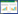文章信息
- 侯兵晓, 刘珊娜, 王斌斌, 朱宏吉, 乔建军.
- HOU Bing-xiao, LIU Shan-na, WANG Bin-bin, ZHU Hong-ji, QIAO Jian-jun.
- 热休克蛋白调控机制
- Advances on Regulatory Mechanism of Heat-shock Proteins
- 中国生物工程杂志, 2016, 36(9): 87-93
- China Biotechnology, 2016, 36(9): 87-93
- http://dx.doi.org/DOI:10.13523/j.cb.20160911
-
文章历史
- 收稿日期: 2016-03-22
- 修回日期: 2016-04-25
热休克蛋白(heat-shock protein)是生物体应对温度、pH、渗透压等不利环境刺激时合成的一种高度保守的保护蛋白。热休克反应的特点是快速诱导编码热休克蛋白基因的表达,这些基因编码的蛋白质可以防止蛋白质的聚集、协助蛋白质跨膜转运、参与寡聚结构的组装和降解等[1],降解不可逆损伤蛋白质[2]。调控因子可以在转录水平上对热休克基因正向调控和逆向调控,在环境应激时调控热休克基因的表达,恢复或加速清除细胞内已经变性的蛋白质,使细胞处于稳态并产生耐受性。
在革兰氏阳性菌中,根据它们的共同调控因子,编码热休克蛋白的基因分成6类[3]。如表 1所示,HrcA是第I类热休克调控因子,是一个抑制因子,可以调控下游基因(包括groEL和dnaK操纵子)的表达,进而抑制异常蛋白聚合和折叠[4]。σB是第II类热休克调控因子,在菌体受到化学、物理、生物等各种各样的胁迫条件时,σB可以调控200多个热休克基因的表达[5]。CtsR是第III类热休克调控因子[6],阻遏热休克基因的表达,主要包括clpB、clpC、clpE、clpX、clpP等clp蛋白家族基因[7],当菌体受到高温胁迫时,CtsR阻遏作用解除,进而调控这类基因的表达。第IV类基因是htpG(high-temperature protein G);第V类基因是由双组分系统CssRS(control secretion stress regulator and sensor)调控的基因[8];第VI类基因包括ftsH、clpX、lon等。这6类基因编码的热休克蛋白在抵御刺激和适应环境中发挥重要作用,可以诱导热休克蛋白基因的转录和表达,进一步调解生理反应和维持细胞稳态。本文综述了热休克转录调控系统的特征和调控功能,并初步概述了转录调控因子的相互作用,为深入构建转录调控网络奠定基础。其中HrcA、σB和CtsR可以响应多种胁迫压力,在转录水平上对热休克基因正向调控和逆向调控,对其进行了重点阐述。
| Class | Heat shock genes | Regulator | Mechanism |
| Class I | groEL operon and dnaK operon | CIRCE-HrcA | Negative control |
| Class II | gspA, csbA, katE, and more | σB | Positive control |
| Class III | clpB, clpC, clpE, clpL, clpP, and more | CtsR | Negative control |
| Class IV | htpG(class IV) | Unknown | Positive control |
| Class V | htrA and htrB(class V) | CssRS | Positive control |
| Class VI | ftsH, clpX, lon, and more (class VI) | Unknown | Negative control |
1 HrcA-CIRCE调控系统 1.1 hrcA基因及CIRCE反向重复序列
在120余种细菌中,hrcA基因可以编码一种与CIRCE(controlling inverted repeat of chaperone expression)操纵基因相互作用的转录抑制蛋白[9],从而发挥下游基因的调控作用,即调控第I类热休克基因,如调控groEL-ES和dnaK操纵子的表达。在细菌中HrcA蛋白的分子质量大小为39~40kDa,序列具有高度的保守性,C端是一个功能特殊的区域[10],N端具有两个DNA结合域。然而,在衣原体菌属编码的HrcA蛋白中,C端有附加序列,所以它的分子质量要比其它菌属的大10%。这段额外的C端区域有三个作用:
①降低HrcA与CIRCE操纵基因的结合能力;②抑制衣原体热休克基因的表达;③降低HrcA抑制转录的能力。研究表明,C端还可以被热休克蛋白GroEL调节[10]。
1989年,在分枝杆菌groEL-ES基因的上游第一次发现CIRCE反向重复序列[11]。1992年,在丙酮丁醇梭菌groEL-ES和dnaK操纵子及枯草芽孢杆菌groEL-ES操纵子的转录起始位点下游,发现了一个几乎相同的反向重复序列[12-13]。随后的研究发现,这个反向重复序列存在于多种细菌中,并且高度保守TTAGCACTC-N9-GAGTGCTAA,被命名为CIRCE[14-15]。同时,CIRCE反向重复序列在40余种真细菌中也确定存在,通常与HrcA转录调控基因相互作用,调控下游热休克操纵子groEL-ES和dnaK的表达。
