseⅠ was loaded in lane 1, 0.2μg reference Alu was loaded in lane 2 (b) IOD values of each lane of the northern blotting results (means of three independent experiments) |
外源RNA有调节基因表达作用[17-20]。Elbashir等[21]证明合成的siRNA转染进入培养的人细胞后,能进入RNA诱导沉默复合体,降解靶mRNA进而沉默基因表达。本实验室最近的研究发现,从细胞中提取的外源性肝RNA能诱导小鼠成纤维细胞白蛋白基因的DNaseⅠ消化敏感性上升,提示外源RNA有改变染色质构象的作用[11]。
外源RNA的获得途径包括:RNA合成仪合成[9]、体外转录[10]和从细胞中提取[11]。BL-21大肠杆菌属于缺陷型细菌,适当质粒转化后,经IPTG诱导能产生大量基因工程蛋白[22-23],是产生基因工程蛋白的常用菌。本文中,建立用BL-21大肠杆菌基因工程方法制备人源Alu RNA的方法,制备出的RNA作为外源RNA用于生物学实验。
DE3菌、pET-DE3菌、pET-Alu×8-DE3菌用LB-卡那霉素培养基培养,不加IPTG诱导,菌液600nm OD值为2.8时达到平台值。三种菌分别用IPTG诱导后,DE3菌的生长曲线与没有诱导的类似;pET-DE3菌、pET-Alu×8-DE3菌在OD值为1.3左右时达到平台期。pET-DE3菌和pET-Alu×8-DE3菌在不加IPTG时,不降低细菌的生长,加入IPTG后降低细菌生长;DE3菌加IPTG和不加IPTG细菌生长曲线类似(图 2)。说明单纯IPTG不影响细菌的生长,IPTG和pET质粒联用才能降低细菌生长;pET和pET-Alu×8质粒转化加IPTG均降低细菌生长,说明降低细菌生长的原因不是来自Alu序列,而是pET质粒的原因。
张树军等[24]证明YfiF融合蛋白质在IPTG诱导9h时蛋白质产量达到高峰,11h和9h的表达量类似。本文证明IPTG诱导4h Alu RNA产量达到高峰,8h以后显著降低,这可能是因为RNA容易降解,蛋白质更容易积累的缘故。张树军等[24]用终浓度为0.1~1mmol/L的IPTG诱导工程菌,测定YfiF融合蛋白质,发现提高IPTG浓度没有增加蛋白质表达量。本文中用终浓度为0.05~1.6mg/ml(0.21~6.72mmol/L)的IPTG诱导pET-Alu×8-DE3菌,发现0.1~0.4mg/ml(0.42~1.68mmol/L)的诱导效率较高,偏离该范围诱导量略有下降(图 4)。
改变串联Alu拷贝数增加RNA量是本文的独特设计。因为产生基因工程蛋白质需要考虑基因的读码框,此外还要考虑5′上游非编码区和3′下游非编码区的结构,不适合通过增加基因拷贝数提高诱导量。本文的结果显示,随Alu拷贝数增加Alu RNA的诱导量明显上升(图 5),pET-Alu×14-DE3菌中Alu RNA的表达量是pET-Alu×8-DE3菌的2倍。
本文中使用三种不同基因型的BL21菌作为宿主菌。DE3适合于pET衍生质粒无毒性基因工程蛋白质的表达,pLysS适合于pET衍生质粒有毒性基因工程蛋白质的表达,TransBL不适合pET衍生质粒的基因工程蛋白质的表达。在本文中pET-Alu×8-DE3菌和pET-Alu×8-pLysS菌在IPTG诱导的情况下均能产生Alu RNA,其中pET-Alu×8-DE3菌产量较高(图 7)。
为了证明Alu RNA诱导的特异性,本文使用pET-Alu×8-TransBL菌株进行IPTG诱导实验,发现没有Alu RNA的产生(图 7泳道1),说明仅有质粒而没有合适的宿主菌,不能诱导Alu RNA的产生。pET-Alu×8-DE3菌如果不用IPTG诱导也不会产生Alu RNA(图 4泳道1),说明IPTG诱导对于Alu RNA的产生是必要的。pET质粒插入序列转录需要T7 RNA聚合酶,该聚合酶属于IPTG诱导型。DE3和pLysS菌中含有IPTG诱导型的T7 RNA聚合酶,TransBL菌不含有IPTG诱导型的T7 RNA聚合酶。pET-Alu×8-DE3菌不加IPTG则不产生T7 RNA聚合酶,Alu RNA诱导为阴性,pET-Alu×8-TransBL菌加IPTG也不能产生Alu RNA,这说明本实验Alu RNA诱导的特异性。本文中经IPTG诱导才能使基因工程菌产生Alu RNA,这与基因工程蛋白质诱导的结果类似[25]。
为了检测DNA和RNA在Northern杂交信号中的作用,本文中进行了核酸酶消化实验。DNaseⅠ消化与不消化的比较,杂交信号区别不显著(图 8泳道1vs泳道2),说明DNA污染在Northern杂交信号强度上所起的作用不大;于DNaseⅠ消化的基础上再用RNase A消化,完全失去杂交信号(图 8泳道3),说明检测到的杂交信号属于RNA。
本文Northern实验中所用的Alu RNA采用热酚法提取,Alu RNA以小片段为主。用Gel-Pro-analyzer和伯乐分子质量分析系统检测,以23S rRNA(2 900nt)与16S rRNA(1 540nt)作为分子质量参照,Alu RNA集中在600bp左右。Northern印记膜次甲基蓝染色显示23S rRNA与16S rRNA的比例大约是2:1(图 3b),说明热酚法提取的RNA并未降解,因此提取的Alu RNA中以小片段为主,不是提取过程中Alu RNA降解的缘故。
本文中用Alu单链DNA作为参照对提取的Alu RNA进行了定量检测。如果忽略DNA单链和RNA单链与探针杂交信号的差异,那么DNA的杂交信号强度就能反应RNA的杂交信号强度。上样总RNA为每孔5μg,单链Alu DNA的量为每孔0.2μg。Alu×14 RNA的信号强度是Alu参照的3.95倍(图 9)。所以5μg上样量中约含有0.79μg Alu RNA,Alu RNA约占总RNA的15.8%。细菌总RNA中以rRNA为主,其次为tRNA,本文中使用T7启动子,IPTG诱导,使纯Alu RNA达到总RNA的15.8%,每100ml菌液可产生纯Alu RNA 0.46mg。综上所述,本文建立了制备基因工程人源Alu RNA的技术。今后的工作设想是使用亲和层析技术进一步提高Alu RNA的纯度。
