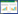文章信息
- 赵秀丽, 周丹丹, 闫晓光, 吴昊, 财音青格乐, 李艳妮, 乔建军.
- ZHAO Xiu-li, ZHOU Dan-dan, YAN Xiao-guang, WU Hao, CAIYIN Qing-gele, LI Yan-ni, QIAO Jian-jun.
- 细菌小RNA的调控及在代谢工程中的应用
- Regulation and Application in Metabolic Engineering of Bacterial Small RNAs
- 中国生物工程杂志, 2017, 37(6): 97-106
- China Biotechnology, 2017, 37(6): 97-106
- http://dx.doi.org/DOI:10.13523/j.cb.20170615
-
文章历史
- 收稿日期: 2017-03-06
- 修回日期: 2017-03-27
细菌代谢工程为环境治理、医学应用,可再生能源的化工生产提供了支持,但是代谢工程也面临诸多问题,如平衡关键代谢物的流量分布、有毒中间体积累和生产环境压力等。为了更好地感知和响应外界环境的变化,同时完成自身的体内循环,使代谢产物的生物合成更高效,越来越多的细菌sRNA应用于代谢工程中。细菌sRNA是一类长度在50~500 nt之间,不编码蛋白质的RNA分子,主要以转录后调控的形式来发挥其调控作用。这样利于较小的靶标mRNA的周转并且可以完成对不同靶标的分级调控[1]。sRNA是从转录水平对菌体生理活动进行调控,设计和合成sRNA策略的优势在于不需要敲除染色体基因组就可以调控微生物代谢网络,避免了突变菌株库的构建过程,省时省力,而且sRNA的表达不会为微生物代谢增加负担。现已发现了超过150多种细菌sRNA[2-3],有关sRNA的研究大多集中于革兰氏阴性菌。近年来伴随着现代生物学和信息学手段、技术的飞速发展,也有越来越多的sRNA在革兰氏阳性菌中(如酿脓链球菌Streptococcus pyogenes[4],乳酸乳球菌Lactococcus lactis[5])被发现,其生物学功能也逐渐被揭示。
1 细菌sRNA靶标基因的预测与鉴定随着大量的sRNA被发掘,识别sRNA的靶标进而探究其功能已成为当今生物学领域研究的热点之一。细菌sRNA靶标基因的识别主要通过基于生物信息学的计算机预测方法与实验分析方法的结合来进行。
生物信息学的计算机预测方法对细菌sRNA靶标的预测主要依据序列长度、碱基匹配、RNA二级结构、序列保守性及其位置等特点。目前常用的sRNA靶标mRNA预测的软件有RNAPredator[6]、sRNATarBase[7]、CopraRNA[8-9]、RNAup[10]、sRNATarget[11]、sTarPicker[12]、IntaRNA[13]、TargetRNA2[14]、RNAcofold[15]、RNAplex[16]和RNAhybrid[17]。CopraRNA、RNAplex、IntaRNA、RNAup和TargetRNA2能够识别最有可能发生相互作用的局部区域并打分;RNAhybrid和RNAcofold更多用于sRNA与较长RNA相互作用的场合。局部相互作用的软件(CopraRNA、IntaRNA、RNAplex、RNAup、TargetRNA2) 比全部相互作用的软件(RNAhybrid和RNAcofold)要好[18]。经实验验证软件的配对率IntaRNA/CopraRNA76.7%,RNAplex 73.6%,TargetRNA2 55.9%,RNAup 78.9%[18]。主要用于预测sRNAs靶标mRNAs计算机工具的特点比较见表 1。

|
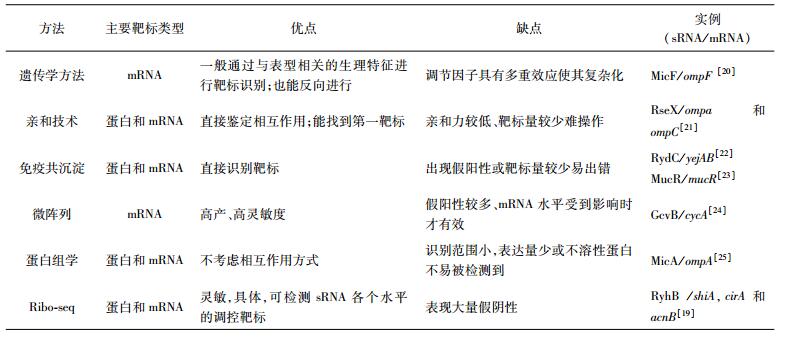
用于验证sRNA靶标基因的方法有很多种,目前比较成熟,使用较多的有:遗传学方法、亲和技术、免疫共沉淀法和微阵列技术。除此之外,ribosome profiling (Ribo-seq)技术是最近开发的基因组规模的方法,可以同时确定体内RNA水平和翻译水平[18],Wang等[19]使用Ribo-seq全面验证大肠杆菌(Escherichia coli)sRNA RyhB已知的靶标,并确定很多新奇的sRNA靶标。实验数据表明,Ribo-seq是一种识别sRNA靶标的高效方法,并且是检测sRNA各级调控(即转录、mRNA稳定性、翻译)的有效方法。各种用于鉴别sRNA靶标的方法之比较见表 2。
2 细菌sRNA与其靶标的相互作用
细菌非编码sRNA按照其在基因组中的位置不同,可以分为顺式编码sRNA(cis-encoded sRNA)和反式编码sRNA(trans-encoded sRNA),顺式编码sRNA由它所调控的mRNA的互补链合成,可以通过与靶标mRNA进行严格的互补配对,抑制其表达或使其基因沉默[26]。细菌sRNA以反式编码sRNA为主,反式编码sRNA大部分都需要RNA分子伴侣Hfq蛋白的协助来发挥作用,主要通过与靶标mRNA上7~12个碱基进行不严格的互补配对,抑制或促进靶mRNA的翻译,加速或减缓靶mRNA的降解。
