文章信息
- 张祎祎, 吴嫣爽, 孙瑞珍, 雷蕾.
- ZHANG Yi-yi, WU Yan-shuang, SUN Rui-zhen, LEI Lei.
- CRISPR/Cas9介导的疾病模型构建与基因修复研究进展
- The Progress of CRISPR/Cas9 for Disease Modeling and Gene Correction
- 中国生物工程杂志, 2016, 36(12): 98-103
- China Biotechnology, 2016, 36(12): 98-103
- http://dx.doi.org/DOI:10.13523/j.cb.20161214
-
文章历史
- 收稿日期: 2016-07-25
- 修回日期: 2016-08-27
CRISPR/Cas (clustered regularly interspaced short palindromic repeats/CRISPR-associated proteins)的全称是成簇规律间隔的短回文重复序列/CRISPR相关蛋白系统[1]。CRISPR系统中包含多个高度保守的重复序列和间隔序列形成交叉结构,在CRISPR前段还有一些保守的Cas蛋白在防御机制中起关键作用,同时Cas基因附近有一段转录活化RNA (trans-activating crRNA, tracrRNA)来促进crRNAs的成熟,帮助crRNAs与Cas蛋白结合形成复合物从而作用于目标DNA,完成整个防御机制[2-3]。
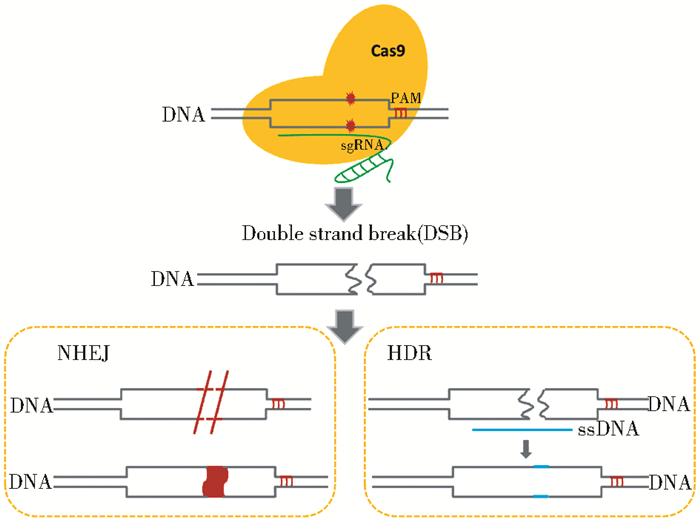
CRISPR系统是细菌和古生菌免疫防御机制的一部分,在外源的质粒或病毒入侵宿主后,CRISPR通过内部的间隔序列来识别这些外源DNA,并形成记忆。当噬菌体再次入侵时,CRISPR区域内一段序列转录成前体的RNA (pre-CRISPR RNA,pre-crRNA),pre-crRNAs受到trancrRNA转录活化成为小的成熟的crRNA,结合相关的Cas蛋白形成crRNA-Cas蛋白复合体,再通过碱基互补配对精确地与目标DNA结合,从而指导Cas蛋白对目标DNA进行切割,并由这些核酸内切酶造成靶向的DNA双链断裂(double-strand breaks,DSBs)。经非同源末端连接(non-homologous end joining,NHEJ)途径修复会出现非特异性的碱基缺失、插入或其他形式突变;DSBs也可利用DNA修复模板,如单链寡聚核苷酸(single-strand oligo DNA,ssODN)在切割位点经同源重组修复(homology-directed repair,HDR)纠正突变或是插入新的基因序列[4-6]。而以Cas9蛋白作用的Ⅱ型CRISPR系统[7],由于不需要复杂的蛋白复合体、且系统组成简单而广泛应用在哺乳动物的基因编辑中(图 1)。
|
| 图 1 CRISPR/Cas9技术的工作原理模式图 Figure 1 Schematic of the CRISPR/Cas9 system |
CRISPR/Cas9在基因编辑平台上展现出明显的技术优势和广阔应用前景。2012年,Jienk等[8]在CRISPR系统中发现了一种双链RNA,并将这种双链RNA改造为一段由回文序列形成的长度约为12~20bp的发卡结构,是能与靶向DNA互补的RNA单链序列,能指导Cas9蛋白对几乎所有DNA序列进行剪切的sgRNA (single-guide RNA)结构,这种结构的发现使CRISPR/Cas9技术实现了进一步的优化。
研究人员在利用CRISPR/Cas9技术进行基因操作过程中发现存在着脱靶问题[9-10],一些实验室分别针对sgRNA长度和Cas9蛋白结构进行改造,优化后的体系能够保证高效特异的基因修饰,并且降低了CRISPR/Cas9的脱靶效应。有研究发现延伸sgRNA的3′端能够提高Cas9对DNA靶位点的识别效率[8]。而Fu等[11]在细胞中证明使用17~18bp缩短的sgRNA也能够降低脱靶效率,并且直接转染合成的sgRNA比转染质粒具有更高的特异性。随后经Ran等[12]证实Cas9内切酶的一种突变体Cas9-D10A配合一对gRNA使用,能够最大限度地降低脱靶效应。Yu等[13]通过加入小分子化合物来提高经HDR或NHEJ的编辑效率,研究中还发现提高经HDR编辑效率的小分子同时对经NHEJ途径的基因编辑起到抑制作用,从而提出两种修复途径间可能存在竞争关系的假设。添加一种抗癌药物Scr7到遗传编辑的受精卵中,能够将Cas9技术的基因编辑效率提高19倍[14]。Gilbert等[15]和Sanjana等[16]将慢病毒载体与CRISPR结合,充分发挥了慢病毒的稳定转染功能和CRISPR的编辑功能,进而建立更加简便、高效的基因编辑方法。
