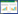文章信息
- 堵晶晶, 谭镇东, 刘辰东, 巫小倩, 张培文, 张顺华, 朱砺.
- DU Jing-jing, TAN Zhen-dong, LIU Chen-dong, WU Xiao-qiao, ZHANG Pei-wen, ZHANG Shun-hua, ZHU Li.
- 长链非编码RNA的研究现状
- Research Progress of Long Non-coding RNAs
- 中国生物工程杂志, 2016, 36(9): 59-74
- China Biotechnology, 2016, 36(9): 59-74
- http://dx.doi.org/DOI:10.13523/j.cb.20160908
-
文章历史
- 收稿日期: 2016-02-01
蛋白质是基因功能的执行者,RNA在蛋白质合成过程中扮演着重要作用。RNA可分为两类,一类为具有蛋白质编码功能的编码RNA(coding RNA),另一类为不具有蛋白质编码功能的非编码RNA(non-coding RNA,ncRNA)。与蛋白质编码RNA相比,ncRNA在很长一段时间里都被当做转录的副产物。直到1990年,RNA干扰(RNAi)的首次发现才引起研究者对ncRNA功能的关注[1]。随着深度测序技术的迅猛发展,人们惊奇地发现,在哺乳动物中编码蛋白质的基因仅约占全基因组的2% ,全基因组的70% 最终转录为无编码蛋白质功能的非编码RNA,如转运RNA(tRNA)、核糖体RNA(rRNA)、small interfering RNA(siRNA)、microRNA(miRNA)、小核RNA(snRNA)和长链非编码RNA(long non-coding RNA,lncRNA)等[2-6]。截至目前,如miRNA、siRNA等ncRNA的生物学特性、功能及其分子作用机制已被基本阐释清楚[7-16]。
lncRNA与蛋白质编码mRNA在结构上存在许多相似之处[17]。尽管许多研究表明lncRNA广泛参与了大量生物学调控过程,如X染色体失活、染色质修饰、转录激活、基因组印迹、核内运输及转录干扰等,但由于技术手段和工具的限制,lncRNA的功能研究一直进展缓慢,人们对其基本特征、分类、功能等方面的认识也较为滞后。为此,本文主要从lncRNA的基本特征、分类、功能、相关的数据库、研究方法及其与癌症之间的关系等进行综述,全面展现lncRNA的研究进展。
1 lncRNA的特征长度大于200bp(200bp~100kbp)、无蛋白质编码功能是lncRNA最基本的特征[18-20]。与编码RNA相比,广泛存在于哺乳动物中的lncRNA拥有更少外显子,缺少开放阅读框(open reading frame,ORF)及典型的起始密码子和终止子。研究者推论这可能是lncRNA不能编码蛋白质的原因[21-26]。lncRNA也有许多类似于mRNA的特征,如绝大多数lncRNA拥有5′帽子结构和polyA尾巴,由RNA转录酶Ⅱ转录,并经多聚腺苷酸化、pre-RNA拼接在内的共转录修饰[27-28]。
lncRNA在细胞质、细胞核、细胞器均有分布,但主要存在细胞核内[20]。哺乳动物全基因组的4% ~9% 最终转录成lncRNA[23, 29]。lncRNA是一类物种内序列进化保守、表达量易受外界环境影响的RNA分子,物种间的序列保守性较差,丰度也低于mRNA[30-31]。lncRNA的组织特异性远强于蛋白质编码RNA,不仅不同组织间的表达量不同,甚至同一组织不同部位的lncRNA都存在不同的表达模式[30, 32-33]。此外,lncRNA具有很强的时空特异性,同一lncRNA的表达量在同一组织或器官的不同发育阶段均有较大差异[34-37]。
2 lncRNA的分类lncRNA二级、三级结构复杂,功能多样,但截至目前,仍缺乏有效的分类方法。目前,根据在基因组中与邻近蛋白质编码基因的位置关系,研究者将lncRNA分为(图 1):①反义lncRNA(antisense lncRNA),由邻近的编码基因反方向转录,与其他蛋白质编码基因存在重叠区域(如共享外显子);②基因内lncRNA(intronic lncRNA),起始于蛋白质编码基因的内含子,但与其无重叠区域,由相对邻近的蛋白质编码基因从任意方向转录;③异质性lncRNA(divergent lncRNA),由邻近的蛋白质编码基因的启动子反方向转录,转录启动时常伴随相邻蛋白质编码基因转录起始位点数百bp的转录;④基因间lncRNA(intergenic lncRNA),由蛋白质编码基因及那些不共享启动子、外显子或内含子的蛋白质编码基因共转录而来;⑤增强子式lncRNA(enhancer lncRNA),由增强子转录,可长距离或短距离介导调控转录lncRNA自身的增强子与基因组内其它转录因子间的相互作用[9]。
|
| 图 1 lncRN分类(基于基因组上的位置) Figure 1 Classification of lncRNA (genomic location) (Marko Koll,et al. Nature Review. 2015,Doi:10.1038/nrendo.2014.229) |
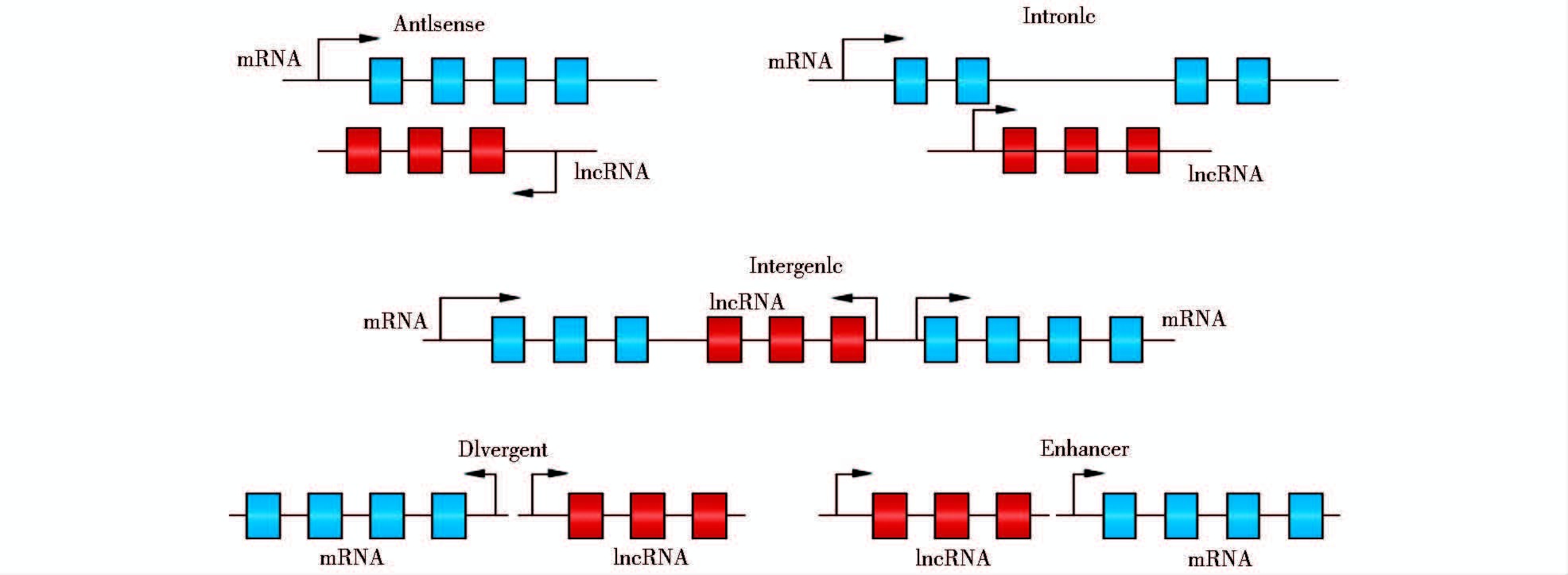
lncRNA具有不同的亚细胞定位[38],部分研究者根据其亚细胞定位差异,将存在于细胞核内的定义为核lncRNA,存在于细胞质内的定义为质lncRNA(图 2)。Derrien等评估了细胞核、细胞质内lncRNA的相对丰度,其中17% 富集在细胞核内,4% 富集在细胞质内[31]。lncRNA的功能与其所处位置息息相关。前者主要通过染色质修饰、转录激活、转录干扰等方式来行使生物学功能[28-43];后者主要充当“诱饵分子”与蛋白质、DNA和RNA相互作用,如H19序列中包含数个let-7的结合位点,可吸附let-7形成复合体,抑制let-7所调控的生物学功能[44-47]。近年来在培养细胞中还观察到核lncRNA被短发夹RNA(shRNA)损耗的现象,故在细胞核与细胞质的间隙可能同样存在lncRNA [31, 38]。
|
| 图 2 lncRNA分类(基于亚细胞定位) Figure 2 Classification of lncRNA (subcellular localization) (Marko Koll,et al. Nature Review. 2015,Doi:10.1038/nrendo.2014.229) |
lncRNA的二级、三级结构复杂,亚细胞定位多样,在RNA加工[48-49]、基因转录调控[50-52]、染色质修饰[53-54]、细胞凋亡[55-56]、端粒维护[57]、蛋白质与蛋白质或DNA与蛋白质交互作用的调控[58-60]等生物学过程中扮演重要角色。但与miRNA相对单一的作用机制不同,lncRNA的功能较复杂,目前主要分为5类。
3.1 转录调节其它基因的表达lncRNA可作为转录因子在转录水平直接或间接地与靶基因相互作用,调控靶基因的转录与表达。例如,位于SER3上游的lncRNA-SRG1,其3′序列能与SER3的对应序列互补形成RNA-DNA复杂结构,lncRNA-SRG1的积极转录或过表达均会抑制SER3的转录[61]。lncRNA-DEANR1可通过激活FOXA2促进人类内胚层分化[62]。部分lncRNA与其他基因存在重叠区域,当重叠区域位于启动子区时,对应基因的转录会因lncRNA竞争性地占据转录起始因子结合位点而受到抑制。此外,lncRNA本身或其产物可直接与转录因子结合,改变转录因子的结构,降低转录因子活性,甚至无法与目标基因结合,从而抑制目标基因的表达。近年来还有研究发现lncRNA能与核糖核蛋白形成复合体来调控基因的表达。例如,由Dlx5和Dlx6之间的序列转录而来的lncRNA-Efv2可与转录因子Dlx2形成复合体,激活Dlx5、Dlx6的增强子,最终促使Dlx5和Dlx6的转录上调。
3.2 调节组蛋白修饰与染色质重塑表观遗传是指DNA序列不变,但基因的表达却发生可遗传的变化,主要涉及DNA甲基化、组蛋白修饰和染色体构象的改变。lncRNA可通过招募染色质或组蛋白修饰相关蛋白形成作用复合体,之后到达目标位点,并促使该位点上的基因发生表观遗传修饰,调控下游基因表达(图 3)。PRC2复合体、CoREST/REST及SCMX蛋白是重要的染色质重塑相关蛋白,全基因组分析表明,大约1/3基因间lncRNA都涉及PRC2复合体、CoREST/REST或SCMX蛋白。最典型的例子是lncRNA-RepA可招募PRC2到Xist基因启动子,引发Xist启动子的组蛋白H3第27位赖氨酸发生甲基化(H3K27me3),最终导致X染色体失活。或lncRNA-Air通过招募组蛋白甲基化转移酶G9a到Igf2启动子区域,使组蛋白H3K9发生甲基化,最终沉默父源染色体提供的相关印迹基因的表达。而lncRNA-HOTAIR则是作为一种反式调控,通过招募PRC2形成复合体,远距离介导HOXD发生H3K27的三甲基化(H3K27me3),抑制相关基因的表达。lncRNA-Firre 可通过绑定CTCF对细胞核周围不活跃的X染色体定位,维持H3K27me3处的甲基化水平[63]。lncRNA-ADINR由C/EBPα基因上游450bp位置转录而来,在脂肪生成过程中可特异性地结合PA1,招募MLL3/4组蛋白甲基转移酶,增加C/EBPα位点的H3K4me3,最终减少H3K27me3组蛋白修饰。它的损耗可导致严重的脂肪形成缺陷[64]。
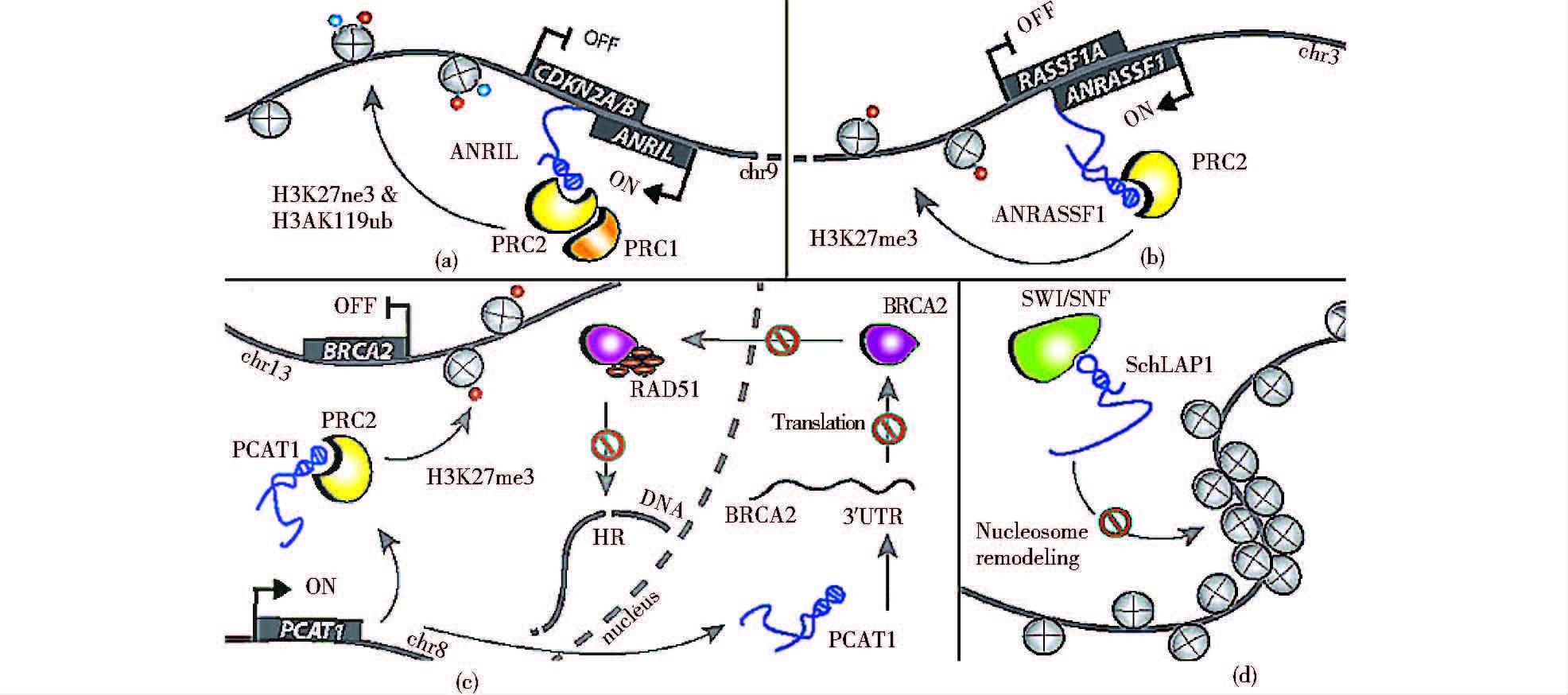
|
| 图 3 lncRNAs介导染色质重塑 Figure 3 IncRNAs mediate chromatin remodeling [Melanie Weiss,et al. Biol Chem. 2014,395(11):1275-1290]There are lots of lncRNA,which are associated with epigenetic silencing,such as ANRIL (a),ANRASSF1 (b),PCAT-1 (c),SChLAP1 (d) |
mRNA由miRNA的前前体(pre-miRNA)及前体(pri-miRNA)剪接而来,许多蛋白质已被证明可直接与Drosha和Dicer互作或绑定miRNA前体去调控miRNA的前体(pri-miRNA)或前前体(pre-miRNA)的生成[65-66]。而lncRNA可通过调节RNA聚合酶Ⅱ的活性来调节RNA的生成[67]。RNA聚合酶Ⅱ是通用转录因子,由SINEs转录而来的lncRNA-Alu和lncRNA-B2可直接抑制RNA聚合酶Ⅱ的活性来改变转录起始复合体的构象[68-69]。lncRNA-FGFR2通过招募多硫蛋白和脱甲基酶KDM2a建立特异性剪接的染色质信号,调控可变剪接[39]。而在RNA结合蛋白HEXIM1的参与下,lncRNA-7SK可与P-TEFb形成复合体,降低P-TEFb活性及磷酸化[70]。此外,源于假基因的转录本也可引起部分具有蛋白质编码功能的mRNA形成小RNA分子,如同样源于假基因的反义lncRNA能和与之相应的、被剪接的mRNA杂交,并经Dicer酶裂解形成内源性siRNAs[71-72]。因此,假基因并非仅扮演非功能性角色,当作为lncRNA转录时,也可作为调控基因表达的关键因子。
3.4 作为竞争性内源RNA(ceRNA)2011年提出的ceRNA假说认为,当circuRNA、mRNA等具有相同的miRNA应答元件(miRNA response element,MRE)时,它们会竞争性地结合MRE而调控相关基因的表达水平[73]。lncRNA也是一种竞争性内源RNA,能与其它RNA转录体竞争相同的miRNA,进而在转录后水平上间接调控miRNA参与的生物学过程[74-75](表 1)。但与其他ceRNA不同的是,lncRNA与miRNA的作用机制主要有两种。最普遍的是lncRNA直接与miRNA种子序列互补配对,吸附miRNA形成复合体,降低细胞内参与下游基因调控的miRNA数量[图 4(a)]。例如,lncRNA-APF与miR-188-3p的种子序列互补,间接调控miR-188-3p的靶标ATG7的表达量,参与细胞自噬和心肌梗死的发生[76];肌肉特异性lncRNA-MD1可吸附miR-133、miR-135,进而调节肌肉特异基因的转录激活因子MAML1、MEF2C,影响肌肉分化[77]。而当lncRNA序列里包含miRNA的种子序列时(lncRNA序列内含有可与miRNA竞争结合靶mRNA的3′UTR),lncRNA也可作为ceRNA发挥功能[图 4(b)],如lncRNA-Sirt1序列包含miR-34a的种子序列(TGGCAGT),可竞争性地结合miR-34a的靶标[78]。
| LncRNA | miRNA | 细胞类型 | 功能 | 年份 | 参考文献 |
| CCAT1 | Let-7 | HCC cell | 促进肝癌的恶化 | 2015 | [79] |