2.1 细菌sRNA与mRNA的相互作用 2.1.1 sRNA调节mRNA翻译和mRNA稳定性通常情况下,反式编码sRNA通过与位于靶标mRNA5′-UTR的核糖体结合位点(RBS)进行不完全互补配对阻止30S核糖体的结合和翻译的起始(图 1a),例如在大肠杆菌中首次发现的主要孔蛋白OmpC合成抑制因子sRNA MicC,MicC通过与核糖体竞争结合位点抑制ompC翻译[27]。还有一种抑制方式是sRNA在Hfq的协助下与正在进行翻译的mRNA结合,导致mRNA发生结构重组,RBS区域进行碱基互补结合形成二级结构阻止了核糖体结合,使得翻译抑制(图 1c)。相反,有些sRNA通过与靶标的5′-UTR区结合激活翻译,通常这些靶标mRNA 5′-UTR区内在的二级结构阻止核糖体结合,当sRNA结合到5′-UTR区,RBS被打开,使翻译起始(图 1d)。例如在大肠杆菌中DsrA和RyhB可分别促进Rpos[28]和shiA[29]的翻译起始,就是这种调节机制在发挥作用。
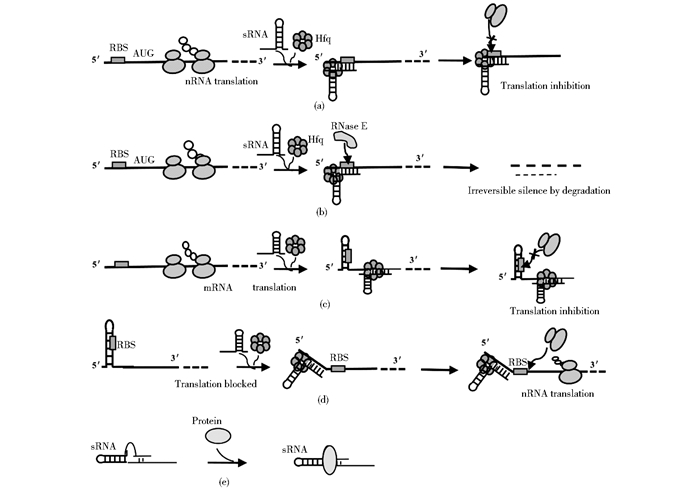
|
| 图 1 反式编码sRNA调控靶标的机制 Figure 1 Regulatory mechanisms of trans-encoded sRNAs (a) Translation inhibition by blocking of RBS (b) sRNAs imperfectly base-pair with target mRNAs in order to repress translation and speed up degradation by RNase (c) Translation inhibition by blocking of RBS (d) Translation activation by mediating mRNA structure change (e) sRNAs act by binding to the target protein to regulate their activities |
RNA稳定性的调节是sRNA主要的调节机制,GadY属于顺式编码的一类sRNA,这种结合通常引导一个负调控,可以积极调节gadW和gadX mRNA水平参与应对酸压力。GadY可以直接与gadX-gadW mRNA基因间区域进行严格的碱基配对,形成双链结构,增加mRNA的稳定性,保护mRNA不被RNaseE降解[30-32](图 2b)。
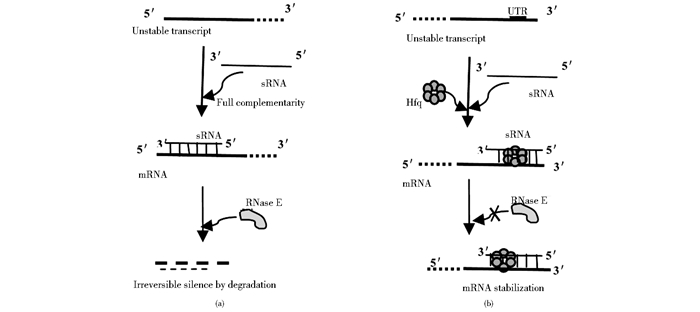
|
| 图 2 顺式编码sRNA调控靶标的机制 Figure 2 Regulatory mechanisms of cis-encoded sRNAs (a) sRNAs perfect complementarity with targets and degraded by an RNase E (b) sRNAs complementarity with mRNA forming duplex structure to increase the stability of mRNA |
sRNA调控靶标mRNA的另一种机制是与mRNA碱基互补配对后,在核酸酶E(RNase E)作用下使mRNA降解。参与靶mRNA降解的RNase E可以特异性地作用于富含AU的单链RNA,将靶mRNA和sRNA同时降解[33-34](图 1b和图 2a)。在这一机制中Hfq参与sRNA诱导的mRNA降解。最先发现的sRNA调控mRNA降解的是RyhB,在铁离子缺乏的情况下,RyhB可导致至少18种编码需铁蛋白的mRNA在10分钟内迅速降解,使铁离子重新分布到细胞重要区域[35]。类似的快速降解mRNA机制也在其他sRNA如OmrA,OmrB[36]和RybB[37]中得到证实。在RNaseE的作用下通过sRNAs/Hfq导致mRNA降解的作用机制还不很清楚,但可以从两种途径来说明mRNA的不稳定性:一种可能是mRNA与sRNA碱基互补配对之后,失去核糖体的保护使其更易受到RNase的攻击;另一种可能是所形成的sRNA/Hfq/RNase E复合物对RNA的降解有促进作用。
2.2 细菌sRNA与蛋白相互作用sRNA可与相应蛋白质结合行使相应的调控功能,sRNA和sRNA结合蛋白的研究均具有重要意义。大多数sRNA需要与蛋白质结合在体内起作用,sRNA和蛋白质相互作用可以分为两大类:一是调控RNA结合蛋白;二是调节酶的活性。
sRNA调节RNA结合蛋白通常是通过模仿作用,如在大肠杆菌中,sRNA基因CsrB和CsrC是CsrA蛋白(全局转录后调控因子)的拮抗物,CsrB和CsrC与CsrA蛋白相互作用可形成一个调控反应回路,从而调控该蛋白的活性[38]。CsrA通过与glgC(糖原代谢基因)转录物的RBS周围序列结合抑制翻译的起始。CsrB和CsrC可通过与CsrA蛋白结合,作为CsrA的拮抗物发挥调节功能[39-40]。
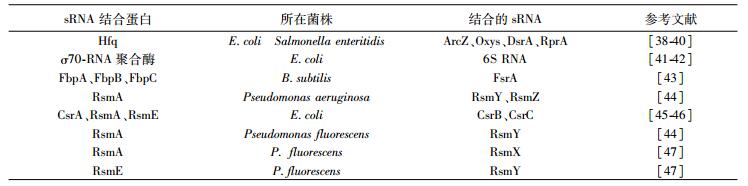
有些sRNA通过结合具有酶活性的蛋白质,有可能抑制、激活或修改蛋白质的活动。在大肠杆菌中,6sRNA可以通过与σ70-RNA聚合酶的相互作用改变σ70-RNA聚合酶对启动子识别的特异性。在转录起始时6sRNA二级结构在很大程度上模仿了DNA构象,与s70-RNA聚合酶全酶结合改变RNA聚合酶的活性,从而抑制启动子区域-10bp上游含TGn序列(TGn TATAAT)基因的转录[41-42]。Smaldone等[43]在枯草芽孢杆菌中发现了FsrA,FsrA能与三种蛋白FbpA、FbpB和FbpC共同作用,抑制包括TCA循环中的琥珀酸脱氢酶和乌头酸酶以及含铁硫簇的谷氨酸合酶等的合成,从而调节细菌体内的铁离子平衡和代谢稳定。目前已经报道的sRNA结合蛋白见表 3所示。
3 细菌sRNA的功能与代谢工程中的应用
sRNA作为一种重要的基因表达调控工具,随着微生物中sRNA越来越多地被发现,它在微生物体内所起的作用以及调控机制也逐渐得到阐明,从而为改变细菌生理特性、改变菌株代谢途径、提高菌株的代谢物产量和微生物细胞工厂构建的方法创新提供了可能。