2 基于CRISPR/Cas9技术的动物疾病模型构建与修复2013年Cong等[17]将CRISPR/Cas9技术应用到精准的哺乳动物基因编辑中,该技术比锌指核酸酶(zinc-finger nuclease, ZFN)与转录激活因子样效应物核酸酶(transcriptionactivator-like effector nuclease, TALEN)技术具有更强的优势。ZFN和TALEN都是通过蛋白去识别特异序列的基因组区域,而CRISPR/Cas9技术以标准的Waston-crick结构作为限制性内切酶Cas9结合支架,利用sgRNA来识别基因组目标的方式更加便捷高效。近年来一些研究将CRISPR/Cas9技术应用到动物疾病模型的构建中,避免了传统的建模耗时长,效率低以及定位不精准等问题。基于CRISPR/Cas9技术的精准编辑这一特征,也使之能够应用于基因修复的研究。表 1总结了近几年利用CRISPR/Cas9技术在疾病模型构建与基因修复上的应用。
| Species | Materials | Target gene | Disease | Methods | References |
| Human Cell | hISCs | APC、p53、KRAS、SMAD4 | colorectal cancer, CRC | NHEJ | [31] |
| iPSCs | HBB | β-thalassemia | NHEJ | [33] | |
| CFTR | cysticfibrosis,CF* | HDR | [35] | ||
| PRPF8 | retinitis pigmentosa,RP* | HDR | [34] | ||
| JAK3 | severe combined immunodeficiency,SCID* | HDR | [36] | ||
| T cell | CXCR4 | acquired immunodeficiency syndrome,AIDS* | HDR | [39] | |
| Mice | ESCs | CDX2、GSK3α | chromosomal translocations | NHEJ | [32] |
| SSCs | Crygc | cataract* | NHEJ、HDR | [37] | |
| Embryo | Crygc | cataract* | HDR | [26] | |
| Liver | Pten Stnnb1 | hepatoma nuclear localization of β-Catenin | NHEJ HDR | [22] | |
| Fah | hereditary tyrosinemia type I,HT1* | HDR | [28] | ||
| Pancreas | APC、Cdkn2a etc | pancreatic ductal adenocarcinoma,PDAC | NHEJ | [23] | |
| Rabbit | Embryo | APOE、CD36、LDLR、SCARB1、RyR2 | lipid metabolisms,atherosclerosis,heart arrhythmia | NHEJ | [24] |
| Pig | Fibroblasts | TYRPARK2、PINK1 | oculo-cutaneous albinism type I,OCA1 parkinsondisease, PD | NHEJ | [18] |
| Embryo | vWF | hemophilia | NHEJ | [20] | |
| Mitf | Waardenburg and Tietz | NHEJ | [21] | ||
| DJ-1、Parkin、PINK2 | parkinson disease, PD | NHEJ | [19] | ||
| Sox10 | Warrdenburg | NHEJ、HDR | [27] | ||
| Monkey | Embryo | Dystrophin | duchenne muscular dystrophy, DMD | NHEJ | [25] |
| *:gene correction | |||||
2.1 构建模拟人类疾病的猪模型
猪是生物医学研究中重要的模式动物,它要比一些小型的啮齿动物更适合于构建模拟人类疾病的动物模型。尤其是模拟人类的神经退行性疾病,基因修饰的小鼠不能够有效地获得与人类神经退行性疾病一致的临床表型,而猪有与人类大脑皮层相似的脑回,更适合模拟人类的神经退行性疾病。2014年,Zhou等[18]在猪胎儿成纤维细胞上利用CRISPR/Cas9技术构建PARK2和PINK1两个基因敲除的细胞,作为体细胞核移植(somatic cell nuclear transfer, SCNT)的供体细胞从而获得了模拟帕金森疾病的动物模型。同时该研究中还针对TYR基因进行敲除,获得了模拟人类Ⅰ型白化病表型的动物。Wang等[19]于2016年用该技术建立3个基因(DJ-1、Parkin、PINK1)同时敲除的巴马小型猪模型,以此来模拟隐性遗传的家族性早发帕金森疾病。
2014年1月,Hai等[20]应用CRISPR/Cas9技术在猪上实现了靶向vWF的基因敲除,获得模拟人类血友病的猪模型。通过原核注射的方式在猪受精卵中共注射了Cas9mRNA和靶向vWF基因第5外显子的sgRNA。对出生的幼崽进行鉴定,检测发现vWF蛋白水平的大幅下降或不表达,血浆内几乎检测不到vWF,凝血因子8活性也显著降低,这种vWF突变的仔猪存在严重的出血倾向,且这种突变还可以遗传给下一代。Wang等[21]通过建立一套sgRNA评估体系在猪的基因组上靶向小眼畸形相关转录因子Mitf,高效地获得明显带有黑色素缺失表型的猪模型,成功模拟了耳聋-色素综合症疾病。以上研究表明CRISPR/Cas9技术可以应用在大动物疾病模型构建中,通过突变特定基因导致其功能性缺失,从而获得与人类疾病表型一致的动物模型。