| MEG3 | miR-29 | HCC cell | 促进肝癌的恶化 | 2011 | [80] |
| MDRL | miR-484 | 心肌细胞 | 抑制线粒体分裂 | 2014 | [46] |
| CARL | miR-539 | 心肌细胞 | 促进线粒体分裂,心肌梗死 | 2014 | [81] |
| UCA1 | miR-16 | UMUC2 and 5637 cells | 促进GLS2代谢,抑制肺癌细胞内的ROS | 2015 | [45] |
| UCA1 | miR-216 | HCC cell | 促进肝癌恶化 | 2015 | [47] |
| HOTAIR | miR-331 | MGC-803,SGC-7901,BGC-823,and AGS cells | 促进胃癌恶化 | 2014 | [34] |
| MALAT1 | miR-22 | HUVECs cell | 抑制内皮细胞功能紊乱 | 2015 | [82] |
| MALAT1 | miR-217 | HBE cell | 对抗上皮细胞功能紊乱 | 2015 | [83] |
| MALAT1 | miR-205 | HK-2和A-498 cells | 促进肾癌 | 2015 | [84] |
| MALAT1 | miR-101,217 | ESCC cell | 促进食管癌恶化 | 2015 | [85] |
| MIAT | miR-150 | HUVECs和HMVECs cells | 对抗心血管疾病 | 2015 | [86] |
| HOTTIP | miR-125b | HCC cell | 促进肝癌恶化 | 2015 | [87] |
| NEAT1 | miR-140 | 3T3-L1 cell | 促进脂质形成 | 2015 | [88] |
| CASC2 | miR-21 | Human U251,U87 glioma cells,HEK-293 cell | 对抗神经胶质瘤 | 2015 | [89] |
| TUG1 | miR-144 | GEC cell | 调节血脑屏障 | 2015 | [90] |
| CHRF | miR-489 | 心肌细胞 | 调节心脏肥大 | 2014 | [91] |
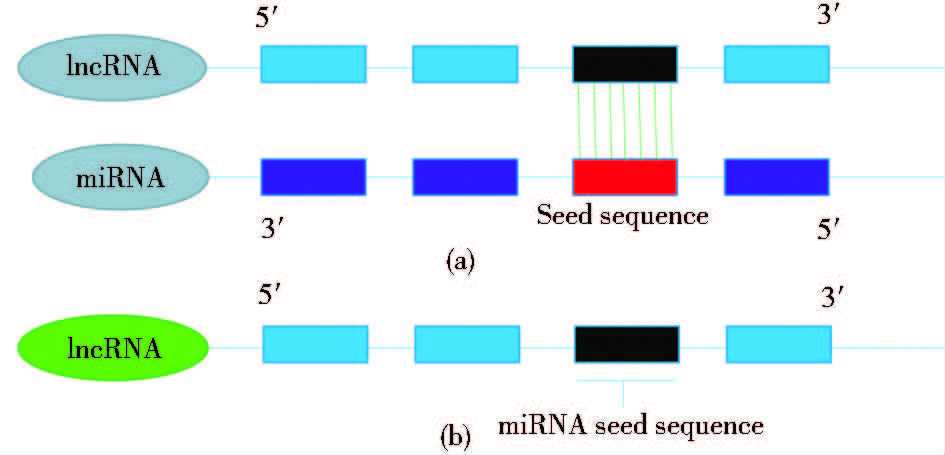
|
| 图 4 lncRNA与miRNA的作用机制 Figure 4 Mechanism of lncRNA mediating miRNA (a) lncRNA as miRNAs sponges (b) The seed sequence of miRNAs exist in the lncRNA sequence |
通常情况下,miRNA由长转录本经Drosha酶和Dicer酶连续不断剪接而来[92-94],Piwi-interacting RNAs(piRNAs)由长的单转录本生成[95],但仍有许多小RNA的功能与作用机制并不特别明晰。随着高通量测序技术的发展,发现部分lncRNA内包含某些小RNA,如Wilusz等从lnc-MALAT1中鉴定出一段长度约为61bp、序列高度保守的小RNA分子[96-97]。部分功能验证性实验已证明lncRNA可以作为某些长度小于200bp的小RNA(如miRNA、piRNAs)的前体分子,参与转录后调控[98-99],如lncRNA-Xist和lncRNA-Tsix可能是通过这种机制调控哺乳动物X染色体失活的[100]。
4 lncRNA的相关数据库随着大量lncRNA的发现,收集、整理、分类lncRNA并构建lncRNA相关信息的数据库成为lncRNA研究的重要任务。近年来,研究人员结合生物信息学技术建立了多个与lncRNA 相关的基础数据库(表 2),同时推出了以lncRNA功能为侧重点的生物信息数据库,如lncRNASNP、StarBase(表 2)。这些数据库的建立不仅提供各物种的lncRNA 的综合信息,还为挖掘lncRNA功能、研究lncRNA与其它分子之间的调控关系提供了一个非常重要的信息参考平台。
| 数据库名称 | 网址 | 相关功能 |
| LNCcipedia | http://www.Lnccipedia.org | 人类lncRNA序列和结构全面注释 |
| Noncode | http://www.Noncode.org/ NONCODEv3 | ncRNA全面注释,可预测lncRNA 功能 |
| ChIPBase | http://deepbase.sysu.edu.cn/chipbase/ | 提供lncRNA的表达图谱及专利调控的全面鉴定、注释。同时整合提供RNA-seq鉴定的lncRNA与CHIP-seq鉴定的转录因子的结合位点 |
| Noncoding RNA database | http://biobases.ibch.poznan.pl/ncRNA/ | 具有生物学功能的ncRNA注释 |
| NRED | http://jsm-research.imb.uq.edu.au/NRED | 提供人和小鼠的lncRNA在芯片数据上的表达信息 |
| ncFANs | http://www.ebiomed.org/ncFANs/ | lncRNA功能注释 |
| DIANA-LncBase | http://www.microrna,gr/lncBase | 整理提供lncRNA上靶miRNA及功能注释等综合信息 |
| lncRNAdb | http://www.lncrnadb.org/ | 提供有生物学功能的lncRNA的全面注释 |
| fRNAdb | http://www.ncrna.org/ | 收集H-inv、Noncode和RNAdb三个数据库已注释或未注释的ncRNA |
| Gencode | http://www.gencodegenes.org/ | 提供人和小鼠已注释lncRNA的比较 |
| NPInter | http://www.bioinfo.org/NPInter/ | 提供已被实验验证的ncRNA与DNA、蛋白质、RNA互作的信息 |
| lncRNA Diease | http://210.73.221 .6 /lncrnadisease | 提供文献已报道的与疾病相关的lncRNA的注释 |
| RNA22 | https://cm.jefferson.edu/rna22/ | lncRNA Sponge miRNA的预测 |
| RNAhybrid | http://bibiserv.techfak.uni-bielefeld.de/ | lncRNA靶标的鉴定 |
| LncRNASNP | http://bioinfo.life.hust.edu.cn/lncRNASNP/ | 提供lncRNA SNP位点及功能预测 |
| StarBase | http://starbase.sysu.edu.cn/mirCircRNA.php | lncRNA调控网络的预测 |
5 lncRNA的研究方法
目前,lncRNA的研究主要集中在lncRNA数据挖掘和lncRNA功能验证两方面。lncRNA芯片和lncRNA-seq是当前lncRNA数据挖掘的重要手段。lncRNA芯片基于已发现的lncRNA数据定制,研究者可通过与数万个芯片位点的结合分析来获知目标样本中的lncRNA表达谱,鉴定与研究目标相关的lncRNA,准确度较高[101-103]。而与传统的lncRNA芯片检测技术相比,基于去除核糖体RNA(rRNA)的建库方法及高通量测序技术的lncRNA-seq不仅可以检测识别基因家族中相似基因或由可变剪切造成的不同转录本的单碱基差异,还可检测到低丰度的稀有转录本,发掘新的lncRNA[30, 104-105]。因此,lncRNA-seq 更适用于大规模测序、发掘新的lncRNA。
lncRNA功能验证应该包括细胞水平验证与动物活体水平验证。但截至目前,尚未有在动物活体水平上对lncRNA进行功能验证的研究报道。在细胞水平上,可运用的研究工具较多,但主要分为以下几类。第一,通过qRT-PCR对目标lncRNA在样本中的变化进行定量分析或对lncRNA-seq测序数据进行验证[30, 40, 106-107],但lncRNA的结构复杂、存在诸多重叠区域,在对lncRNA引物设计时需要综合考虑众多因素。第二,lncRNA的作用机制与其位置密切相关,因此lncRNA核质定位信息可为lncRNA功能预测提供有力参考。
目前,研究者既可通过原位杂交技术、lncRNA-FISH对lncRNA定位分析,也可基于核质RNA分离抽提技术,通过qRT-PCR对细胞核与细胞质内目标lncRNA定量来对lncRNA进行定位分析[35, 101]。第三,一直以来,细胞内目的片段的缺失或获得是研究目的片段功能最为直接的手段,lncRNA也不例外。目前,研究者可通过siRNA、反义寡核苷酸链(ASO)等工具抑制细胞内目标lncRNA的表达,或构建lncRNA载体,对lncRNA进行功能缺失或获得性研究[76, 101, 108-109]。但lncRNA较长,作用机制复杂,其载体的构建存在一定难度,目前仅有少数研究者运用了这种研究方法[81]。一般情况下,功能验证性lncRNA的过表达载体应为全长,而在验证lncRNA与miRNA的靶标关系时,构建与miRNA种子序列互补区域的上下游200~500bp即可。第四,可以通过RNA pull down、CHIRP-seq、RNA-IP等方法来确定能与目的lncRNA互作的蛋白质、DNA、RNA[26, 104, 110-113]。第五,身处大数据时代,生物信息技术已成为分析目标数据的必备手段,lncRNA也不例外。通过生物信息技术与lncRNA-seq(或芯片)的结合分析,研究者可筛选出差异表达的lncRNA、构建共表达网络,并对lncRNA的功能进行预测,为后续的功能性细胞实验提供强有力的科学依据(图 5)[28, 40]。
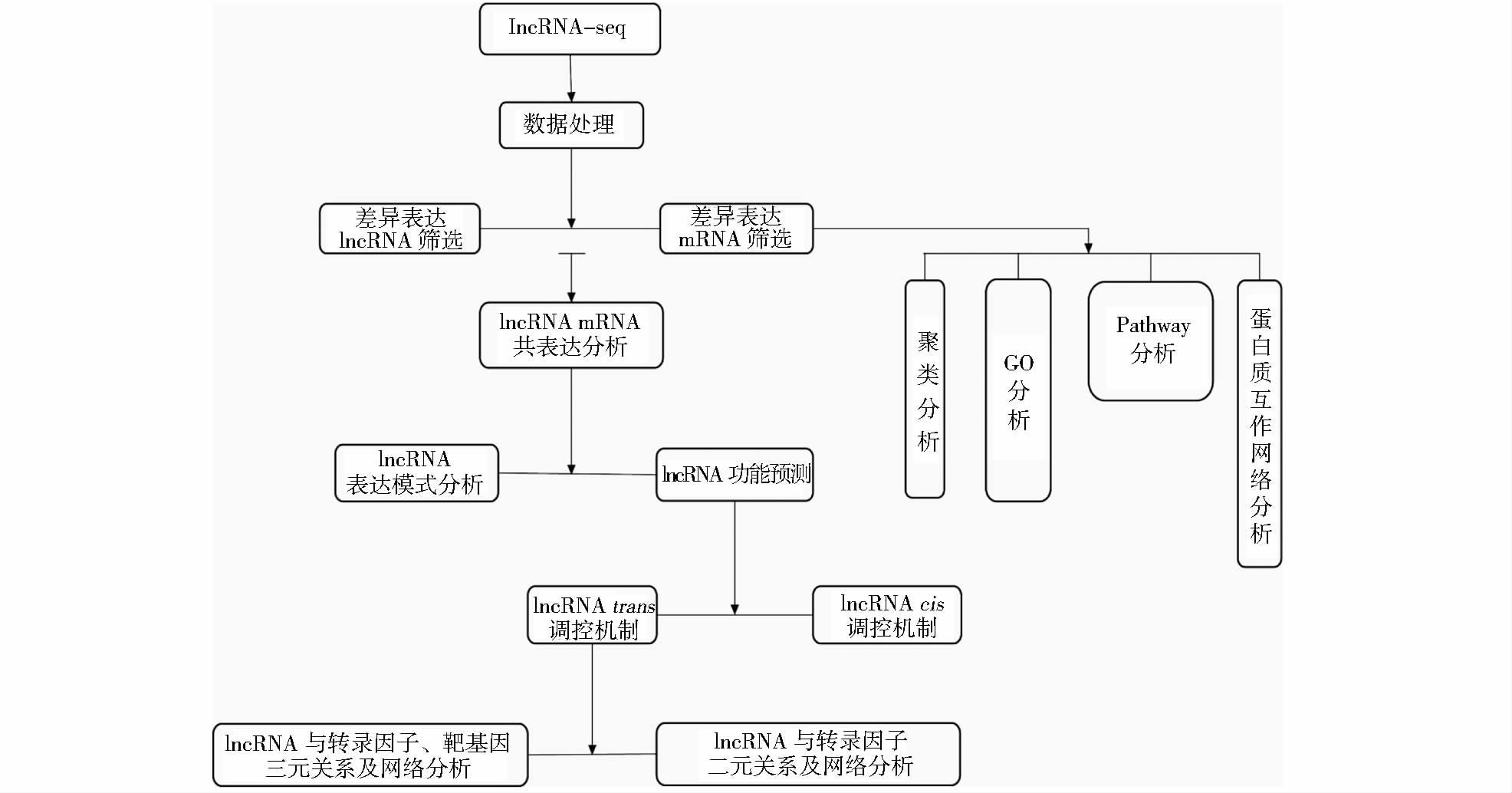
|
| 图 5 lncRNA的数据分析及研究思路 Figure 5 Data analysis and research ideas of lncRNA |
癌症的发生受多种因素调控,如遗传。癌症的发生常伴随着大量异常基因的出现,但目前的证据倾向于癌症并非由蛋白质编码序列的改变而引发。最近,越来越多的lncRNA被发现可作为致癌或抑癌因子(表 3),许多致癌的基因组位点也主要是由那些在癌症诱发过程中扮演着重要角色的lncRNA转录而来。随着高通量技术的迅猛发展,大量与肿瘤发生相关的lncRNA被鉴定出。例如,Nicole等鉴定了111种与肺癌相关的基因间lncRNA[114],Matthew等利用高通量测序确定了58 000个涉及正常组织和常见类型癌症的lncRNA[115]。
| 类型 | lncRNA | 相关功能 | 年份 | 参考文献 |
| 前列腺癌 | SChLAP1 | 与前列腺癌发生呈正相关 | 2015,2014 | [123-125] |
| PCAT29 | 抑制前列腺癌细胞的增殖、迁移 | 2014 | [126] | |
| NEAT1 | 促前列腺恶化 | 2014 | [127] | |
| PCAT-1 | 促前列腺癌细胞增殖,破坏DNA修复 | 2015,2014 | [128-129] | |
| H19 | 抑制前列腺癌细胞转移 | 2014 | [130] | |
| PCA3 | 2015 | [131] | ||
| HOTAIR | 调节雄激素,促进癌细胞增殖、迁移 | 2015 | [132-133] | |
| PCG | 调控一个涉及前列腺癌发生的网络 | 2015 | [134] | |
| P21 | 有助于区分前列腺癌和良性疾病 | 2015 | [135] | |
| Linc00963 | 抑制前列腺癌细胞迁移 | 2014 | [136] | |
| 乳腺癌 | HIT | 抑制TGF-β诱导的癌细胞迁移、侵袭 | 2015 | [137] |
| M41 | 提高癌细胞的侵袭能力及其三苯氧胺耐药性 | 2015 | [138] | |
| BC200 | 抑制乳腺癌细胞凋亡 | 2015 | [56] | |
| Smad7 | 抑制乳腺癌细胞凋亡 | 2014 | [120] | |
| BCAR4 | 调节乳腺癌细胞内信号转导 | 2015 | [139] | |
| SPRY-1T1 | 促进乳腺癌细胞增殖 | 2015 | [140] | |
| AK023948 | 促进乳腺癌的发生 | 2015 | [141] | |
| HOTAIR | 增强ER信号,促进乳腺癌细胞增殖 | 2015 | [142] | |
| Hh | 促进乳腺癌的发生 | 2015 | [143] | |
| ATB | 促进乳腺癌的发生 | 2015 | [144] | |
| 膀胱癌 | ZEB2NAT | 促进膀胱癌细胞侵袭 | 2015 | [145] |
| UCA1 | 增强膀胱癌细胞耐药性 | 2014 | [146] | |
| HOTAIR | 调节介导膀胱癌恶化的exosome | 2015 | [147] | |