3.1 细菌sRNA的功能随着越来越多的细菌sRNA被识别,其在菌株中所起的作用也逐渐被挖掘出来。细菌sRNA可通过调控细菌在宿主细胞中的毒力启动细菌细胞内部的相关机制,使细菌对生长条件的变化做出反应来适应感染细胞内的环境变化[48],也可以通过调节群体感应系统的感应调节蛋白的表达来控制菌体的密度[49]。在生长环境中营养物质不足时,细菌sRNA可通过抗营养应激(碳源、氮源)发挥重要的调控作用,在大肠杆菌中,sRNA基因CsrB和CsrC既调节糖原代谢[50]也可以调控氨基酸的代谢过程[51]。
细菌细胞中的微量元素是细菌代谢过程中不可缺少的一部分,有些参与辅助酶促反应,sRNA可以调节微量元素在细胞中的含量使其相对平衡,如sRNA基因RyhB在大肠杆菌中能调节Fur基因的表达,在减缓细菌的铁饥饿中起着重要作用[52]。而sRNA基因RyhB在福氏志贺氏菌可以调节ydeP基因的表达水平从而调控耐酸性[53]这点也说明了同种sRNA在不同细菌中可能有不同的调控机制,导致不同的功能效应。除了可以调控酸耐受之外,sRNA也可以调节其他环境应激,如温度[54]、氧气胁迫[55]。一种反式编码sRNA基因DsrA在大肠杆菌中可以调节菌体对温度变化的感应。当温度较低时,DsrA可以在Hfq蛋白协助下与RpoS mRNA的前导序列碱基配对,激活mRNA的翻译,DsrA大量表达,促进RpoS基因调控细菌RNA聚合酶σ因子的表达[56-58]。sRNA的不同调控功能见图 3。
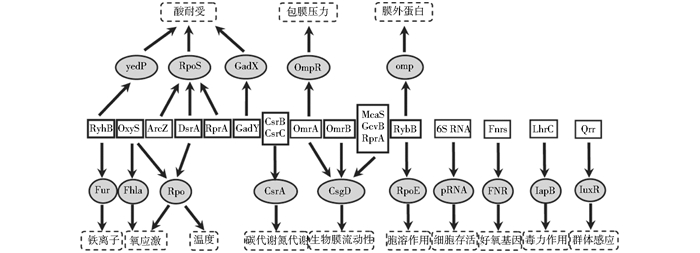
|
| 图 3 细菌sRNA对多种胁迫的调控作用 Figure 3 Bacterial sRNA modulate cells to resist various stresses |
由于sRNA在许多环境压力中扮演至关重要的角色,如在发酵过程中,对保持鲁棒性和长期在压力条件下生产力起重要作用。因此,提高菌株的耐受性将有利于生物工艺应用。sRNA基因DsrA被认为是大肠杆菌中的一个多功能的基因调节器,全局转录调控因子rpoS是由sRNA DsrA,RprA和ArcZ转录后的调节,由OxyS下调表达[59]。最近,Gaida等[60]报道,同时过表达DsrA RprA,ArcZ可大幅增加酸耐受并在对数生长期对羧酸和氧化应激提供保护。同样,应用这种多种转录后调控策略,Kang等[61]建造了一个压力诱导系统来生产高级化学品、包括聚羟基丁酸酯和1,3-丙二醇。因此,用sRNA来提高压力耐受是一种有效的策略。
本实验室以乳酸乳球菌F44为研究对象,在酸胁迫下转录组测序和差异基因表达分析挖掘乳酸乳球菌的sRNA,运用sRNA过表达的方法实验验证并筛选能明显提高乳酸乳球菌F44的sRNA。由预测验证得到的sRNA真正靶标来分析sRNA作用的耐酸机制。通过人工设计合成sRNA,然后将其导入乳酸乳球菌F44中构建出新的高耐酸菌株,从而提高菌株酸耐受性和生产nisin的能力(未发表)。利用sRNA在乳酸乳球菌F44中介导的耐酸调控对构建乳酸乳球菌的耐酸网络具有重要的意义。
3.3 利用反义sRNA“knock down”调控代谢通路反式作用调控小RNA(sRNA)由与mRNA互补结合的靶向性反义RNA和一个召集宿主Hfq的支架组成,随后可以通过核糖核酸酶E诱导mRNA降解[62]。人工合成反义sRNA现已广泛用于基因沉默,sRNA介导的基因沉默是利用天然RNA辅助骨架构建了人工sRNA,afsRNA与mRNA复合物在Hfq协助下实现对内源基因表达进行微调和动态控制[63-64]。
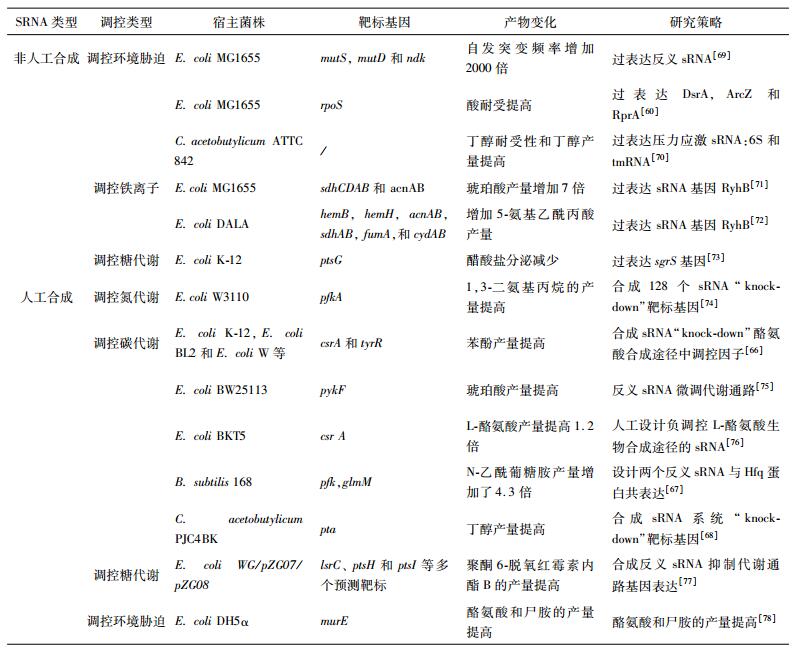
以天然sRNA为依据,合理设计人工sRNA并将其连至载体质粒上,可以在不改变染色质基因的前提下,实现快速高通量的“knock-down”研究,这在代谢工程中得到了广泛应用[65](表 4)。如Lee等[66]通过在18株大肠杆菌菌株中合成调控sRNA将酪氨酸生物合成途径中的碳存储调节器(csrA)和酪氨酸抑制因子(tyrR)两个调控因子“knock-down”。然后在E.coil中过表达酪氨酸生物合成路径中基因和酪氨酸酚裂合酶(TPL)基因,从葡萄糖出发生产苯酚,最后在18株工程菌株中筛选得到了苯酚产量为3.79 g/L的PheBL21菌株,达到了微生物发酵的最高效价。在大肠杆菌中利用sRNA策略来提高目标产品的产量已得到广泛使用,近来,在其他菌株中sRNA策略也成功实现了运用,如枯草芽孢杆菌(Bacillus subtilis)中,Liu等[67]应用sRNA策略提高N-乙酰葡萄糖胺(GlcNAc)的产量、GlcNAc合成、GlcNA糖酵解和肽聚糖合成模块竞争相同的前体。为了直接进行GlcNAc合成,设计了两个靶向于pfk(编码6-磷酸果糖激酶)和glmM(编码磷酸葡糖胺变位酶)的sRNA分别用于限制糖酵解和肽聚糖合成。为了达到更佳抑制效果,对Hfq蛋白进行了共表达。和sRNA一起同时抑制pfk和glmM表达,优化菌株的GlcNAc产量增加了4.3倍。在丙酮丁醇梭菌(Clostridium acetobutylicum PJC4BK)中,Cho等[68]使用合成sRNA敲除pta基因使丁醇的产量增加。sRNA在代谢工程中的应用实例见表 4。
4 小结与展望
RNA深度测序(RNAseq)技术的发展和生物信息学预测以及实验分析手段的完善为细菌sRNA的发现提供了支持。预测并挖掘出更多天然sRNA可以更好地推动代谢工程和合成生物学的发展。但目前仍有很多sRNA功能还未通过实验确定,识别细菌sRNA的靶标,研究sRNA与其靶标之间的相互作用,可以极大地丰富调节代谢工程的方法。