1.2 HrcA-CIRCE热休克调控系统HrcA的活性取决于groEL-ES的构象变化[16]。HrcA-CIRCE热休克调控系统见图 1,在正常情况下,GroEL稳定的构象维持HrcA处于抑制活性状态,阻遏CIRCE调控的groEL-ES操纵子表达。应激状态时,伴侣蛋白groEL-ES修复折叠和变性蛋白质,进而构象改变,HrcA丧失抑制活性导致靶基因的诱导转录[17]。缺失hrcA基因的链霉菌菌株groES操纵子和groEL基因的转录量增加,对缺失菌株进行hrcA基因回补后发现,转录量减少。HrcA-CIRCE热休克调控系统广泛存在于真核细菌中,它通常调控groES-groEL和dnaK操纵子。但是,最近对李斯特氏菌研究发现,HrcA可以直接调控被groES-groEL操纵子趋异转录的lmo2070基因;间接激活氧化应激的基因(lmo0669、lmo2159和lmo0641)[18]、冷应激的基因cspD[19-20]、酸应激的基因gadC/gadB[21]和渗透压应激的基因cysK[22];在肺炎支原体中还调控热休克基因clpC的表达。因此,调控因子HrcA,在氧化、低温、高温、低pH、高渗透压等应激下都能调控相关基因表达,提高菌株耐受性[4, 23]。近年来,随着对HrcA-CIRCE调控系统的解析,HrcA与groEL-ES之间是如何作用的,响应应激信号的调控机制也逐渐被解析,为人们深入了解HrcA-CIRCE的调控机制,进一步改造菌株提供了非常有利的支持。

|
| 图 1 HrcA-CIRCE热休克调控机制 Figure 1 Heat-shock regulatory mechanism of HrcA-CIRCE (-)Represent negative regulation |
σB调控因子是sigB基因编码的产物,其活性主要通过一系列rsb基因编码产生的Rsb蛋白来调节[24]。在热休克调控系统中σB调控因子调控的基因最多,在枯草芽孢杆菌中调控200多个基因,这些基因编码不同功能的蛋白质,如氧化应激的保护、适应盐胁迫和水分胁迫、氧化还原平衡、调解细胞膜功能及调节代谢的蛋白质。金黄色葡萄球菌和枯草芽孢杆菌σB调控蛋白相似[5]。在炭疽杆菌中,σB因子是一个调控毒力的转录因子,并在稳定生长期和热休克之后被激活[25]。
在革兰氏阳性菌中σB的调控是通过一个所谓的伴侣切换机制实现的[26],如图 2所示。在无应激条件下,σB处于非激活状态被抗σ因子RsbW(an anti-sigma factor)封存。当面对应激时,一个抗σ因子拮抗剂RsbV和RsbW结合,这导致σB从被RsbW封存状态中释放,处于激活状态。随后,激活的σB可以与RNA核心酶结合,进而诱导依赖σB的基因转录。RsbW不仅是抗σ因子,也是RsbV磷酸化的一种激酶。RsbV磷酸化形式是无法结合RsbW并激活σB的,然而,在胁迫条件下,RsbV的磷酸化形式可由PP2C型的磷酸酶作用而脱磷酸,进而形成RsbVRsbW蛋白复合体和激活σB[24, 27]。在蜡样芽孢杆菌应激条件下,PP2C磷酸rsbY基因似乎是唯一负责σB活性的磷酸酶,rsbY基因的缺失导致σB调控的热休克系统摧毁[28]。
|
| 图 2 σB因子的热休克调控机制 Figure 2 Heat-shock regulatory mechanism of σB |
σB可以响应不同的应激条件,在枯草芽孢杆菌中有两种不同的途径导致σB激活。第一个途径是在环境应激条件下诱发σB(如乙醇应激和渗透压应激),第二个途径是降低细胞内ATP浓度激活σB[29]。对蜡样芽孢杆菌30℃升温至42℃后,蛋白质印迹法显示,σB的活性提高20.1倍;渗透压和乙醇应激后σB的活性同样也提高;暴露蜡样芽胞杆菌于羰基氰化物间氯苯腙(CCCP)降低细胞内ATP浓度,σB的活性只是有限的提高,表明对于蜡样芽胞杆菌降低细胞内ATP浓度,不是激活σB的重要因素[30]。P.McGann等[31]研究发现,单核细胞增生李斯特菌σB基因缺失后,表现出适应高渗透压的能力降低;单核细胞增生李斯特菌野毒株和缺失株分别在高温(T=42℃)、强酸(pH=3) 、强碱(pH=9) 条件下应激,发现缺失菌株耐受能力均弱于野生株;随后发现,σB基因的缺失使得单核细胞增生李斯特菌毒力明显下降[32]。因此,σB是对环境胁迫产生应答反应的主要调控因子。
3 CtsR调控系统 3.1 CtsRCtsR(class three stress gene repressor)是在枯草芽孢杆菌中最早被发现的高度保守的转录抑制子[33],随后也在大部分低GC含量的革兰氏阳性菌中被发现。CtsR通过识别并结合一个保守的重复的七核苷酸操纵子序列(5'-RGTCADN NAN RGTCADN-3')来抑制和调节第III类热休克基因的表达(clpP、clpE和clpC操纵子)[6, 34]。研究表明,CtsR主要调控ClpATP酶和ClpP蛋白酶基因的表达;然而,在葡萄球菌中CtsR也参与调控groEL-ES和dnaK操纵子的表达[35]。在植物乳杆菌中,CtsR调节clpC操纵子、ftsH的转录[13-14],ftsH基因编码一种膜结合的金属蛋白酶,是一种新颖的CtsR应激反应调节子[36]。在应激状态下,细胞中缺失CtsR基因将导致热敏感和细胞表面形态的改变,因此,CtsR在应激耐受性和参与蛋白质质量控制方面起关键作用[37]。
3.2 ClpATP酶ClpATP酶是在原核生物和真核生物中高度保守的的一种酶类,许多Clp蛋白(ClpA、ClpX和ClpC)具有ATP依赖性。在低GC含量的革兰氏阳性菌中CtsR调控Clp基因的表达。CtsR的DNA结合活性是温度依赖性的,调控clp表达的核心问题是CtsR如何监测温度。最近研究发现,ctsR操纵子中的翼状螺旋-转角-螺旋结构域具有四-甘氨酸环钩,可以将DNA钩住[38],利用定点诱变技术证实CtsR存在感温位点,该位点可以自身“感受”热量和反映周围环境的温度,造就了蛋白质热失活的易感性[39]。CtsR与DNA结合抑制其转录时,Clp复合体非常稳定;当外界刺激出现时,Clp复合体则会降解CtsR,稳定状况被破坏。金黄色葡萄球菌中的研究证实了上述研究结果,在正常生长条件下,CtsR抑制了clpP、clpB基因的转录,但热激不久后抑制作用解除[40-41]。在枯草芽孢杆菌中,应激后,CtsR从DNA上解离,会立即被ClpEP蛋白酶识别,被ClpCP降解[38]。众多研究证实,ClpCP介导的CtsR降解有助于解除CtsR调控子的阻遏作用[42-43]。