| H19 | 促进膀胱癌细胞的增殖、迁移 | 2015 | [148] | |
| TUG1 | 促进膀胱癌中上皮细胞向间充质细胞转化 | 2015 | [149] | |
| 直肠癌 | ATB | 涉及直肠癌的预后 | 2015 | [150] |
| MALAT1 | 促进直肠癌的恶化 | 2014 | [151] | |
| CCAL | 促进直肠癌的恶化 | 2015 | [152] | |
| RP11-462C24 | 调节直肠癌细胞的转移 | 2014 | [153] | |
| GAS5 | 抑制直肠癌细胞增殖 | 2014 | [154] | |
| 肺癌 | AK126698 | 增强肺癌细胞对顺氯氨铂的耐药性 | 2013 | [155] |
| SCAL1 | 调节肺癌细胞的恶化 | 2013 | [156] | |
| GAS5 | 调节肺癌细胞的增殖、凋亡 | 2013 | [157] | |
| CCAT2 | 促进肺癌细胞的侵袭 | 2014 | [158] | |
| 甲状腺癌 | PTCSC3 | 抑制甲状腺癌恶化 | 2013 | [159] |
| 胃癌 | H19 | 促进胃癌细胞的增殖 | 2012,2014 | [160-161] |
| AC096655 | 调节胃癌细胞的转移 | 2013 | [162] | |
| MEG3 | 促进胃癌细胞增殖、胃癌恶化 | 2014 | [163] | |
| HULC | 促进胃癌细胞的增殖、迁移,抑制胃癌细胞的凋亡 | 2014 | [164] | |
| 胰腺癌 | HOTAIR | 促进胰腺癌细胞的增殖 | 2013 | [165] |
| H19 | 促进胰腺癌细胞的迁移 | 2014 | [166] | |
| 肝癌 | MEG3 | 促进肝癌细胞增殖 | 2011 | [80] |
| TCF7 | 促进肝癌细胞自我更新 | 2015 | [167] | |
| HOTTIP | 促进肝癌恶化 | 2015 | [168] | |
| UCA1 | 促进肝癌恶化 | 2015 | [47] |
在常见肿瘤里,lncRNA的功能并不单一,它们促进或抑制肿瘤的发生。例如,参与了转录调控和转录后调控的lncRNA-MALAT1在许多肿瘤组织或细胞内都高表达,与癌细胞的转移、复发相关[116-117];又如,Lai等[118]通过对9种肝癌细胞系、112个肝癌病例分析发现,MALAT1可以作为一个独立的肝癌复发的预测因子,它的抑制可显著降低HepG2的生存率、迁移能力。Liu等[119]发现,MALAT1于胰腺癌组织高表达,且其表达水平随着肿瘤的不断增大、恶化而逐渐升高。Arase等[120]通过对小鼠乳腺上皮细胞测序鉴定出lncRNA-Smad7,并发现其可以靶向调控TGF-β,增强乳腺癌细胞抗凋亡作用。lncRNA-H19位于染色体11p15.5,可与EZH2结合,抑制E-cad的表达,提高膀胱癌细胞的新陈代谢能力与迁移能力,其被认为是膀胱癌早期复发的一个标志物[121]。
lncRNA在癌症抑制过程也扮演着重要角色。COX-2介导的炎症会在癌症的早期阶段促进肿瘤的生长和扩散,但在肿瘤晚期COX-2却会将扩散中的肿瘤细胞暴露给免疫系统。因此,COX-2的活性在癌症晚期往往会被抑制。2014年Krawcczyk等[122]在Elife上发文表示,一种被命名为PACER的lncRNA能抑制COX-2基因的启动抑制因子p50,从而增强COX-2在晚期癌症细胞内的活性以便协助免疫系统对抗癌症。p53被认为与癌症之间存在不容质疑的相关性,有研究者通过全基因组测序和CHIP-seq分析鉴定出18种可受p53直接调控的、在癌细胞里低表达的lncRNA,p53可通过调节这些lncRNA建立一个监管循环反馈系统,促进p53肿瘤抑制因子的活化[28, 40]。如图 6所示,有研究者将lncRNA涉及癌症发生的普遍机制进行了归纳。
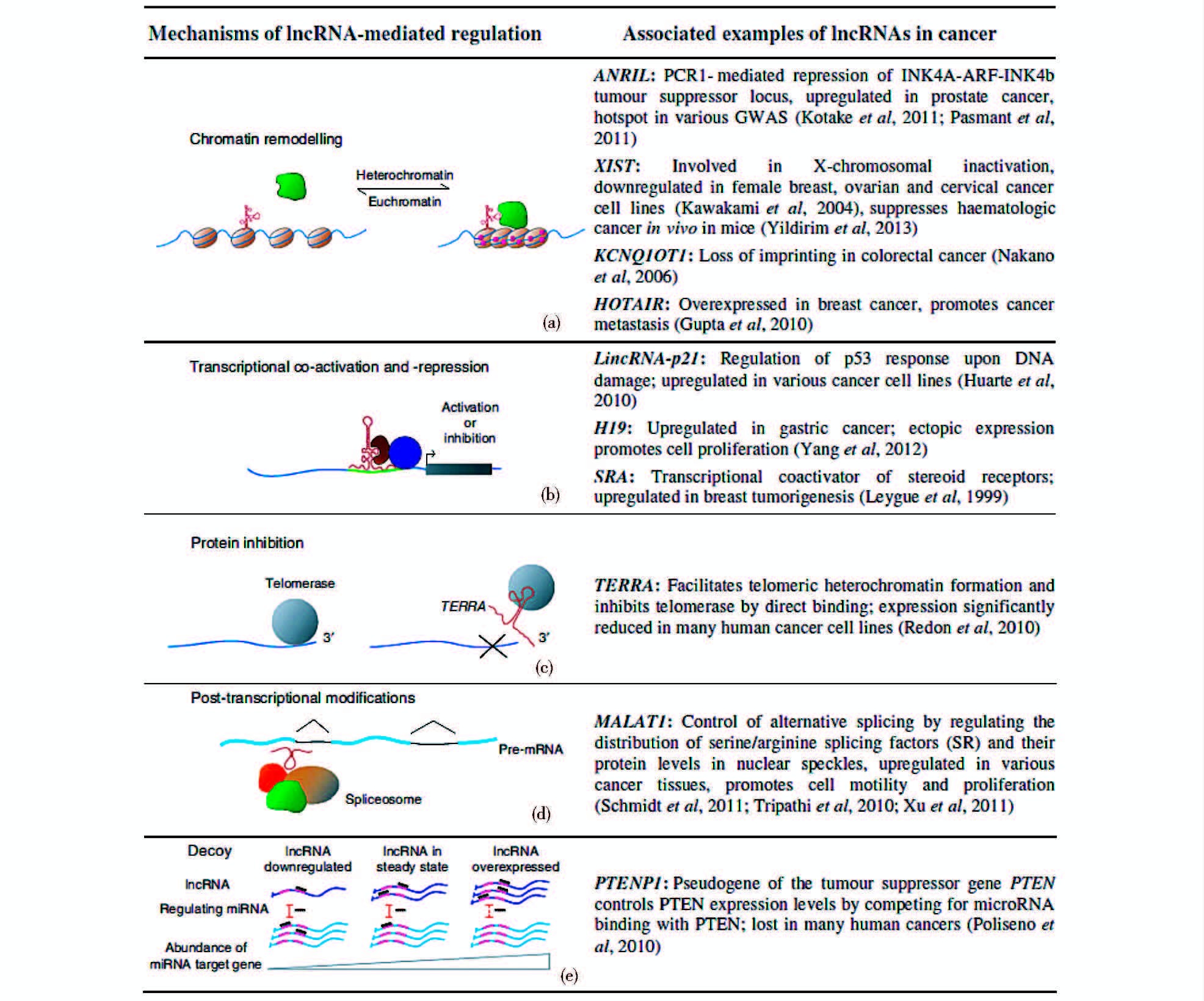
|
| 图 6 lncRNA涉及癌症发生的普遍机制及其相关实例 Figure 6 Universal mechanisms and examples that lncRNAs mediate cancer There are various mechanisms of lncRNAs,such as charomatin remodeling(a),transcriptional co-activator and -repression(b),protein inhibition(c),post-transcriptional modifications(d),decoy(e) (Cheetham S W,et al. British Journal of Cancer,2013,108:2419-2425) |
无蛋白质编码功能的lncRNA已成为时下核酸研究的热点领域。从目前的研究证据来看,lncRNA结构复杂,作用机制多样,参与了表观遗传调控、转录调控、转录后调控,牵涉细胞的增殖、分化、凋亡等生命环节,在遗传信息逐级流动过程中扮演着不可或缺的监管角色。越来越多的证据表明lncRNA与疾病的发生息息相关,如参与癌症的诱发或抑制,未来可被作为癌症的潜在生物学靶标或开发作为分子类药物运用于疾病的诊断与治疗。此外,基础研究是推动科学文明发展的基石,lncRNA的基础研究亟须更多的研究团队加入,更多、更快地研究、阐释lncRNA在生命体发育过程扮演的角色及其作用机制。
尽管如此,由于研究手段或技术的欠缺,成本高昂等原因,lncRNA的研究还处于起步阶段,特别是在利用高通量测序数据挖掘涉及生物学调控的lncRNA和lncRNA的功能性研究方面。想全面了解lncRNA介导生物学调控的分子作用机制仍有很长一段路要走。而身处大数据时代,我们急需构建高质量的lncRNA数据库,对lncRNA进行细致、精确的数据收集、整理、分类,而且需要对已知信息进行更精准的关联分析、预测,推进lncRNA的功能性研究;亟须开发并完善lncRNA的研究工具和手段。只有切实可行、成本低廉的实验方法、工具及高质量可靠的数据库之间的协同作用才能最终铺平对lncRNA的研究道路。
| [1] | Hannon G J. RNA interference. Nature , 2002, 418 (6894) : 244–251. DOI:10.1038/418244a |
| [2] | Huang B, Zhang R. Regulatory non-coding RNAs:revolutionizing the RNA world. Molecular Biology Reports , 2014, 41 (6) : 3915–3923. DOI:10.1007/s11033-014-3259-6 |
| [3] | Dong P, Yu F, Fan X, et al. Inhibition of ATIR by shRNA prevents collagen synthesis in hepatic stellate cells. Molecular and Cellular Biochemistry , 2010, 344 (1-2) : 195–202. DOI:10.1007/s11010-010-0542-2 |
| [4] | Gehrau R, Mas V, Villamil F, et al. MicroRNA signature at the time of clinical HCV recurrence associates with aggressive fibrosis progression post-liver transplantation. American Journal of Transplantation , 2013, 13 (3) : 729–737. DOI:10.1111/ajt.2013.13.issue-3 |
| [5] | Ohtani M, Takebayashi A, Hiroyama R, et al. Cell dedifferentiation and organogenesis in vitro require more snRNA than does seedling development in Arabidopsis thaliana. Journal of Plant Research , 2015, 128 (3) : 371–380. DOI:10.1007/s10265-015-0704-0 |
| [6] | Hutten S, Chachami G, Winter U, et al. A role for the Cajal-body-associated SUMO isopeptidase USPL1 in snRNA transcription mediated by RNA polymerase Ⅱ. Journal of Cell Science , 2014, 127 (5) : 1065–1078. DOI:10.1242/jcs.141788 |
| [7] | Matzke M A, Mosher R A. RNA-directed DNA methylation:an epigenetic pathway of increasing complexity. Nature Reviews Genetics , 2014, 15 (6) : 394–408. DOI:10.1038/nrg3683 |
| [8] | Porrua O, Libri D. Transcription termination and the control of the transcriptome:why, where and how to stop. Nature Reviews Molecular Cell Biology , 2015, 16 (3) : 190–202. DOI:10.1038/nrm3943 |
| [9] | Lam M T, Li W, Rosenfeld M G, et al. Enhancer RNAs and regulated transcriptional programs. Trends in Biochemical Sciences , 2014, 39 (4) : 170–182. DOI:10.1016/j.tibs.2014.02.007 |
| [10] | Vance K W, Ponting C P. Transcriptional regulatory functions of nuclear long noncoding RNAs. Trends in Genetics , 2014, 30 (8) : 348–355. DOI:10.1016/j.tig.2014.06.001 |
| [11] | Natoli G, Andrau J C. Noncoding transcription at enhancers:general principles and functional models. Annual Review of Genetics , 2012, 46 (7) : 1–19. |
| [12] | Lee J T. Epigenetic regulation by long noncoding RNAs. Science , 2012, 338 (6113) : 1435–1439. DOI:10.1126/science.1231776 |
| [13] | Guttman M, Rinn J L. Modular regulatory principles of large non-coding RNAs. Nature , 2012, 482 (7385) : 339–346. DOI:10.1038/nature10887 |
| [14] | Wang K C, Chang H Y. Molecular mechanisms of long noncoding RNAs. Molecular Cell , 2011, 43 (6) : 904–914. DOI:10.1016/j.molcel.2011.08.018 |
| [15] | Rinn J L, Chang H Y. Genome regulation by long noncoding RNAs. Annual Review of Biochemistry , 2012, 81 (7) : 145–166. |
| [16] | Ulitsky I, Bartel D P. lincRNAs:genomics, evolution, and mechanisms. Cell , 2013, 154 (1) : 26–46. DOI:10.1016/j.cell.2013.06.020 |
| [17] | Guttman M, Amit I, Garber M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature , 2009, 458 (7235) : 223–227. DOI:10.1038/nature07672 |