通常加强调控和优化基因电路和代谢途径主要是集中在转录和翻译水平,转录后的方法显示许多优点,如反应迅速、灵活、调控精确,没有代谢负担。通过合理设计sRNA来动态调节代谢途径省去了敲除染色体基因和构建突变菌株库的繁琐步骤,可以使菌株快速发展为理想型菌株;另外,在这一领域的研究将有助于对自然生物网络控制机制的理解。但是,细菌体内的调控机制十分复杂,一个sRNA通常调控多个靶标基因,天然或人工合成sRNA在工程菌株中所调控的不同路径是否存在相互影响?人工sRNA与天然sRNA之间是否有相互作用?而且sRNA上游调控机制尚未清楚,这些问题的解决可以帮助我们更好地利用sRNA来调节工程菌株的代谢途径。目前,细菌sRNA的研究与应用大多集中在大肠杆菌等模式生物中,对致病菌的sRNA研究还有待进一步深入。随着对sRNA与靶标mRNA或靶标蛋白作用机制的进一步深入研究,一定会有更多的sRNA功能被发现,这也为工程菌株的改造、致病微生物的预防和工业微生物育种等提供了保障。
| [1] | Mitarai N. Efficient degradation and expression prioritization with small regulatory RNAs. Physical Biology, 2007, 4(3) : 164–171. DOI:10.1088/1478-3975/4/3/003 |
| [2] | Abu-Qatouseh L F, Chinni S V, Seggewiβ J, et al. Identification of differentially expressed small non-protein-coding RNAs in Staphylococcus aureus displaying both the normal and the small-colony variant phenotype. Journal of Molecular Medicine, 2010, 88(6) : 565–575. DOI:10.1007/s00109-010-0597-2 |
| [3] | Raabe C A, Hoe C H, Randau G, et al. The rocks and shallows of deep RNA sequencing:Examples in the Vibrio cholera RNome. RNA, 2011, 17(7) : 1357–1366. DOI:10.1261/rna.2682311 |
| [4] | Perez N, Trevio J, Liu Z, et al. A genome-wide analysis of small regulatory RNAs in the human pathogen group A Streptococcus. PloS One, 2009, 4(11) : e7668. DOI:10.1371/journal.pone.0007668 |
| [5] | van der Meulen S B, de Jong A, Kok J. Transcriptome landscape of Lactococcus lactis reveals many novel RNAs including a small regulatory RNA involved in carbon uptake and metabolism. RNA biology, 2016, 13(3) : 353–366. DOI:10.1080/15476286.2016.1146855 |
| [6] | Eggenhofer F, Tafer H, Stadler P F, et al. RNA predator:fast accessibility-based prediction of sRNA targets. Nucleic Acids Research, 2011, 39(suppl-2) : W149–154. |
| [7] | Wang J, Liu T, Zhao B, et al. sRNATarBase 3.0:an updated database for sRNA-target interactions in bacteria. Nucleic Acids Research, 2015, 44(D1) : D248–D253. |
| [8] | Wright P R, Richter A S, Papenfort K, et al. Comparative genomics boosts target prediction for bacterial small RNAs. Proceedings of the National Academy of Sciences, 2013, 110(37) : 3487–3496. DOI:10.1073/pnas.1303248110 |
| [9] | Wright P R, Georg J, Mann M, et al. CopraRNA and IntaRNA:predicting small RNA targets, networks and interaction domains. Nucleic Acids Research, 2014, 42(W1) : 119–123. DOI:10.1093/nar/gku359 |