| [1] |
Wang X, Ma Z, Cheng J, et al. A genetic program theory of aging using an RNA population model. Ageing Research Reviews, 2014, 13: 46-54. DOI:10.1016/j.arr.2013.11.001 |
| [2] |
Chen G, Gao X, Wang J, et al. Hypoxia-induced microRNA-146a represses Bcl-2 through Traf6/IRAK1 but not Smad4 to promote chondrocyte autophagy. Biological Chemistry, 2017, 398(4): 499-507. |
| [3] |
Zhang Z, Ran Y, Shaw T S, et al. MicroRNAs 10a and 10b regulate the expression of human platelet glycoprotein Ibα for normal megakaryopoiesis. International Journal of Molecular Sciences, 2016, 17(11): 1873. DOI:10.3390/ijms17111873 |
| [4] |
Li L C, Okino S T, Zhao H, et al. Small dsRNAs induce transcriptional activation in human cells. Proceedings of the National Academy of Sciences of the United States of America, 2006, 103(46): 17337-17342. DOI:10.1073/pnas.0607015103 |
| [5] |
Voutila J, Strom P, Mintz P, et al. Gene expression profile changes after short-activating RNA-mediated induction of endogenous pluripotency factors in human mesenchymal stem cells. Molecular Therapy Nucleic Acids, 2012, 1(8): e35. |
| [6] |
Sun C C, Li S J, Li G, et al. Long intergenic noncoding RNA 00511 acts as an oncogene in non-small-cell lung cancer by binding to EZH2 and suppressing p57. Molecular Therapy Nucleic Acids, 2016, 5(11): e385. |
| [7] |
Ahn R, Gupta R, Lai K, et al. Network analysis of psoriasis reveals biological pathways and roles for coding and long non-coding RNAs. BMC Genomics, 2016, 17(1): 841. DOI:10.1186/s12864-016-3188-y |
| [8] |
Hélène C, Toulmé J J. Specific regulation of gene expression by antisense, sense and antigene nucleic acids. Biochimica et Biophysica Acta, 1990, 1049(2): 99-125. DOI:10.1016/0167-4781(90)90031-V |
| [9] |
Wickstrom E. DNA and RNA derivatives to optimize distribution and delivery. Advanced Drug Delivery Reviews, 2015, 87: 25-34. DOI:10.1016/j.addr.2015.04.012 |
| [10] |
Voloudakis A E, Holeva M C, Sarin L P, et al. Efficient double-stranded RNA production methods for utilization in plant virus control. Methods in Molocular Biology, 2015, 1236: 255-274. DOI:10.1007/978-1-4939-1743-3 |
| [11] |
Wang X, Ma Z, Kong X, et al. Effects of RNAs on chromatin accessibility and gene expression suggest RNA-mediated activation. International Journal of Biochemistry and Cell Biology, 2016, 79: 24-32. DOI:10.1016/j.biocel.2016.08.004 |
| [12] |
Sela N, Mersch B, Gal-Mark N, et al. Comparative analysis of transposed element insertion within human and mousegenomes reveals Alu's unique role in shaping the human transcriptome. Genome Biology, 2007, 8(6): R127. DOI:10.1186/gb-2007-8-6-r127 |