| [18] | Qiu M T, Hu J W, Yin R, et al. Long noncoding RNA:an emerging paradigm of cancer research. Tumor Biology , 2013, 34 (2) : 613–620. DOI:10.1007/s13277-013-0658-6 |
| [19] | Birney E, Stamatoyannopoulos J A, Dutta A, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature , 2007, 447 (7146) : 799–816. DOI:10.1038/nature05874 |
| [20] | Djebali S, Davis C A, Merkel A, et al. Landscape of transcription in human cells. Nature , 2012, 489 (7414) : 101–108. DOI:10.1038/nature11233 |
| [21] | Danko C G, Hah N, Luo X, et al. Signaling pathways differentially affect RNA polymerase Ⅱ initiation, pausing, and elongation rate in cells. Molecular Cell , 2013, 50 (2) : 212–222. DOI:10.1016/j.molcel.2013.02.015 |
| [22] | Conley A B, Miller W J, Jordan I K. Human cis natural antisense transcripts initiated by transposable elements. Trends in Genetics , 2008, 24 (2) : 53–56. DOI:10.1016/j.tig.2007.11.008 |
| [23] | Dinger M E, Amaral P P, Mercer T R, et al. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Research , 2008, 18 (9) : 1433–1445. DOI:10.1101/gr.078378.108 |
| [24] | Ponting C P, Oliver P L, Reik W. Evolution and functions of long noncoding RNAs. Cell , 2009, 136 (4) : 629–641. DOI:10.1016/j.cell.2009.02.006 |
| [25] | Ramskold D, Wang E T, Burge C B, et al. An abundance of ubiquitously expressed genes revealed by tissue transcriptome sequence data. PLoS Comput Biol , 2009, 5 (12) : e1000598. DOI:10.1371/journal.pcbi.1000598 |
| [26] | Hu X, Feng Y, Zhang D, et al. A functional genomic approach identifies FAL1 as an oncogenic long noncoding RNA that associates with BMI1 and represses p21 expression in cancer. Cancer Cell , 2014, 26 (3) : 344–357. DOI:10.1016/j.ccr.2014.07.009 |
| [27] | Mikkelsen T S, Ku M, Jaffe D B, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature , 2007, 448 (7153) : 553–560. DOI:10.1038/nature06008 |
| [28] | Sánchez Y, Segura V, Marín-Béjar O, et al. Genome-wide analysis of the human p53 transcriptional network unveils a lncRNA tumour suppressor signature. Nature Communications , 2014, 5 (12) : 1–13. |
| [29] | Zhang Q, Chen C Y, Yedavalli V S, et al. NEAT1 long noncoding RNA and paraspeckle bodies modulate HIV-1 posttranscriptional expression. MBio , 2013, 4 (1) : e00596–00512. |
| [30] | Zhao W, Mu Y, Ma L, et al. Systematic identification and characterization of long intergenic non-coding RNAs in fetal porcine skeletal muscle development. Scientific Reports , 2015, 5 : 1–8. DOI:10.9734/JSRR |
| [31] | Derrien T, Johnson R, Bussotti G, et al. The GENCODE v7 catalog of human long noncoding RNAs:analysis of their gene structure, evolution, and expression. Genome Research , 2012, 22 (9) : 1775–1789. DOI:10.1101/gr.132159.111 |
| [32] | Han L, Zhang K, Shi Z, et al. LncRNA profile of glioblastoma reveals the potential role of lncRNAs in contributing to glioblastoma pathogenesis. International Journal of Oncology , 2012, 40 (6) : 2004–2012. |
| [33] | Cabili MN, Trapnell C, Goff L, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes & Development , 2011, 25 (18) : 1915–1927. |
| [34] | Liu X, Sun M, Nie F, et al. Lnc RNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR-331-3p in gastric cancer. Mol Cancer , 2014, 13 (1) : 92. DOI:10.1186/1476-4598-13-92 |
| [35] | Wang L, Zhao Y, Bao X, et al. LncRNA Dum interacts with Dnmts to regulate Dppa2 expression during myogenic differentiation and muscle regeneration. Cell Research , 2015, 25 (3) : 335–350. DOI:10.1038/cr.2015.21 |
| [36] | Giannakakis A, Zhang J, Jenjaroenpun P, et al. Contrasting expression patterns of coding and noncoding parts of the human genome upon oxidative stress. Scientific Reports , 2015, 5 : 1–16. DOI:10.9734/JSRR |
| [37] | Kim D H, Marinov G K, Pepke S, et al. Single-cell transcriptome analysis reveals dynamic changes in lncRNA expression during reprogramming. Cell Stem Cell , 2015, 16 (1) : 88–101. DOI:10.1016/j.stem.2014.11.005 |
| [38] | van Heesch S, van Iterson M, Jacobi J, et al. Extensive localization of long noncoding RNAs to the cytosol and mono-and polyribosomal complexes. Genome Biol , 2014, 15 (1) : 1–12. DOI:10.1186/gb-2014-15-1-r1 |
| [39] | Gonzalez I, Munita R, Agirre E, et al. A lncRNA regulates alternative splicing via establishment of a splicing-specific chromatin signature. Nature Structural & Molecular Biology , 2015, 22 (5) : 370–376. |
| [40] | Léveillé N, Melo C A, Rooijers K, et al. Genome-wide profiling of p53-regulated enhancer RNAs uncovers a subset of enhancers controlled by a lncRNA. Nature Communications , 2015, 6 : 1–12. |
| [41] | ØromU, DerrienT, GuigoR, 等. Long noncoding RNAs as enhancers of gene expression. Cold Spring Harbor Symposia on Quantitative Biology , 2011, 75 (58) : 325–331. |
| [42] | van Dijk M, Visser A, Buabeng K M, et al. Mutations within the LINC-HELLP non-coding RNA differentially bind ribosomal and RNA splicing complexes and negatively affect trophoblast differentiation. Human Molecular Genetics , 2015, 24 (19) : 5475–5485. DOI:10.1093/hmg/ddv274 |
| [43] | Han P, Li W, Lin C H, et al. A long noncoding RNA protects the heart from pathological hypertrophy. Nature , 2014, 514 (7520) : 102–106. DOI:10.1038/nature13596 |
| [44] | Yan L, Zhou J, Gao Y, et al. Regulation of tumor cell migration and invasion by the H19/let-7 axis is antagonized by metformin-induced DNA methylation. Oncogene , 2014, 34 (23) : 3076–3084. |
| [45] | Li H J, Li X, Pang H, et al. Long non-coding RNA UCA1 promotes glutamine metabolism by targeting miR-16 in human bladder cancer. Japanese Journal of Clinical Oncology , 2015, 45 (11) : 1055–1063. DOI:10.1093/jjco/hyv132 |