| [10] | Mückstein U, Tafer H, Hackermüller J, et al. Thermodynamics of RNA-RNA binding. Bioinformatics, 2006, 22(10) : 1177–1182. DOI:10.1093/bioinformatics/btl024 |
| [11] | Ying X, Cao Y, Wu J, et al. STarPicker:a method for efficient prediction of bacterial sRNA targets based on a two-step model for hybridization. PLoS One, 2011, 6(7) : e22705. DOI:10.1371/journal.pone.0022705 |
| [12] | Busch A, Richter A S, Backofen R. IntaRNA:efficient prediction of bacterial sRNA targets incorporating target site accessibility and seed regions. Bioinformatics, 2008, 24(24) : 2849–2856. DOI:10.1093/bioinformatics/btn544 |
| [13] | Lorenz R, Bernhart S H, Zu Siederdissen C H, et al. ViennaRNA Package 2.0. Algorithms for Molecular Biology, 2011, 6(1) : 26. DOI:10.1186/1748-7188-6-26 |
| [14] | Tafer H, Hofacker I L. RNAplex:a fast tool for RNA-RNA interaction search. Bioinformatics, 2008, 24(22) : 2657–2663. DOI:10.1093/bioinformatics/btn193 |
| [15] | Kery M B, Feldman M, Livny J, et al. TargetRNA2:identifying targets of small regulatory RNAs in bacteria. Nucleic Acids Research, 2014, 42(W1) : 124–129. DOI:10.1093/nar/gku317 |
| [16] | Krüger J, Rehmsmeier M. RNAhybrid:microRNA target prediction easy, fast and flexible. Nucleic Acids Research, 2006, 34(suppl 2) : 451–454. |
| [17] | Papenfort K, Vanderpool C K. Target activation by regulatory RNAs in bacteria. FEMS Microbiology Reviews, 2015, 39(3) : 362–378. DOI:10.1093/femsre/fuv016 |
| [18] | Ingolia N T, Ghaemmaghami S, Newman J R, et al. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science, 2009, 324(5924) : 218–223. DOI:10.1126/science.1168978 |
| [19] | Wang J, Rennie W, Liu C, et al. Identification of bacterial sRNA regulatory targets using ribosome profiling. Nucleic Acids Research, 2015, 43(21) : 10308–10320. |
| [20] | Delihas N, Forst S. MicF:an anti sense RNA gene involved in response of Escherichia coil to global stress factors. Journal of Molecular Biology, 2001, 313(l) : 1–12. |
| [21] | Douchin V, Bohn C, Bouloc P. Down-regulation of porins by a small RNA bypasses the essentiality of the regulated intramembrane proteolysis protease RseP in Escherichia coli. Journal of Biological Chemistry, 2006, 281(18) : 12253–12259. DOI:10.1074/jbc.M600819200 |
| [22] | Antal M, Borteau V, Douchin V, et al. A small bacterial RNA regulates a putative ABC transporter. Journal of Biological Chemistry, 2005, 280(9) : 7901–7908. DOI:10.1074/jbc.M413071200 |
| [23] |
董浩, 彭小薇, 王晓英, 等.