3.3 CtsR热休克调控系统CtsR热休克调控系统见图 3,在正常情况下,CtsR和DNA操纵子序列结合抑制其活性。McsB激酶与McsA相互作用并与ClpC结合,处于失活状态[44]。应激状态时,CtsR由于构象改变丧失与DNA结合的能力,进而与McsB激酶结合,ClpCP降解CtsR,CtsR丧失抑制活性导致靶基因的诱导转录[45]。McsB和McsA作为衔接蛋白直接参与第III类热休克反应的调节,被称为CtsR的调节剂。McsB[44]包含一个高度保守的ATP结构域胍磷酸转移激酶(guanidino phospho transferase),可以抑制CtsR与DNA结合能力,调节ClpCP降解CtsR。McsA作为McsB的抑制剂,两者一旦相互作用,就会防止McsB与失活的CtsR DNA结合[44]。研究表明,不同菌株CtsR在特殊应激反应中的失活机制完全取决于McsB[46]。然而,并不是所有的革兰氏阳性菌中都存在应激传感复合体McsA/ McsB,如在金黄色葡萄球菌中这类反应可能是由McsB和McsA的同系物参与的[47]。另外,在链球菌和乳酸菌中也没有发现特定的mcsB和mcsA基因,CtsR的活性由ClpE伴侣进行调节,表明它们的调控模式不同于生物模式菌株枯草芽孢杆菌[48]。调控因子CtsR,在高温、低pH、氧化应激、嘌呤霉素、羰基亲电子、不饱和长链游离脂肪酸酸和低温等环境应激后,阻遏作用解除,可以诱导相关耐受基因表达,提高菌株耐受性。
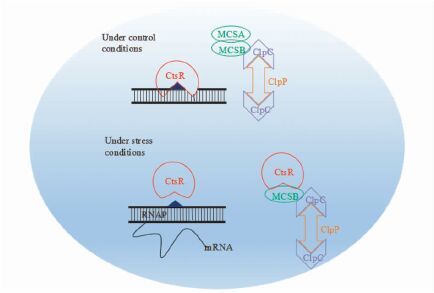
|
| 图 3 CtsR热休克调控机制 Figure 3 Heat-shock regulatory mechanism of CtsR |
第V类热休克调控系统CssRS双组分系统介导的信号诱导htrA和htrB基因的表达[49],在枯草芽孢杆菌中,htrA和htrB基因编码涉及质量控制的丝氨酸型表面蛋白酶,这些蛋白酶的编码严格依赖于CssRS。当外界应激时,细胞膜和细胞壁界面就会聚集错误折叠的蛋白质,CssRS是降解这些蛋白质的一个关键因素[50]。但是CssRS双组分调控系统的应激信号是如何传递的,如何识别并调控这些基因表达,不同菌株的调控模式是否相同,还有待于进一步研究。第IV类基因是htpG,是热休克90蛋白家族中的一员,参与多种细胞过程,包括蛋白质折叠修复和信号转导。第VI类基因ftsH是作用在膜上的蛋白酶,分解异常的膜蛋白和部分胞质蛋白沉淀。但是这两类具体的调控系统还不明确,还有待于进一步研究。
5 HrcA、σB和CtsR的相互调控关系为适应无穷变幻的环境,微生物演变出非常复杂的调控网络,快速的调解生理反应以恢复细胞稳态。HrcA和σB之间存在复杂的相互影响[18]:σB和HrcA都会自动调节各自的转录;σB也会直接调控一些HrcA依赖型的基因;σB和HrcA两者转录后都会被调节。例如,在枯草芽孢杆菌中,依赖HrcA的groEL-ES操纵子编码的GroE反过来调节HrcA活性[51]。总体来说,σB和HrcA依赖的调节机制在转录和转录后出现交互,在应激反应时,σB和HrcA相互作用共同调控相应基因的表达。σB和CtsR的调控子也存在相当大一部分重叠,在热应激中扮演很重要角色的clpC、clpP和clpB[52],以及在酸应激中扮演很重要角色的gadC和gadB,均被σB和CtsR共同调控。σB有助于CtsR的转录后调控,在一些应激条件下,σB通过mcsA上游的σB依赖型启动子调控mcsA-mcsB-clpC的转录[53]。mcsA-mcsB-clpC编码的蛋白质对CtsR的转录后调控起很重要的作用。HrcA、CtsR之间存在复杂的相互影响,HrcA和CtsR依赖的调节机制在转录和转录后也出现交互。在链球菌类(肺炎链球菌、化脓性链球菌、变形链球菌、无乳链球菌、乳酸乳球菌和唾液链球菌)中,groEL-ES操纵子呈现串联的CtsR靶向位点序列和CIRCE序列[54]。Chastanet等[55]的研究表明,HrcA和CtsR协同调控dnaK和groEL-ES操纵子的低水平表达,而HrcA调节子被完全嵌入在CtsR调节子内。CssRS双组分系统与其他热休克调控系统之间的联系研究较少,有待于进一步研究。
6 结论和展望面对环境变化所带来的挑战及胁迫,微生物已经进化出错综复杂的调控网络。热休克调控网络是从总体上深入了解热休克调控不可或缺的一部分。李斯特菌HrcA、σB和PrfA构成的调控网络,有助于毒力基因和应激基因的转录表达,提高菌株耐受不同应激压力[55]。谷氨酸棒状杆菌HspR、HrcA和Sigma因子构成的调控网络,应激情况下,157个热休克基因被正向调控和逆向调控[56]。热休克调控系统的分类现已成熟,但是这些调控系统之间的内在机制联系有待于进一步研究。例如,htpG和ftsH受哪些系统调控;CssRS双组分系统与HrcA-CIRCE等系统之间如何联系,不同应激情况对于各调控系统内在联系有何影响,这些问题解读对构建热休克调控网络具有一定的启示作用。另外,随着系统生物学及代谢工程的发展,在微生物中越来越多的调控因子已经被发现,除了本文所述的调节子外,SOS反应调节子LexA、氧化还原传感器Rex、过氧化物传感器PerR、精氨酸生物合成和代谢调节剂ArgR、组氨酸合成稳压器HisR、半胱氨酸代谢抑制子CymR和嘌呤代谢抑制子PurR等也已被发现[57]。如果将不同种调控系统集成一个完整的应激调控网络,针对不同的应激条件激活调控因子,将有望进一步提高菌株对胁迫环境的适应能力。
| [1] | Hartl F U, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature , 2011, 475 (7356) : 324–332. DOI:10.1038/nature10317 |