2.2 其他哺乳动物的疾病模型在进行人类肿瘤疾病的相关研究中,利用CRISPR/Cas9技术能够在成体动物器官内直接导致功能性的缺失突变。Xue等[22]针对小鼠肝脏特异性地敲除了Pten基因以构建脂质堆积、晚发型肝癌的疾病模型。通过尾静脉高压注射的方式将靶向Pten基因的sgRNA和Cas9共表达的质粒注入野生型FVB小鼠中,使肝细胞瞬时表达CRISPR组件,从而获得小鼠肝脏上Pten基因的特异性敲除,并经基因型鉴定证实Pten基因只在肝脏中特异性突变,出现了肝癌的典型表型特征;研究组同样通过尾静脉高压注射的方式,共注射了表达靶向编码β链蛋白的Ctnnb1基因的sgRNA表达质粒以及带4个点突变(丝氨酸/苏氨酸)的ssODN模板序列,经HDR可以导致β链蛋白去磷酸化并定位在核内,引起肝细胞的功能性异常。证明CRISPR技术既可以在体内导致功能缺失性突变也能实现功能获得性突变。Maresch等[23]在成体小鼠胰腺的特定位点通过电转的方式,将13个靶向不同抑癌基因的sgRNA表达载体转入胰腺细胞内,实现多个抑癌基因的突变,进而获得胰腺导管腺癌(pancreatic ductal adenocarcinoma,PDAC)小鼠模型。同时还对这类转移性肿瘤病灶进行了来源追踪,CRISPR/Cas9能够引起染色体重排包括大片段缺失或易位,造成非整倍体细胞的出现进而形成肿瘤。
Yang等[24]利用CRISPR/Cas9技术在家兔的受精卵上靶向APOE、CD36、LDLR等基因的突变,以深入研究脂质代谢异常、动脉粥样硬化症以及心律失常等疾病。在人类临床上的杜氏肌萎缩症疾病(Duchenne muscular dystrophy, DMD)患者都存在肌萎缩蛋白基因突变,Chen等[25]在猕猴上就Dystrophin基因进行了打靶突变,突变效率达到87%,并成功获得模拟人类DMD疾病的猕猴模型。
2.3 基于动物模型的基因修复与应用利用CRISPR/Cas9技术经HDR途径纠正部分由定点突变导致的疾病,这对人类疾病临床治疗的相关研究具有深远意义。中国科学院李劲松团队Wu等[26]针对由crygc基因定点突变导致的白内障小鼠,在受精卵胞质共注射Cas9mRNA及sgRNA的同时还加入了同源的ssODN序列,以其为模板经HDR途径来修复这种点突变导致的眼部疾病。Zhou等[27]用Cas9技术在猪的受精卵上也通过HDR的方式,纠正先天性耳聋及色素分布异常疾病主要致病基因Sox10的错义突变,并进一步探讨了HDR与NHEJ运作间的竞争关系。Yin等[28]在模拟人遗传性酪氨酸血症的小鼠模型中纠正Fah基因的突变,通过尾静脉高压注射的方式将Cas9蛋白和sgRNA表达的载体以及野生型Fah序列的ssODN注入肝细胞中,并扩增这种Fah阳性的肝细胞来修复体重减轻的疾病表型。
在动物模型上研究某个特定基因的作用,为研究人类疾病开发新的治疗靶点及临床应用提供新途径。作为重要的大动物模型,小型猪也可以作为异种器官移植的器官来源,而组织相容性复合体(MHC)分子在传染病和移植免疫排斥反应中有特殊作用。Reyes等[29]利用CRISPR/Cas9技术并通过SCNT成功地构建了细胞表面缺乏MHC I类分子的猪模型。Yang等[30]也利用慢病毒载体与CRISPR/Cas9技术相结合的方式,在猪的肾脏上皮细胞系PK15上成功删除了包含猪内源性逆转录病毒(PERV)的62个拷贝,并将修饰后的PK15细胞与人源细胞共培养,在细胞上清中未检测到逆转录酶活性。表明修饰后的细胞只产生少量或几乎不产生病毒颗粒。这些研究都为异种器官移植提供了基础条件,对攻克异种移植研究的难关有重要意义。
3 细胞水平上的疾病建模与修复 3.1 构建模拟人类疾病的细胞模型研究人员利用CRISPR/Cas9技术在干细胞、成体细胞和肿瘤细胞等多种细胞系上进行基因操作模拟多种人类临床疾病。Drost等[31]通过CRISPR/Cas9基因编辑技术在人肠道干细胞(human intestinal stem cells,hISCs)中靶向常见的结直肠癌基因(APC、P53、KRAS、SMAD4)进行编辑,4个基因突变的hISCs具有肠道肿瘤的生物学特征,能够模拟人类结直肠癌的发生。Jiang等[32]诱导了特定位点染色体易位的小鼠胚胎干细胞,用来建立细胞或动物模型,以便后续研究染色体易位相关的先天性遗传疾病、不育和癌症等。
3.2 基于细胞模型的基因修复及应用CRISPR/Cas9技术高效精准的特性正是在细胞模型水平上实施基因治疗所必需的关键。许多研究团队利用CRISPR/Cas9技术经HDR途径在病人特异的干细胞上修复了单基因的遗传性疾病。Xie等[33]在β地中海贫血病人特异的诱导多能干细胞(induced pluripotent stem cells,iPSCs)上,运用Cas9技术与piggyBac转座子结合的方式,纠正了人β血红蛋白(HBB)基因的突变。Howden等[34]对视网膜色素变性病人的成纤维细胞在进行iPS诱导的同时,通过CRISPR/Cas9技术对PRPF8基因的点突变进行了纠正。Firth等[35]在体外培养的肠囊性纤维化(cysticfibrosis,CF)患者干细胞上利用Cas9技术进行基因操作,生理状态下cAMP浓度的增加会导致细胞器的迅速膨胀并打开囊性纤维化跨膜导体受体(CFTR),而从CF患者分离出来的细胞则是缺失这种反应的。通过插入正常CFTR基因序列经HDR来修复这种缺陷,纠正后的细胞有正确的基因表达且具备功能性。