染色质免疫共沉淀技术对羊种布鲁氏菌转录调控因子MucR靶基因的筛选. 中国农业大学学报, 2016, 21(4) : 102–106.
Dong H, Peng X W, Wang X Y, et al. Identification of the targets of MucR by chromatin immunop recip itation in Brucella melitensis. Journal of China Agricultural University, 2016, 21(4) : 102–106. |
| [24] | Pulvermacher S C, Stauffer L T, Stauffer G V. Role of the sRNA GcvB in regulation of cycA in Escherichia coli. Microbiology, 2009, 155(1) : 106–114. DOI:10.1099/mic.0.023598-0 |
| [25] | Rasmussen A A, Eriksen M, Gilany K, et al. Regulation of ompA mRNA stability the role of a small regulatory RNA in growth phase-dependent control. Molecular Microbiology, 2005, 58(5) : 1421–1429. DOI:10.1111/j.1365-2958.2005.04911.x |
| [26] | Cho K H, Kim J H. Cis-encoded non-coding antisense RNAs in streptococci and other low GC Gram (+) bacterial pathogens. Frontiers in Genetics, 2015, 6 : 110. |
| [27] | Davanloo P, Rosenberg A H, Dunn J J, et al. Cloning and expression of the gene for bacteriophage T7 RNA polymerase. Proceedings of the National Academy of Sciences, 1984, 81(7) : 2035–2039. DOI:10.1073/pnas.81.7.2035 |
| [28] | McCullen C A, Benhammou J N, Majdalani N, et al. Mechanism of positive regulation by DsrA and RprA small noncoding RNAs:pairing increases translation and protects rpoS mRNA from degradation. Journal of Bacteriology, 2010, 192(21) : 5559–5571. DOI:10.1128/JB.00464-10 |
| [29] | Salvail H, Lanthier-Bourbonnais P, Sobota J M, et al. A small RNA promotes siderophore production through transcriptional and metabolic remodeling. Proceedings of the National Academy of Sciences, 2010, 107(34) : 15223–15228. DOI:10.1073/pnas.1007805107 |
| [30] | Opdyke J A, Fozo E M, Hemm M R, et al. RNase Ⅲ participates in GadY-dependent cleavage of the gadX-gadW mRNA. Journal of Molecular Biology, 2011, 406(1) : 29–43. DOI:10.1016/j.jmb.2010.12.009 |
| [31] | Opdyke J A, Fozo E M, Hemm M R, et al. RNase Ⅲ participates in GadY-dependent cleavage of the gadX-gadW mRNA. Journal of Molecular Biology, 2011, 406(1) : 29–43. DOI:10.1016/j.jmb.2010.12.009 |
| [32] | Tramonti A, De Canio M, De Biase D. GadX/GadW-dependent regulation of the Escherichia coli acid fitness island:transcriptional control at the gadY-gadW divergent promoters and identification of four novel 42 bp GadX/GadW-specific binding sites. Molecular Microbiology, 2008, 70(4) : 965–982. |
| [33] | Mackie G A. RNase E:at the interface of bacterial RNA processing and decay. Nature Reviews Microbiology, 2012, 11(1) : 45–57. DOI:10.1038/nrmicro2930 |
| [34] | Saramago M, Bárria C, dos Santos R F, et al. The role of RNases in the regulation of small RNAs. Current Opinion in Microbiology, 2014, 18(4) : 105–115. |
| [35] | Wang J, Rennie W, Liu C, et al. Identification of bacterial sRNA regulatory targets using ribosome profiling. Nucleic Acids Research, 2015, 43(21) : 10308–10320. |
| [36] | Smirnov A, F rstner K U, Holmqvist E, et al. Grad-seq guides the discovery of ProQ as a major small RNA-binding protein. Proceedings of the National Academy of Sciences, 2016, 113(41) : 11591–11596. DOI:10.1073/pnas.1609981113 |
| [37] | Zhang A, Schu D J, Tjaden B C, et al. Mutations in interaction surfaces differentially impact E. coli Hfq association with small RNAs and their mRNA targets. Journal of Molecular Biology, 2013, 425(19) : 3678–3697. DOI:10.1016/j.jmb.2013.01.006 |
| [38] | Majdalani N, Cunning C, Sledjeski D, et al. DsrA RNA regulates translation of RpoS message by an anti-antisense mechanism, independent of its action as an antisilencer of transcription. Proceedings of the National Academy of Sciences, 1998, 95(21) : 12462–12467. DOI:10.1073/pnas.95.21.12462 |
| [39] | Majdalani N, Hernandez D, Gottesman S. Regulation and mode of action of the second small RNA activator of RpoS translation, RprA. Molecular Microbiology, 2002, 46(3) : 813–826. DOI:10.1046/j.1365-2958.2002.03203.x |
| [40] | Mandin P, Gottesman S. Integrating anaerobic/aerobic sensing and the general stress response through the ArcZ small RNA. The EMBO Journal, 2010, 29(18) : 3094–3107. DOI:10.1038/emboj.2010.179 |