| [2] | Sghaier H, Ai T L H, Horiike T, et al. Molecular chaperones:proposal of a systematic computer-oriented nomenclature and construction of a centralized database. In Silico Biology , 2004, 4 (3) : 311–322. |
| [3] | Sugimoto S, Sonomoto K. Molecular chaperones in lactic acid bacteria:physiological consequences and biochemical properties. Journal of bioscience and bioengineering , 2008, 106 (4) : 324–336. DOI:10.1263/jbb.106.324 |
| [4] | Selby K, Lindström M, Somervuo P, et al. Important role of class I heat shock genes hrcA and dnaK in the heat shock response and the response to pH and NaCl stress of group I Clostridium botulinum strain ATCC 3502. Applied and Environmental Microbiology , 2011, 77 (9) : 2823–2830. DOI:10.1128/AEM.02633-10 |
| [5] | Pané-Farré J, Jonas B, Förstner K, et al. The σB regulon in Staphylococcus aureus and its regulation. International Journal of Medical Microbiology , 2006, 296 (4) : 237–258. |
| [6] | Derré I, Rapoport G, Msadek T. CtsR, a novel regulator of stress and heat shock response, controls clp and molecular chaperone gene expression in Gram-positive bacteria. Molecular Microbiology , 1999, 31 (1) : 117–131. DOI:10.1046/j.1365-2958.1999.01152.x |
| [7] | Varmanen P, Ingmer H, Vogensen F K. ctsR of Lactococcus lactis encodes a negative regulator of clp gene expression. Microbiology , 2000, 146 (6) : 1447–1455. DOI:10.1099/00221287-146-6-1447 |
| [8] | Noone D, Botella E, Butler C, et al. Signal perception by the secretion stress-responsive CssRS two-component system in Bacillus subtilis. Journal of Bacteriology , 2012, 194 (7) : 1800–1814. DOI:10.1128/JB.05767-11 |
| [9] | Wilson A C, Tan M. Stress response gene regulation in Chlamydia is dependent on HrcA-CIRCE interactions. Journal of Bacteriology , 2004, 186 (11) : 3384–3391. DOI:10.1128/JB.186.11.3384-3391.2004 |
| [10] | Chen A L, Wilson A C, Tan M. A Chlamydia-specific C-terminal region of the stress response regulator HrcA modulates its repressor activity. Journal of Bacteriology , 2011, 193 (23) : 6733–6741. DOI:10.1128/JB.05792-11 |
| [11] | Baird P N, Hall L M C, Coates A R M. Cloning and sequence analysis of the 10kDa antigen gene of Mycobacterium tuberculosis. Microbiology , 1989, 135 (4) : 931–939. DOI:10.1099/00221287-135-4-931 |
| [12] | Narberhaus F, Bahl H. Cloning, sequencing, and molecular analysis of the groESL operon of Clostridium acetobutylicum. Journal of Bacteriology , 1992, 174 (10) : 3282–3289. |
| [13] | Wetzstein M, Völker U, Dedio J, et al. Cloning, sequencing, and molecular analysis of the dnaK locus from Bacillus subtilis. Journal of Bacteriology , 1992, 174 (10) : 3300–3310. |