Chang等[36]将重度联合免疫缺陷病(severe combined immunodeficiency,SCID)病人的体细胞重编程为iPS细胞,并在iPS细胞上进行JAK3基因第14外显子上C>T突变的纠正,经修复后重新诱导分化成造血干细胞(hematopoietic stem cell,HSC),HSC能够分化成有功能性的成熟T细胞,纠正了SCID患者的T细胞缺陷。Wu等[37]从Crygc基因纯合突变的白内障小鼠上建立了精原干细胞(spermatogonial stem cells,SSCs)系,并通过电转的方式将共表达Cas9蛋白和Crygc-sgRNA转入SSCs系中,鉴定有细胞经NHEJ修复了Crygc基因。随后研究组又尝试经HDR途径来修复该基因的突变,通过加入野生序列的ssODN建立Crygc基因修复的SSCs。修复后的SSCs分化成球形精子细胞,利用球形精子细胞注射的方法最终获得Crygc基因修复的后代。
Beil-Wagner等[38]通过CRISPR/Cas9技术靶向小鼠CD2进行条件性敲除,造成了T细胞的特异性失活。Schumann等[39]利用Cas9:sgRNA核糖核蛋白来精准改造人类T细胞,靶向人类免疫缺陷病毒1型(HIV-1)感染的协同受体CXCR4,能够降低CXCR4在细胞表面的表达,同时通过HDR来修复T细胞。目前对于慢性乙型肝炎患者的抗病毒疗法大多利用核苷类似物或干扰素,这类药物仅能控制已感染的肝细胞,在停止抗病毒治疗后病毒血症就会复发,这是由于HBV复制的原始模板超螺旋的共价闭合环状DNA分子(covalently closed circular DNA,cccDNA)未被消除,它就能够不断地实现HBV复制,持续感染肝细胞[40]。Lin等[41]在人的肝癌细胞系Huh7上共同转染HBV表达载体和特异的CRISPR/Cas9载体,并检测细胞核内和表面抗原的表达。为了证明HBV特异的sgRNAs也可以在体内帮助清除HBV模版cccDNA,该研究还在HBV小鼠模型中通过尾静脉高压注射HBV表达载体与CRISPR/Cas9表达载体,检测结果显示血清中抗原水平明显降低,肝内病毒表达水平也有所下降。以上研究都为治疗癌症、艾滋病、原发性免疫缺陷及自身免疫性疾病等提供了新途径,是CRISPR/Cas9基因编辑技术向人类生物学和医学应用延伸的里程碑。
4 展望近年来,CRISPR/Cas9技术广泛应用在生物学、基础医学等研究领域中,充分证明了该技术具有巨大的发展潜能。CRISPR技术不仅能用于人类疾病模型构建,还能够通过纠正致病性突变基因来修复疾病,利用该技术构建的大动物疾病模型能够推动发病机制的探索和新药研发等方面的研究进展。但是如何通过CRISPR技术获得具有与人类疾病表型一致的大动物模型,以及在疾病治疗中的安全性等问题还有待深入研究。CRISPR技术的创新和发展,也对于更好地推进人类临床疾病研究具有积极作用。Zetsche[42]等在CRISPR系统的2类效应蛋白中发现了Cpf1蛋白酶。较之产生平末端结构的Cas9核酸酶,Cpf1能够靶向地切割DNA并产生粘末端结构,这种粘末端序列能够保证DNA以特定的方向插入基因组。我国Gao[43]等将由单链DNA指导的NgAgo核酸酶应用到人类细胞的基因组编辑中,这种技术简化了基因编辑的步骤,使操作更加便捷。CRISPR技术精准性及高效性等方面的优化对于人类单基因遗传病和肿瘤的基因治疗具有重要意义,该技术的多方位发展将为基因治疗与精准医疗做出贡献。
致谢 本文感谢哈尔滨医科大学创新科学研究基金(2016JCZX36、2016JCZX38)的资助,以及东北农业大学生命科学院刘忠华教授的支持。| [1] | Coffey A, Ross R P. Bacteriophage-resistance systems in dairy starter strains:molecular analysis to application. Antonie Van Leeuwenhoek , 2002, 82 (1-4) : 303–321. |
| [2] | Doudna J A, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science , 2014, 346 (621) : 1258096. |
| [3] | Mojica F J. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology , 2009, 155 . |
| [4] | Jiang W. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol , 2013, 31 (3) : 233–239. DOI:10.1038/nbt.2508 |