| [41] | Lee C A, Fournier M J, Beckwith J. Escherichia coli 6S RNA is not essential for growth or protein secretion. J Bacteriol, 1985, 161(3) : 1156–1161. |
| [42] | Willkomm D K, Hartmann R K. 6S RNA:an ancient regulator of bacterial RNA polymerase rediscovered. Biol Chem, 2005, 386(12) : 1273–1277. |
| [43] | Smaldone G T, Revelles O, Gaballa A, et al. A global investigation of the Bacillus subtilis iron-sparing response identifies major changes in metabolism. Journal of Bacteriology, 2012, 194(10) : 2594–2605. DOI:10.1128/JB.05990-11 |
| [44] | Richter A S, Schleberger C, Backofen R. Seed-based INTARNA prediction combined with GFP-reporter system identifies mRNA targets of the small RNA Yfr1. Bioinformatics, 2010, 26(1) : 1–5. DOI:10.1093/bioinformatics/btp609 |
| [45] | Romeo A, Sonnleitner E, Sorger-Domenigg T, et al. Transcriptional regulation of nitrate assimilation in Pseudomonas aeruginosa occurs via transcriptional antitermination within the nirBD-PA1779-cobA operon. Microbiology, 2012, 158(Pt 6) : 1543–1552. |
| [46] | Romeo T. Global regulation by the small RNA-binding protein CsrA and the non-coding RNA molecule CsrB. Molecular Microbiology, 1998, 29(6) : 1321–1330. DOI:10.1046/j.1365-2958.1998.01021.x |
| [47] | Wilderman P J, Sowa N A, David J. Identification of tandem duplicate regulatory small RNAs in Pseudomonas aeruginosa involved in iron homeostasis. PNAS, 2004, 101(26) : 9792–9797. DOI:10.1073/pnas.0403423101 |
| [48] | Khan M A, G pel Y, Milewski S, et al. Two Small RNAs Conserved in Enterobacteriaceae Provide Intrinsic Resistance to Antibiotics Targeting the Cell Wall Biosynthesis Enzyme Glucosamine-6-Phosphate Synthase. Frontiers in Microbiology, 2016, 7 : 908. |
| [49] | Rutherford S T, van Kessel J C, Shao Y, et al. AphA and LuxR/HapR reciprocally control quorum sensing in vibrios. Genes & Development, 2011, 25(4) : 397–408. |
| [50] | Rutherford S T, van Kessel J C, Shao Y, et al. AphA and LuxR/HapR reciprocally control quorum sensing in vibrios. Genes & Development, 2011, 25(4) : 397–408. |
| [51] | Romeo A, Sonnleitner E, Sorger-Domenigg T, et al. Transcriptional regulation of nitrate assimilation in Pseudomonas aeruginosa occurs via transcriptional antitermination within the nirBD-PA1779-cobA operon. Microbiology, 2012, 158(Pt 6) : 1543–1552. |
| [52] | Jonas K, Melefors O. The Escherichia coli CsrB and CsrC small RNAs are strongly induced during growth in nutrient-poor medium. FEMS Microbiology Letters, 2009, 297(1) : 80–86. DOI:10.1111/fml.2009.297.issue-1 |
| [53] | Oglesby A G, Murphy E R, Iyer V R, et al. Fur regulates acid resistance in Shigella flexneri via RyhB and ydeP. Molecular Microbiology, 2005, 58(5) : 1354–1367. DOI:10.1111/j.1365-2958.2005.04920.x |
| [54] | Dudin O, Lacour S, Geiselmann J. Expression dynamics of Rpo S/Crl-dependent genes in Escherichia coli. Research in Microbiology, 2013, 164(8) : 838–847. DOI:10.1016/j.resmic.2013.07.002 |
| [55] | Updegrove T B, Wartell R M. The influence of Escherichia coli Hfq mutations on RNA binding and sRNAo mRNA duplex formation in rpoS ribo regulation. Biochimica et Biophysica Acta (BBA)-Gene Regulatory Mechanisms, 2011, 1809(10) : 532–540. DOI:10.1016/j.bbagrm.2011.08.006 |
| [56] | Majdalani N, Hernandez D, Gottesman S. Regulation and mode of action of the second small RNA activator of RpoS translation, RprA. Molecular Microbiology, 2002, 46(3) : 813–826. DOI:10.1046/j.1365-2958.2002.03203.x |
| [57] | Papenfort K, Said N, Welsink T, et al. Specific and pleiotropic patterns of mRNA regulation by ArcZ, a conserved, Hfq-dependent small RNA. Molecular Microbiology, 2009, 74(1) : 139–158. DOI:10.1111/mmi.2009.74.issue-1 |
| [58] | Sledjeski D D, Gupta A, Gottesman S. The small RNA, DsrA, is essential for the low temperature expression of RpoS during exponential growth in Escherichia coli. The EMBO Journal, 1996, 15(15) : 3993. |
| [59] | Zhang A, Altuvia S, Tiwari A, et al. The OxyS regulatory RNA represses rpoS translation and binds the Hfq (HF-Ⅰ) protein. The EMBO Journal, 1998, 17(20) : 6061–6068. DOI:10.1093/emboj/17.20.6061 |