| [14] | Segal G, Ron E Z. Regulation and organization of the groE and dnaK operons in Eubacteria. FEMS Microbiology Letters , 1996, 138 (1) : 1–10. DOI:10.1111/fml.1996.138.issue-1 |
| [15] | Zuber U, Schumann W. CIRCE, a novel heat shock element involved in regulation of heat shock operon dnaK of Bacillus subtilis. Journal of Bacteriology , 1994, 176 (5) : 1359–1363. |
| [16] | Narberhaus F. Negative regulation of bacterial heat shock genes. Molecular Microbiology , 1999, 31 (1) : 1–8. DOI:10.1046/j.1365-2958.1999.01166.x |
| [17] | Inoue M, Mitarai N, Trusina A. Circuit architecture explains functional similarity of bacterial heat shock responses. Physical biology , 2012, 9 (6) : 066003. DOI:10.1088/1478-3975/9/6/066003 |
| [18] | Hu Y, Oliver H F, Raengpradub S, et al. Transcriptomic and phenotypic analyses suggest a network between the transcriptional regulators HrcA and σB in Listeria monocytogenes. Applied and Environmental Microbiology , 2007, 73 (24) : 7981–7991. DOI:10.1128/AEM.01281-07 |
| [19] | Bayles D O, Annous B A, Wilkinson B J. Cold stress proteins induced in Listeria monocytogenes in response to temperature downshock and growth at low temperatures. Applied and Environmental Microbiology , 1996, 62 (3) : 1116–1119. |
| [20] | Uppal S, Shetty D M, Jawali N. Cyclic AMP receptor protein regulates cspD, a bacterial toxin gene, in Escherichia coli. Journal of Bacteriology , 2014, 196 (8) : 1569–1577. DOI:10.1128/JB.01476-13 |
| [21] | Cotter P D, Gahan C G M, Hill C. A glutamate decarboxylase system protects Listeria monocytogenes in gastric fluid. Molecular Microbiology , 2001, 40 (2) : 465–475. DOI:10.1046/j.1365-2958.2001.02398.x |
| [22] | Duché O, Trémoulet F, Glaser P, et al. Salt stress proteins induced in Listeria monocytogenes. Applied and Environmental Microbiology , 2002, 68 (4) : 1491–1498. DOI:10.1128/AEM.68.4.1491-1498.2002 |
| [23] | Van Bokhorst-van de Veen H, Bongers R S, Wels M, et al. Transcriptome signatures of class I and Ⅲ stress response deregulation in Lactobacillus plantarum reveal pleiotropic adaptation. Microbial Cell Factories , 2013, 12 (1) : 1. DOI:10.1186/1475-2859-12-1 |
| [24] | van Schaik W, Abee T. The role of σ B in the stress response of Gram-positive bacteria-targets for food preservation and safety. Current Opinion in Biotechnology , 2005, 16 (2) : 218–224. DOI:10.1016/j.copbio.2005.01.008 |