| [13] |
Häsler J, Strub K. Alu RNP and Alu RNA regulate translation initiation in vitro. Nucleic Acids Research, 2006, 34(8): 2374-2385. DOI:10.1093/nar/gkl246 |
| [14] |
Ma Z, Jing X, Cheng J, et al. The effects of a short sequence enhancer (5'-GTGAAATAAATGCAAATAAAGT) and its derived sequences on green fluorescent protein expression. Genes & Genomics, 2014, 36: 455-464. |
| [15] |
Lv Z, Cheng J, Xie Y, et al. Finding of IFNγ gene enhancers and their core sequences. Genome, 2013, 56: 147-154. DOI:10.1139/gen-2012-0178 |
| [16] |
段肖翠, 靳霞, 谢英, 等. L1-ORF2不同片段对报告基因表达产生不同影响. 遗传, 2009, 31(1): 50-56. Duan X C, Jin X, Xie Y, et al. Different effects on reporter gene expression by distinct L1-ORF2 segments. Hereditas, 2009, 31(1): 50-56. |
| [17] |
Cheng J, Wang X, Cai N, et al. RNAs specifically affect gene expression in a length, position and sequence dependent manner. International Journal of Clinical and Experimental Pathology, 2014, 7(3): 48-58. |
| [18] |
Zheng L, Wang L, Gan J, et al. RNA activation: promise as a new weapon against cancer. Cancer Letter, 2014, 355(1): 18-24. DOI:10.1016/j.canlet.2014.09.004 |
| [19] |
Morris K V. RNA-directed transcriptional gene silencing and activation inhuman cells. Oligonucleotides, 2009, 19(4): 299-306. DOI:10.1089/oli.2009.0212 |
| [20] |
Janowski B A, Younger S T, Hardy D B, et al. Activating gene expression in mammalian cells with promoter-targetedduplex RNAs. Nature Chemical Biology, 2007, 3(3): 166-173. DOI:10.1038/nchembio860 |
| [21] |
Elbashir S M, Harborth J, Lendeckel W, et al. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature, 2001, 411(6836): 494-498. DOI:10.1038/35078107 |
| [22] |
Tsai W C, Wu T C, Chiang B L, et al. Cloning, expression, and purification of recombinant major mango allergen Mani1 in Escherichia coli. Protein Expression and Purification, 2016, 130: 35-43. |
| [23] |
Nasiri K, Zibaee S, Nassiri M, et al. Production of specific IgY antibody to the recombinant FanC protein produced in Escherichia coli. Iranian Journal of Basic Medical Sciences, 2016, 19(8): 883-889. |
| [24] |
张树军, 狄建军, 张国文. 大肠杆菌yfiF基因原核表达系统构建、表达条件优化及蛋白纯化. 生物技术, 2016, 26(3): 229-233. Zhang S J, Di J J, Zhang G W, et al. Construction of prokaryotic expression system of Escherichia coli yfiF gene, optimization of expression conditions and purification of protein. Biotechnology, 2016, 26(3): 229-233. |
| [25] |
李佳楠, 杨薇, 吴红梅, 等. 人β-NGF在大肠杆菌中的表达、纯化及活性测定. 中国药学杂志, 2014, 49(20): 1785-1790. Li J N, Yang W, Wu H M, et al. Expression, purification and anctivity determination of human-NGF in Escheeichia coli. Chinese Pharmaceutical Journal, 2014, 49(20): 1785-1790. |
 2017, Vol. 37
2017, Vol. 37