| [46] | Wang K, Sun T, Li N, et al. MDRL lncRNA regulates the processing of miR-484 primary transcript by targeting miR-361. PLoS Genetics , 2014, 10 (7) : 1–12. |
| [47] | Wang F, Ying H, He B, et al. Upregulated lncRNA-UCA1 contributes to progression of hepatocellular carcinoma through inhibition of miR-216b and activation of FGFR1/ERK signaling pathway. Oncotarget , 2015, 6 (10) : 7899–7917. DOI:10.18632/oncotarget |
| [48] | Gong C, Maquat L E. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3[prime] UTRs via Alu elements. Nature , 2011, 470 (7333) : 284–288. DOI:10.1038/nature09701 |
| [49] | Wilusz J E. Long noncoding RNAs:re-writing dogmas of RNA processing and stability. Biochimica et Biophysica Acta (BBA)-Gene Regulatory Mechanisms , 2015, 1859 (2016) : 128–138. |
| [50] | Li W, Notani D, Ma Q, et al. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature , 2013, 498 (7455) : 516–520. DOI:10.1038/nature12210 |
| [51] | Kim T, Cui R, Jeon Y J, et al. Long-range interaction and correlation between MYC enhancer and oncogenic long noncoding RNA CARLo-5. Proceedings of the National Academy of Sciences , 2014, 111 (11) : 4173–4178. DOI:10.1073/pnas.1400350111 |
| [52] | Lai F, Orom U A, Cesaroni M, et al. Activating RNAs associate with Mediator to enhance chromatin architecture and transcription. Nature , 2013, 494 (7438) : 497–501. DOI:10.1038/nature11884 |
| [53] | Mathiyalagan P, Keating S T, Du X J, et al. Chromatin modifications remodel cardiac gene expression. Cardiovascular Research , 2014, 103 (1) : 7–16. DOI:10.1093/cvr/cvu122 |
| [54] | Böhmdorfer G, Wierzbicki A T. Control of chromatin structure by long noncoding RNA. Trends in Cell Biology , 2015, 25 (10) : 623–632. DOI:10.1016/j.tcb.2015.07.002 |
| [55] | Bida O, Gidoni M, Ideses D, et al. A novel mitosis-associated lncRNA, MA-linc1, is required for cell cycle progression and sensitizes cancer cells to Paclitaxel. Oncotarget , 2015, 6 (29) : 27880–27890. DOI:10.18632/oncotarget |
| [56] | Singh R, Mo Y Y. LncRNA BC200 is induced by estradiol and regulates apoptosis in MCF-7 breast cancer cells. Cancer Research , 2015, 75 (15 Supplement) : 156–156. DOI:10.1158/1538-7445.AM2015-156 |
| [57] | Feng J, Funk W D, Wang S S, et al. The RNA component of human telomerase. Science , 1995, 269 (5228) : 1236–1241. DOI:10.1126/science.7544491 |
| [58] | Yang L, Lin C, Liu W, et al. ncRNA-and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell , 2011, 147 (4) : 773–788. DOI:10.1016/j.cell.2011.08.054 |
| [59] | Yang L, Lin C, Jin C, et al. lncRNA-dependent mechanisms of androgen-receptor-regulated gene activation programs. Nature , 2013, 500 (7464) : 598–602. DOI:10.1038/nature12451 |
| [60] | Tsai M C, Manor O, Wan Y, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science , 2010, 329 (5992) : 689–693. DOI:10.1126/science.1192002 |
| [61] | Martens J A, Laprade L, Winston F. Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature , 2004, 429 (6991) : 571–574. DOI:10.1038/nature02538 |
| [62] | Jiang W, Liu Y, Liu R, et al. The lncRNA DEANR1 facilitates human endoderm differentiation by activating FOXA2 expression. Cell Reports , 2015, 11 (1) : 137–148. DOI:10.1016/j.celrep.2015.03.008 |
| [63] | Yang F, Deng X, Ma W, et al. The lncRNA Firre anchors the inactive X chromosome to the nucleolus by binding CTCF and maintains H3K27me3 methylation. Genome Biology , 2015, 16 (1) : 1–17. DOI:10.1186/s13059-014-0572-2 |
| [64] | Xiao T, Liu L, Li H, et al. Long Noncoding RNA ADINR Regulates Adipogenesis by Transcriptionally Activating C/EBPα. Stem Cell Reports , 2015, 5 (5) : 856–865. DOI:10.1016/j.stemcr.2015.09.007 |
| [65] | Winter J, Jung S, Keller S, et al. Many roads to maturity:microRNA biogenesis pathways and their regulation. Nature Cell Biology , 2009, 11 (3) : 228–234. DOI:10.1038/ncb0309-228 |
| [66] | Lauressergues D, Couzigou J M, San Clemente H, et al. Primary transcripts of microRNAs encode regulatory peptides. Nature , 2015, 520 (7545) : 1–15. DOI:10.1038/nature14036 |
| [67] | Pefanis E, Wang J, Rothschild G, et al. RNA exosome-regulated long non-coding RNA transcription controls super-enhancer activity. Cell , 2015, 161 (4) : 774–789. DOI:10.1016/j.cell.2015.04.034 |
| [68] | Yakovchuk P, Goodrich J A, Kugel J F. B2 RNA and Alu RNA repress transcription by disrupting contacts between RNA polymerase Ⅱ and promoter DNA within assembled complexes. Proceedings of the National Academy of Sciences , 2009, 106 (14) : 5569–5574. DOI:10.1073/pnas.0810738106 |
| [69] | Rinn J L, Kertesz M, Wang J K, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell , 2007, 129 (7) : 1311–1323. DOI:10.1016/j.cell.2007.05.022 |
| [70] | Goodrich J A, Kugel J F. Non-coding-RNA regulators of RNA polymerase Ⅱ transcription. Nature Reviews Molecular Cell Biology , 2006, 7 (8) : 612–616. DOI:10.1038/nrm1946 |
| [71] | Watanabe T, Totoki Y, Toyoda A, et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature , 2008, 453 (7194) : 539–543. DOI:10.1038/nature06908 |
| [72] | Tam O H, Aravin A A, Stein P, et al. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature , 2008, 453 (7194) : 534–538. DOI:10.1038/nature06904 |
| [73] | Salmena L, Poliseno L, Tay Y, et al. A ceRNA hypothesis:the Rosetta Stone of a hidden RNA language?. Cell , 2011, 146 (3) : 353–358. DOI:10.1016/j.cell.2011.07.014 |
| [74] | Paraskevopoulou M D, Georgakilas G, Kostoulas N, et al. DIANA-LncBase:experimentally verified and computationally predicted microRNA targets on long non-coding RNAs. Nucleic Acids Research , 2013, 41 (D1) : 239–245. DOI:10.1093/nar/gks1246 |