| [25] | Fouet A, Namy O, Lambert G. Characterization of the operon encoding the alternative σB factor from Bacillus anthracis and its role in virulence. Journal of Bacteriology , 2000, 182 (18) : 5036–5045. DOI:10.1128/JB.182.18.5036-5045.2000 |
| [26] | Utratna M, Cosgrave E, Baustian C, et al. Effects of growth phase and temperature on activity within a Listeria monocytogenes population:evidence for rsbV-independent activation of at refrigeration temperatures. BioMed Research International , 2013, 2013 (17) : 641647. |
| [27] | Kazmierczak M J, Wiedmann M, Boor K J. Alternative sigma factors and their roles in bacterial virulence. Microbiology and Molecular Biology Reviews , 2005, 69 (4) : 527–543. DOI:10.1128/MMBR.69.4.527-543.2005 |
| [28] | van Schaik W, Tempelaars M H, Zwietering M H, et al. Analysis of the role of RsbV, RsbW, and RsbY in regulating σB activity in Bacillus cereus. Journal of Bacteriology , 2005, 187 (16) : 5846–5851. DOI:10.1128/JB.187.16.5846-5851.2005 |
| [29] | Sonenshein A L, Hoch J A, Losick R. Bacillus subtilis and its closest relatives:from genes to cells. Nature , 2002, 415 (6869) : 263–264. |
| [30] | van Schaik W, Tempelaars M H, Wouters J A, et al. The alternative sigma factor σB of Bacillus cereus:response to stress and role in heat adaptation. Journal of bacteriology , 2004, 186 (2) : 316–325. DOI:10.1128/JB.186.2.316-325.2004 |
| [31] | McGann P, Wiedmann M, Boor K J. The alternative sigma factor σB and the virulence gene regulator PrfA both regulate transcription of Listeria monocytogenes internalins. Applied and Environmental Microbiology , 2007, 73 (9) : 2919–2930. DOI:10.1128/AEM.02664-06 |
| [32] | Yang L H, Meng Q L, Qiao J. Effect of SigmaB gene deletion on virulence of Listeria monocytogene. Shihezi University, College of Animal Science a nd Technology , 2013 . |
| [33] | Krüger E, Hecker M. The first gene of the Bacillus subtilis clpC operon, ctsR, encodes a negative regulator of its own operon and other class Ⅲ heat shock genes. Journal of Bacteriology , 1998, 180 (24) : 6681–6688. |
| [34] | Derré I, Rapoport G, Msadek T. The CtsR regulator of stress response is active as a dimer and specifically degraded in vivo at 37℃. Molecular Microbiology , 2000, 38 (2) : 335–347. DOI:10.1046/j.1365-2958.2000.02124.x |
| [35] | Chastanet A, Fert J, Msadek T. Comparative genomics reveal novel heat shock regulatory mechanisms in Staphylococcus aureus and other Gram-positive bacteria. Molecular Microbiology , 2003, 47 (4) : 1061–1073. DOI:10.1046/j.1365-2958.2003.03355.x |