| [5] | Mali P. RNA-guided human genome engineering via Cas9. Science , 2013, 339 (6121) : 823–826. DOI:10.1126/science.1232033 |
| [6] | Garneau J E. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature , 2010, 468 (7320) : 67–71. DOI:10.1038/nature09523 |
| [7] | Makarova K S. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol , 2011, 9 (6) : 467–477. DOI:10.1038/nrmicro2577 |
| [8] | Jinek M. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science , 2012, 337 (6096) : 816–821. DOI:10.1126/science.1225829 |
| [9] | Fu Y. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol , 2013, 31 (9) : 822–826. DOI:10.1038/nbt.2623 |
| [10] | Pattanayak V. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat Biotechnol , 2013, 31 (9) : 839–843. DOI:10.1038/nbt.2673 |
| [11] | Fu Y, Sander D. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol , 2014, 32 (3) : 279–284. DOI:10.1038/nbt.2808 |
| [12] | Ran F A, Hsu P D, Lin C Y, et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell , 2013, 154 (6) : 1380–1389. DOI:10.1016/j.cell.2013.08.021 |
| [13] | Yu C, Liu Y, Ma T, et al. Small molecules enhance CRISPR genome editing in pluripotent stem cells. Cell Stem Cell , 2015, 16 (2) : 142–147. DOI:10.1016/j.stem.2015.01.003 |
| [14] | Maruyama T. Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat Biotechnol , 2015, 33 (5) : 538–542. DOI:10.1038/nbt.3190 |
| [15] | Gilbert L A, Horlbeck M A, Adamson B, et al. Genome-scale CRISPR-mediated control of gene repression and activation. Cell , 2014, 159 (3) : 647–661. DOI:10.1016/j.cell.2014.09.029 |
| [16] | Sanjana N E, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods , 2014, 11 (8) : 783–784. DOI:10.1038/nmeth.3047 |
| [17] | Cong L, Ran F A, Cox D, et al. Multiplex genome engineering using CRISPR/Cas systems. Science , 2013, 339 (6121) : 819–823. DOI:10.1126/science.1231143 |
| [18] | Zhou X, Xin J, Fan N, et al. Generation of CRISPR/Cas9-mediated gene-targeted pigs via somatic cell nuclear transfer. Cell Mol Life Sci , 2015, 72 (6) : 1175–1184. DOI:10.1007/s00018-014-1744-7 |
| [19] | Wang X, Huang J, Cao C, et al. One-step generation of triple gene-targeted pigs using CRISPR/Cas9 system. Sci Rep , 2016, 6 : 20620. DOI:10.1038/srep20620 |