| [75] | Wang J, Liu X, Wu H, et al. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Research , 2010, 38 (16) : 5366–5383. DOI:10.1093/nar/gkq285 |
| [76] | Wang K, Liu C Y, Zhou L Y, et al. APF lncRNA regulates autophagy and myocardial infarction by targeting miR-188-3p. Nature Communications , 2015, 6 (6779) : 1–11. |
| [77] | Cesana M, Cacchiarelli D, Legnini I, et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell , 2011, 147 (2) : 358–369. DOI:10.1016/j.cell.2011.09.028 |
| [78] | Wang Y, Pang W J, Wei N, et al. Identification, stability and expression of Sirt1 antisense long non-coding RNA. Gene , 2014, 539 (1) : 117–124. DOI:10.1016/j.gene.2014.01.037 |
| [79] | Deng L, Yang S B, Xu F F, et al. Long noncoding RNA CCAT1 promotes hepatocellular carcinoma progression by functioning as let-7 sponge. Journal of Experimental & Clinical Cancer Research , 2015, 34 (18) : 1–10. |
| [80] | Braconi C, Kogure T, Valeri N, et al. microRNA-29 can regulate expression of the long non-coding RNA gene MEG3 in hepatocellular cancer. Oncogene , 2011, 30 (47) : 4750–4756. DOI:10.1038/onc.2011.193 |
| [81] | Wang K, Long B, Zhou L Y, et al. CARL lncRNA inhibits anoxia-induced mitochondrial fission and apoptosis in cardiomyocytes by impairing miR-539-dependent PHB2 downregulation. Nature Communications , 2014, 5 (3539) : 1–13. |
| [82] | Tang Y, Jin X, Xiang Y, et al. The lncRNA MALAT1 protects the endothelium against ox-LDL-induced dysfunction via upregulating the expression of the miR-22-3p target genes CXCR2 and AKT. FEBS Letters , 2015, 589 (20) : 3189–3196. |
| [83] | Lu L, Luo F, Liu Y, et al. Posttranscriptional silencing of the lncRNA MALAT1 by miR-217 inhibits the epithelial-mesenchymal transition via enhancer of zeste homolog 2 in the malignant transformation of HBE cells induced by cigarette smoke extract. Toxicology and Applied Pharmacology , 2015, 289 (2) : 276–285. DOI:10.1016/j.taap.2015.09.016 |
| [84] | Hirata H, Hinoda Y, Shahryari V, et al. Long Noncoding RNA MALAT1 Promotes Aggressive Renal Cell Carcinoma through Ezh2 and Interacts with miR-205. Cancer Research , 2015, 75 (7) : 1322–1331. DOI:10.1158/0008-5472.CAN-14-2931 |
| [85] | Wang X, Li M, Wang Z, et al. Silencing of long noncoding rna malat1 by mir-101 and mir-217 inhibits proliferation, migration, and invasion of esophageal squamous cell carcinoma cells. Journal of Biological Chemistry , 2015, 290 (7) : 3925–3935. DOI:10.1074/jbc.M114.596866 |
| [86] | Yan B, Yao J, Liu J Y, et al. lncRNA-MIAT regulates microvascular dysfunction by functioning as a competing endogenous RNA. Circulation Research , 2015, 116 (7) : 1143–1156. DOI:10.1161/CIRCRESAHA.116.305510 |
| [87] | Tsang F H, Au S L, Wei L, et al. Long non-coding RNA HOTTIP is frequently up-regulated in hepatocellular carcinoma and is targeted by tumour suppressive miR-125b. Liver International , 2015, 35 (5) : 1597–1606. DOI:10.1111/liv.2015.35.issue-5 |
| [88] | Gernapudi R, Wolfson B, Zhang Y, et al. miR-140 Promotes Expression of long non-coding RNA NEAT1 in Adipogenesis. Molecular and Cellular Biology , 2015, 36 (1) : 30–38. |
| [89] | Wang P, Liu Y H, Yao Y l, et al. Long non-coding RNA CASC2 suppresses malignancy in human gliomas by miR-21. Cellular Signalling , 2015, 27 (2) : 275–282. DOI:10.1016/j.cellsig.2014.11.011 |
| [90] | Cai H, Xue Y, Wang P, et al. The long noncoding RNA TUG1 regulates blood-tumor barrier permeability by targeting miR-144. Oncotarget , 2015, 6 (23) : 19759–19779. DOI:10.18632/oncotarget |
| [91] | Wang K, Liu F, Zhou L Y, et al. The long noncoding RNA CHRF regulates cardiac hypertrophy by targeting miR-489. Circulation Research , 2014, 114 (9) : 1377–1388. DOI:10.1161/CIRCRESAHA.114.302476 |
| [92] | Cai X, Hagedorn C H, Cullen B R. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. Rna , 2004, 10 (12) : 1957–1966. DOI:10.1261/rna.7135204 |
| [93] | Liz J, Portela A, Soler M, et al. Regulation of pri-miRNA processing by a long noncoding RNA transcribed from an ultraconserved region. Molecular Cell , 2014, 55 (1) : 138–147. DOI:10.1016/j.molcel.2014.05.005 |
| [94] | Lee Y, Kim M, Han J, et al. MicroRNA genes are transcribed by RNA polymerase Ⅱ. The EMBO Journal , 2004, 23 (20) : 4051–4060. DOI:10.1038/sj.emboj.7600385 |
| [95] | Aravin A A, Hannon G J, Brennecke J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science , 2007, 318 (5851) : 761–764. DOI:10.1126/science.1146484 |
| [96] | Wilusz J E, Freier S M, Spector D L. 3' end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell , 2008, 135 (5) : 919–932. DOI:10.1016/j.cell.2008.10.012 |
| [97] | Ganesan G, Rao S M. A novel noncoding RNA processed by Drosha is restricted to nucleus in mouse. Rna , 2008, 14 (7) : 1399–1410. DOI:10.1261/rna.838308 |
| [98] | Fejes-Toth K, Sotirova V, Sachidanandam R, et al. Post-transcriptional processing generates a diversity of 5'-modified long and short RNAs. Nature , 2009, 457 (7232) : 1028–1032. DOI:10.1038/nature07759 |
| [99] | Wilusz J E, Sunwoo H, Spector D L. Long noncoding RNAs:functional surprises from the RNA world. Genes & Development , 2009, 23 (13) : 1494–1504. |
| [100] | Ogawa Y, Sun B K, Lee J T. Intersection of the RNA interference and X-inactivation pathways. Science , 2008, 320 (5881) : 1336–1341. DOI:10.1126/science.1157676 |
| [101] | Xiao T, Liu L, Li H, et al. Long Noncoding RNA ADINR Regulates Adipogenesis by Transcriptionally Activating C/EBPα. Stem Cell Reports , 2015, 5 (5) : 856–865. DOI:10.1016/j.stemcr.2015.09.007 |
| [102] | Yang F, Zhang L, Huo X S, et al. Long noncoding RNA high expression in hepatocellular carcinoma facilitates tumor growth through enhancer of zeste homolog 2 in humans. Hepatology , 2011, 54 (5) : 1679–1689. DOI:10.1002/hep.24563 |