| [36] | Fiocco D, Collins M, Muscariello L, et al. The Lactobacillus plantarum ftsH gene is a novel member of the CtsR stress response regulon. Journal of Bacteriology , 2009, 191 (5) : 1688–1694. DOI:10.1128/JB.01551-08 |
| [37] | Russo P, De La Luz Mohedano M, Capozzi V, et al. Comparative proteomic analysis of Lactobacillus plantarum WCFS1 and ΔctsR mutant strains under physiological and heat stress conditions. International Journal of Molecular Sciences , 2012, 13 (9) : 10680–10696. |
| [38] | Fuhrmann J, Schmidt A, Spiess S, et al. McsB is a protein arginine kinase that phosphorylates and inhibits the heat-shock regulator CtsR. Science , 2009, 324 (5932) : 1323–1327. DOI:10.1126/science.1170088 |
| [39] | Elsholz A K W, Gerth U, Hecker M. Regulation of CtsR activity in low GC, Gram+ bacteria. Advances in Microbial Physiology , 2010, 57 : 119–144. DOI:10.1016/B978-0-12-381045-8.00003-5 |
| [40] | Fleury B, Kelley W L, Lew D, et al. Transcriptomic and metabolic responses of Staphylococcus aureus exposed to supra-physiological temperatures. BMC Microbiology , 2009, 9 (1) : 1. DOI:10.1186/1471-2180-9-1 |
| [41] | Frees D, Chastanet A, Qazi S, et al. Clp ATPases are required for stress tolerance, intracellular replication and biofilm formation in Staphylococcus aureus. Molecular Microbiology , 2004, 54 (5) : 1445–1462. DOI:10.1111/j.1365-2958.2004.04368.x |
| [42] | Miethke M, Hecker M, Gerth U. Involvement of Bacillus subtilis ClpE in CtsR degradation and protein quality control. Journal of Bacteriology , 2006, 188 (13) : 4610–4619. DOI:10.1128/JB.00287-06 |
| [43] | Krüger E, Zühlke D, Witt E, et al. Clp-mediated proteolysis in Gram-positive bacteria is autoregulated by the stability of a repressor. The EMBO Journal , 2001, 20 (4) : 852–863. DOI:10.1093/emboj/20.4.852 |
| [44] | Kirstein J, Zühlke D, Gerth U, et al. A tyrosine kinase and its activator control the activity of the CtsR heat shock repressor in B. subtilis. The EMBO Journal , 2005, 24 (19) : 3435–3445. DOI:10.1038/sj.emboj.7600780 |
| [45] | Elsholz A K W, Michalik S, Zühlke D, et al. CtsR, the Gram-positive master regulator of protein quality control, feels the heat. The EMBO Journal , 2010, 29 (21) : 3621–3629. DOI:10.1038/emboj.2010.228 |
| [46] | Elsholz A K W, Hempel K, Pöther D C, et al. CtsR inactivation during thiol-specific stress in low GC, Gram+ bacteria. Molecular Microbiology , 2011, 79 (3) : 772–785. DOI:10.1111/j.1365-2958.2010.07489.x |
| [47] | Wozniak D J, Tiwari K B, Soufan R, et al. The mcsB gene of the clpC operon is required for stress tolerance and virulence in Staphylococcus aureus. Microbiology , 2012, 158 (10) : 2568–2576. |