| [60] | Gaida S M, Al-Hinai M A, Indurthi D C, et al. Synthetic tolerance:three noncoding small RNAs, DsrA, ArcZ and RprA, acting supra-additively against acid stress. Nucleic Acids Research, 2013, 41(18) : 8726–8737. DOI:10.1093/nar/gkt651 |
| [61] | Kang Z, Wang Q, Zhang H, et al. Construction of a stress-induced system in Escherichia coli for efficient polyhydroxyalkanoates production. Applied Microbiology and Biotechnology, 2008, 79(2) : 203–208. DOI:10.1007/s00253-008-1428-z |
| [62] | Aiba H. Mechanism of RNA silencing by Hfq-binding small RNAs. Curr Opin Microbiol, 2007, 10(2) : 134–139. DOI:10.1016/j.mib.2007.03.010 |
| [63] | Park H, Bak G, Kim S C, et al. Exploring sRNA-mediated gene silencing mechanisms using artificial small RNAs derived from a natural RNA scaffold in Escherichia coli. Nucleic Acids Research, 2013, 41(6) : 3787–3804. DOI:10.1093/nar/gkt061 |
| [64] | Sharma V, Yamamura A, Yokobayashi Y. Engineering artificial small RNAs for conditional gene silencingin Escherichia coli. ACS Synthetic Biology, 2011, 1(1) : 6–13. |
| [65] | Yoo S M, Na D, Lee S Y, et al. Design and use of synthetic regulatory small RNAs to control gene expression in Escherichia coli. Nature Protocols, 2013, 8(9) : 1694–1707. DOI:10.1038/nprot.2013.105 |
| [66] | Lee S Y, Na D, Kim B, et al. Metabolic engineering of Escherichia coli for the production of phenol from glucose. Nature Biotechnology, 2014, 9(5) : 621–629. |
| [67] | Liu Y, Zhu Y, Li J, et al. Modular pathway engineering of Bacillus subtilis for improved N-acetyl glucosamine production. Metabolic Engineering, 2014, 23 : 42–52. DOI:10.1016/j.ymben.2014.02.005 |
| [68] | Cho C, Lee S Y. Efficient gene knockdown in Clostridium acetobutylicum by synthetic small regulatory RNAs. Biotechnology and Bioengineering, 2017, 114(2) : 374–383. DOI:10.1002/bit.v114.2 |
| [69] | Nakashima N, Tamura T. Conditional gene silencing of multiple genes with antisense RNAs and generation of a mutator strain of Escherichia coli. Nucleic Acids Research, 2009, 37(15) : e103. DOI:10.1093/nar/gkp498 |
| [70] | Jones A J, Venkataramanan K P, Papoutsakis T. Overexpression of two stress-responsive, small, non-coding RNAs, 6S and tmRNA, imparts butanol tolerance in Clostridium acetobutylicum. FEMS Microbiology Letters, 2016, 363(8) : 63. |
| [71] | Kang Z, Wang X, Li Y, et al. Small RNA RyhB as a potential tool used for metabolic engineering in Escherichia coli. Biotechnology Letters, 2012, 34(3) : 527–531. DOI:10.1007/s10529-011-0794-2 |
| [72] | Li F, Wang Y, Gong K, et al. Constitutive expression of RyhB regulates the heme biosynthesis pathway and increases the 5-aminolevulinic acid accumulation in Escherichia coli. FEMS Microbiology Letters, 2014, 350(2) : 209–215. DOI:10.1111/fml.2014.350.issue-2 |
| [73] | Negrete A, Majdalani N, Phue J N, et al. Reducing acetate excretion from E. coli K-12 by over-expressing the small RNA SgrS. New Biotechnology, 2013, 30(2) : 269–273. DOI:10.1016/j.nbt.2011.11.007 |
| [74] | Chae T U, Kim W J, Choi S, et al. Metabolic engineering of Escherichia coli for the production of 1, 3-diaminopropane, a three carbon diamine. Scientific Reports, 2015, 5 : 13040–13053. DOI:10.1038/srep13040 |
| [75] | Zhao Y, Wang C S, Li F F, et al. Targeted optimization of central carbon metabolism for engineering succinate production in Escherichia coli. BMC Biotechnology, 2016, 16(1) : 52. DOI:10.1186/s12896-016-0284-7 |
| [76] |
姚元锋, 赵莹, 赵广荣.
人工sRNA沉默csrA基因以优化大肠杆菌生产L-酪氨酸. 中国生物工程杂志, 2013, 33(8) : 60–65.
Yao Y F, Zhao Y, Zhao G R. Artificial sRNAs silencing csrA to optimize the production of L-tyrosine in Escherichia coli. China Biotechnology, 2013, 33(8) : 60–65. |
| [77] | Meng H L, Xiong Z Q, Song S J, et al. Construction of polyketide overproducing Escherichia coli strains via synthetic antisense RNAs based on in silico fluxome analysis and comparative transcriptome analysis. Biotechnology Journal, 2016, 11(4) : 530–541. DOI:10.1002/biot.201500351 |
| [78] | Yoo S M, Na D, Lee S Y. Design and use of synthetic regulatory small RNAs to control gene expression in Escherichia coli. Nat Protoc, 2013, 8 : 1694–1707. DOI:10.1038/nprot.2013.105 |
 2017, Vol. 37
2017, Vol. 37