| [103] | Zhao X Y, Li S, Wang G X, et al. A long noncoding RNA transcriptional regulatory circuit drives thermogenic adipocyte differentiation. Molecular Cell , 2014, 55 (3) : 372–382. DOI:10.1016/j.molcel.2014.06.004 |
| [104] | Ⅱott N E, Heward J A, Roux B, et al. Long non-coding RNAs and enhancer RNAs regulate the lipopolysaccharide-induced inflammatory response in human monocytes. Nature Communications , 2014, 5 (3979) : 1–13. |
| [105] | Yu Y, Fuscoe J C, Zhao C, et al. A rat RNA-Seq transcriptomic BodyMap across 11 organs and 4 developmental stages. Nature Communications , 2014, 5 (3230) : 1–11. |
| [106] | Sun K, Zhao Y, Wang H, et al. Sebnif:An Integrated Bioinformatics Pipeline for the Identification of Novel Large Intergenic Noncoding RNAs (lincRNAs)-Application in Human Skeletal Muscle Cells. PloS one , 2014, 9 (1) : 1–9. |
| [107] | Divoux A, Karastergiou K, Xie H, et al. Identification of a novel lncRNA in gluteal adipose tissue and evidence for its positive effect on preadipocyte differentiation. Obesity , 2014, 22 (8) : 1781–1785. DOI:10.1002/oby.v22.8 |
| [108] | Sun L, Goff L A, Trapnell C, et al. Long noncoding RNAs regulate adipogenesis. Proceedings of the National Academy of Sciences , 2013, 110 (9) : 3387–3392. DOI:10.1073/pnas.1222643110 |
| [109] | Li P, Ruan X, Yang L, et al. A liver-enriched long non-coding RNA, lncLSTR, regulates systemic lipid metabolism in mice. Cell Metabolism , 2015, 21 (3) : 455–467. DOI:10.1016/j.cmet.2015.02.004 |
| [110] | Leucci E, Patella F, Waage J, et al. microRNA-9 targets the long non-coding RNA MALAT1 for degradation in the nucleus. Scientific Reports , 2013, 3 (2535) : 1–6. |
| [111] | Imam H, Bano A S, Patel P, et al. The lncRNA NRON modulates HIV-1 replication in a NFAT-dependent manner and is differentially regulated by early and late viral proteins. Scientific Reports , 2015, 5 (8639) : 1–10. |
| [112] | Krol J, Krol I, Alvarez C P P, et al. A network comprising short and long noncoding RNAs and RNA helicase controls mouse retina architecture. Nature Communications , 2015, 6 (7305) : 1–13. |
| [113] | Spurlock Ⅲ C F, Tossberg J T, Guo Y, et al. Expression and functions of long noncoding RNAs during human T helper cell differentiation. Nature Communications , 2015, 6 (6932) : 1–12. |
| [114] | White N M, Cabanski C R, Silva-Fisher J M, et al. Transcriptome sequencing reveals altered long intergenic non-coding RNAs in lung cancer. Genome Biol , 2014, 15 (8) : 1–16. |
| [115] | Iyer M K, Niknafs Y S, Malik R, et al. The landscape of long noncoding RNAs in the human transcriptome. Nature Genetics , 2015 . DOI:10.1038/ng.3192 |
| [116] | Imamura K, Imamachi N, Akizuki G, et al. Long noncoding RNA NEAT1-dependent SFPQ relocation from promoter region to paraspeckle mediates IL8 expression upon immune stimuli. Molecular Cell , 2014, 53 (3) : 393–406. DOI:10.1016/j.molcel.2014.01.009 |
| [117] | Yang F, Yi F, Han X, et al. MALAT-1 interacts with hnRNP C in cell cycle regulation. FEBS Letters , 2013, 587 (19) : 3175–3181. DOI:10.1016/j.febslet.2013.07.048 |
| [118] | Lai M C, Yang Z, Zhou L, et al. Long non-coding RNA MALAT-1 overexpression predicts tumor recurrence of hepatocellular carcinoma after liver transplantation. Medical Oncology , 2012, 29 (3) : 1810–1816. DOI:10.1007/s12032-011-0004-z |
| [119] | Liu J H, Chen G, Dang Y W, et al. Expression and prognostic significance of lncRNA MALAT1 in pancreatic cancer tissues. Asian Pacific journal of cancer prevention:APJCP , 2013, 15 (7) : 2971–2977. |
| [120] | Arase M, Horiguchi K, Ehata S, et al. Transforming growth factor-β-induced lncRNA-Smad7 inhibits apoptosis of mouse breast cancer JygMC (A) cells. Cancer Science , 2014, 105 (8) : 974–982. DOI:10.1111/cas.2014.105.issue-8 |
| [121] | Luo M, Li Z, Wang W, et al. Long non-coding RNA H19 increases bladder cancer metastasis by associating with EZH2 and inhibiting E-cadherin expression. Cancer Letters , 2013, 333 (2) : 213–221. DOI:10.1016/j.canlet.2013.01.033 |
| [122] | Krawczyk M, Emerson B M. p50-associated COX-2 extragenic RNA (PACER) activates COX-2 gene expression by occluding repressive NF-κB complexes. Elife , 2014 . |
| [123] | Prensner J R, Mehra R, Chinnaiyan A M, et al. SChLAP1:a newly validated lncRNA biomarker for aggressive prostate cancer. RNA & DISEASE , 2015, 2 (2) : 1–4. |
| [124] | Mehra R, Shi Y, Udager A M, et al. A novel RNA in situ hybridization assay for the long noncoding RNA SChLAP1 predicts poor clinical outcome after radical prostatectomy in clinically localized prostate cancer. Neoplasia , 2014, 16 (12) : 1121–1127. DOI:10.1016/j.neo.2014.11.006 |
| [125] | Sahu A, Iyer M K, Prensner J R, et al. The role of long noncoding RNA SChLAP1 in prostate cancer. Cancer Research , 2014, 74 (19 Supplement) : 541–541. DOI:10.1158/1538-7445.AM2014-541 |
| [126] | Malik R, Patel L, Prensner J R, et al. The lncRNA PCAT29 inhibits oncogenic phenotypes in prostate cancer. Molecular Cancer Research , 2014, 12 (8) : 1081–1087. DOI:10.1158/1541-7786.MCR-14-0257 |
| [127] | Chakravarty D, Sboner A, Nair S S, et al. The oestrogen receptor alpha-regulated lncRNA NEAT1 is a critical modulator of prostate cancer. Nature Communications , 2014, 5 (5383) : 1–16. |
| [128] | Prensner J R, Feng F Y C. The role of the PCAT-1 lncRNA in prostate cancer tumorigenesis. RNA & Disease , 2015, 2 (1) : 900–908. |
| [129] | Prensner J R, Chen W, Han S, et al. The long non-coding RNA PCAT-1 promotes prostate cancer cell proliferation through cMyc. Neoplasia , 2014, 16 (11) : 900–908. DOI:10.1016/j.neo.2014.09.001 |