| [48] | Elsholz A K W, Michalik S, Zühlke D, et al. CtsR, the Gram-positive master regulator of protein quality control, feels the heat. The EMBO Journal , 2010, 29 (21) : 3621–3629. DOI:10.1038/emboj.2010.228 |
| [49] | Hyyryläinen H L, Bolhuis A, Darmon E, et al. A novel two-component regulatory system in Bacillus subtilis for the survival of severe secretion stress. Molecular Microbiology , 2001, 41 (5) : 1159–1172. |
| [50] | Hyyryläinen H L, Pietiäinen M, Lunden T, et al. The density of negative charge in the cell wall influences two-component signal transduction in Bacillus subtilis. Microbiology , 2007, 153 (7) : 2126–2136. DOI:10.1099/mic.0.2007/008680-0 |
| [51] | Chaturongakul S, Boor K J. RsbT and RsbV contribute to σB-dependent survival under environmental, energy, and intracellular stress conditions in Listeria monocytogenes. Applied and Environmental Microbiology , 2004, 70 (9) : 5349–5356. DOI:10.1128/AEM.70.9.5349-5356.2004 |
| [52] | Gaillot O, Pellegrini E, Bregenholt S, et al. The ClpP serine protease is essential for the intracellular parasitism and virulence of Listeria monocytogenes. Molecular Microbiology , 2000, 35 (6) : 1286–1294. |
| [53] | Chaturongakul S, Boor K J. σB activation under environmental and energy stress conditions in Listeria monocytogenes. Applied and Environmental Microbiology , 2006, 72 (8) : 5197–5203. DOI:10.1128/AEM.03058-05 |
| [54] | Chastanet A, Prudhomme M, Claverys J P, et al. Regulation of Streptococcus pneumoniae clp genes and their role in competence development and stress survival. Journal of Bacteriology , 2001, 183 (24) : 7295–7307. DOI:10.1128/JB.183.24.7295-7307.2001 |
| [55] | Hu Y, Raengpradub S, Schwab U, et al. Phenotypic and transcriptomic analyses demonstrate interactions between the transcriptional regulators CtsR and Sigma B in Listeria monocytogenes. Applied and Environmental Microbiology , 2007, 73 (24) : 7967–7980. DOI:10.1128/AEM.01085-07 |
| [56] | Ehira S, Teramoto H, Inui M, et al. Regulation of Corynebacterium glutamicum heat shock response by the extracytoplasmic-function sigma factor SigH and transcriptional regulators HspR and HrcA. Journal of Bacteriology , 2009, 191 (9) : 2964–2972. DOI:10.1128/JB.00112-09 |
| [57] | Wang Q, Venkataramanan K P, Huang H, et al. Transcription factors and genetic circuits orchestrating the complex, multilayered response of Clostridium acetobutylicum to butanol and butyrate stress. BMC Systems Biology , 2013, 7 (1) : 1. DOI:10.1186/1752-0509-7-1 |
 2016, Vol. 36
2016, Vol. 36