| [20] | Hai T. One-step generation of knockout pigs by zygote injection of CRISPR/Cas system. Cell Res , 2014, 24 (3) : 372–375. DOI:10.1038/cr.2014.11 |
| [21] | Wang X, Zhou J, Cao C, et al. Efficient CRISPR/Cas9-mediated biallelic gene disruption and site-specific knockin after rapid selection of highly active sgRNAs in pigs. Sci Rep , 2015, 5 : 13348. DOI:10.1038/srep13348 |
| [22] | Xue W, Chen S, Yin H, et al. CRISPR-mediated direct mutation of cancer genes in the mouse liver. Nature , 2014, 514 (7522) : 380–384. DOI:10.1038/nature13589 |
| [23] | Maresch R, Mueller S, Veltkamp C, et al. Multiplexed pancreatic genome engineering and cancer induction by transfection-based CRISPR/Cas9 delivery in mice. Nat Commun , 2016, 7 : 10770. DOI:10.1038/ncomms10770 |
| [24] | Yang D, Xu J, Zhu T, et al. Effective gene targeting in rabbits using RNA-guided Cas9 nucleases. J Mol Cell Biol , 2014, 6 (1) : 97–99. DOI:10.1093/jmcb/mjt047 |
| [25] | Chen Y, Zheng Y, Kang Y, et al. Functional disruption of the dystrophin gene in rhesus monkey using CRISPR/Cas9. Hum Mol Genet , 2015, 24 (13) : 3764–3774. |
| [26] | Wu Y, et al. Correction of a genetic disease in mouse via use of CRISPR-Cas9. Cell Stem Cell , 2013, 13 (6) : 659–662. DOI:10.1016/j.stem.2013.10.016 |
| [27] | Zhou X, Wang L, Du Y. Efficient generation of gene-modified pigs harboring precise orthologous human mutation via CRISPR/Cas9-induced homology-directed repair in zygotes. Hum Mutat , 2016, 37 (1) : 110–118. DOI:10.1002/humu.22913 |
| [28] | Yin H, Xue W, Chen S, et al. Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat Biotechnol , 2014, 32 (6) : 551–553. DOI:10.1038/nbt.2884 |
| [29] | Reyes L M, Estvada J L, Wang Z Y, et al. Creating class I MHC-null pigs using guide RNA and the Cas9 endonuclease. J Immunol , 2014, 193 (11) : 5751–5757. DOI:10.4049/jimmunol.1402059 |
| [30] | Yang L, Guell M, Niu D, et al. Genome-wide inactivation of porcine endogenous retroviruses (PERVs). Science , 2015, 350 (6264) : 1101–1104. DOI:10.1126/science.aad1191 |
| [31] | Drost J, Jaarsveld R H, Ponsioen B, et al. Sequential cancer mutations in cultured human intestinal stem cells. Nature , 2015, 521 (7550) : 43–47. DOI:10.1038/nature14415 |
| [32] | Jiang J, Zhang L, Zhou X, et al. Induction of site-specific chromosomal translocations in embryonic stem cells by CRISPR/Cas9. Sci Rep , 2016, 6 : 21918. DOI:10.1038/srep21918 |
| [33] | Xie F, Ye L, Chang J C, et al. Seamless gene correction of beta-thalassemia mutations in patient-specific iPSCs using CRISPR/Cas9 and piggyBac. Genome Res , 2014, 24 (9) : 1526–1533. DOI:10.1101/gr.173427.114 |
| [34] | Howden S E, Maufort J P, Duffin B M, et al. Simultaneous reprogramming and gene correction of patient fibroblasts. Stem Cell Reports , 2015, 5 (6) : 1109–1118. DOI:10.1016/j.stemcr.2015.10.009 |
| [35] | Firth A L, Menson T, Parker G S, et al. Functional gene correction for cystic fibrosis in lung epithelial cells generated from patient iPSCs. Cell Rep , 2015, 12 (9) : 1385–1390. DOI:10.1016/j.celrep.2015.07.062 |
| [36] | Chang C W, Lai Y S, Westin E, et al. Modeling human severe combined immunodeficiency and correction by CRISPR/Cas9-enhanced gene targeting. Cell Rep , 2015, 12 (10) : 1668–1677. DOI:10.1016/j.celrep.2015.08.013 |
| [37] | Wu Y, Zhou H, Fan X, et al. Correction of a genetic disease by CRISPR-Cas9-mediated gene editing in mouse spermatogonial stem cells. Cell Res , 2015, 25 (1) : 67–79. DOI:10.1038/cr.2014.160 |
| [38] | Beil-Wagner J, Dossinger G, Schober K, et al. T cell-specific inactivation of mouse CD2 by CRISPR/Cas9. Sci Rep , 2016, 6 : 21377. DOI:10.1038/srep21377 |
| [39] | Schumann K, Lin S, Boyer E, et al. Generation of knock-in primary human T cells using Cas9 ribonucleoproteins. Proc Natl Acad Sci U S A , 2015, 112 (33) : 10437–10442. DOI:10.1073/pnas.1512503112 |
| [40] | Seeger C, Sohn J A. Complete spectrum of CRISPR/Cas9-induced mutations on HBV cccDNA. Mol Ther , 2016, 24 (7) : 1258–1266. DOI:10.1038/mt.2016.94 |
| [41] | Lin S R, Yang H C, Kuo Y T, et al. The CRISPR/Cas9 system facilitates clearance of the intrahepatic HBV templates in vivo. Mol Ther Nucleic Acids , 2014, 3 : e186. DOI:10.1038/mtna.2014.38 |
| [42] | Zetsche B, Gootenberg J S, Abudayyeh O, et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell , 2015, 163 (3) : 759–771. DOI:10.1016/j.cell.2015.09.038 |
| [43] | Gao F, Shen X Z, Jiang F, et al. DNA-guided genome editing using the Natronobacterium gregoryi Argonaute. Nat Biotech, 2016, advance online publication. |
 2016, Vol. 36
2016, Vol. 36