| [130] | Zhu M, Chen Q, Liu X, et al. lncRNA H19/miR-675 axis represses prostate cancer metastasis by targeting TGFBI. FEBS Journal , 2014, 281 (16) : 3766–3775. DOI:10.1111/febs.2014.281.issue-16 |
| [131] | Salameh A, Lee A K, Cardó-Vila M, et al. PRUNE2 is a human prostate cancer suppressor regulated by the intronic long noncoding RNA PCA3. Proceedings of the National Academy of Sciences , 2015, 112 (27) : 8403–8408. DOI:10.1073/pnas.1507882112 |
| [132] | Li L, Dang Q, Xie H, et al. Infiltrating mast cells enhance prostate cancer invasion via altering LncRNA-HOTAIR/PRC2-androgen receptor (AR)-MMP9 signals and increased stem/progenitor cell population. Oncotarget , 2015, 6 (16) : 14179–14190. |
| [133] | Zhang A, Zhao J C, Kim J, et al. LncRNA HOTAIR enhances the androgen-receptor-mediated transcriptional program and drives castration-resistant prostate cancer. Cell Reports , 2015, 13 (1) : 209–221. DOI:10.1016/j.celrep.2015.08.069 |
| [134] | Liu Y, Zhang R, Qiu F, et al. Construction of a lncRNA-PCG bipartite network and identification of cancer-related lncRNAs:a case study in prostate cancer. Molecular Bio Systems , 2015, 11 (2) : 384–393. |
| [135] | Isin M, Uysaler E, Özgür E, et al. Exosomal lncRNA-p21 levels may help to distinguish prostate cancer from benign disease. Frontiers in Genetics , 2015, 6 (168) : 1–5. |
| [136] | Wang L, Han S, Jin G, et al. Linc00963:A novel, long non-coding RNA involved in the transition of prostate cancer from androgen-dependence to androgen-independence. International Journal of Oncology , 2014, 44 (6) : 2041–2049. |
| [137] | Richards E, Zhang G, Permuth-Wey J, et al. Identification of TGFβ-regulated long noncoding RNAs in mammary epithelia:lncRNA-HIT mediated TGFβ-induced EMT and breast cancer metastasis. Cancer Research , 2015, 75 (15 Supplement) : 154–154. DOI:10.1158/1538-7445.AM2015-154 |
| [138] | Feng FY, Ma T, Speers C, et al. Abstract PD6-1:The long noncoding RNA M41 promotes aggressiveness and tamoxifen resistance in ER-positive breast cancers. Cancer Research , 2015, 75 (9) : 1–6. |
| [139] | Xing Z, Park P K, Lin C, et al. LncRNA BCAR4 wires up signaling transduction in breast cancer. RNA Biology , 2015, 12 (7) : 681–689. DOI:10.1080/15476286.2015.1053687 |
| [140] | Shi Y, Li J, Liu Y, et al. The long noncoding RNA SPRY4-IT1 increases the proliferation of human breast cancer cells by upregulating ZNF703 expression. Molecular Cancer , 2015, 14 (51) : 1–13. |
| [141] | Koirala P, Mo Y Y. LncRNA AK023948 promotes breast tumorigenesis by enhancing AKT phosphorylation. Cancer Research , 2015, 75 (15 Supplement) : 155–155. DOI:10.1158/1538-7445.AM2015-155 |
| [142] | Xue X, Yang Y, Zhang A, et al. LncRNA HOTAIR enhances ER signaling and confers tamoxifen resistance in breast cancer. Oncogene , 2016, 35 (21) : 2746–2755. DOI:10.1038/onc.2015.340 |
| [143] | Zhou M, Hou Y, Yang G, et al. LncRNA-Hh strengthen cancer stem cells generation in twist-positive breast cancer via activation of hedgehog signaling pathway. Stem Cells , 2015, 34 (1) : 55–66. |
| [144] | Shi S, Wang L, Yu B, et al. LncRNA-ATB promotes trastuzumab resistance and invasion-metastasis cascade in breast cancer. Oncotarget , 2015, 6 (13) : 11652–11663. DOI:10.18632/oncotarget |
| [145] | Zhuang J, Lu Q, Shen B, et al. TGFβ1 secreted by cancer-associated fibroblasts induces epithelial-mesenchymal transition of bladder cancer cells through lncRNA-ZEB2NAT. Scientific Reports , 2015, 5 (11924) : 1–13. |
| [146] | Fan Y, Shen B, Tan M, et al. Long non-coding RNA UCA1 increases chemoresistance of bladder cancer cells by regulating Wnt signaling. FEBS Journal , 2014, 281 (7) : 1750–1758. DOI:10.1111/febs.12737 |
| [147] | Berrondo C, Flax J, Messing E M, et al. The long non-coding RNA HOTAIR affects exosome-mediated bladder cancer progression. Cancer Research , 2015, 75 (15 Supplement) : 152–152. DOI:10.1158/1538-7445.AM2015-152 |
| [148] | Li S, Yu Z, Li F, et al. The YAP1 Oncogene Contributes to Bladder Cancer Cell Proliferation and Migration by Regulating the H19 Long Noncoding RNA. In:Urologic Oncology:Seminars and Original Investigations; 2015:Elsevier , 2015, 33 (427) : 1–10. |
| [149] | Tan J, Qiu K, Li M, et al. Double-negative feedback loop between long non-coding RNA TUG1 and miR-145 promotes epithelial to mesenchymal transition and radioresistance in human bladder cancer cells. FEBS Letters , 2015, 589 (20) : 3175–3181. |
| [150] | Iguchi T, Uchi R, Nambara S, et al. A long noncoding RNA, lncRNA-ATB, is involved in the progression and prognosis of colorectal cancer. Anticancer Research , 2015, 35 (3) : 1385–1388. |
| [151] | Zheng H T, Shi D B, Wang Y W, et al. High expression of lncRNA MALAT1 suggests a biomarker of poor prognosis in colorectal cancer. International Journal of Clinical and Experimental Pathology , 2014, 7 (6) : 3174–3181. |
| [152] | Ma Y, Yang Y, Wang F, et al. Long non-coding RNA CCAL regulates colorectal cancer progression by activating Wnt/β-catenin signalling pathway via suppression of activator protein 2α. Gut , 2016, 65 (9) : 1494–1504. DOI:10.1136/gutjnl-2014-308392 |
| [153] | Shi D, Zheng H, Zhuo C, et al. Low expression of novel lncRNA RP11-462C24. 1 suggests a biomarker of poor prognosis in colorectal cancer. Medical Oncology , 2014, 31 (7) : 1–9. |
| [154] | Yin D, He X, Zhang E, et al. Long noncoding RNA GAS5 affects cell proliferation and predicts a poor prognosis in patients with colorectal cancer. Medical Oncology , 2014, 31 (11) : 1–8. |
| [155] | Yang Y, Li H, Hou S, et al. The noncoding RNA expression profile and the effect of lncRNA AK126698 on cisplatin resistance in non-small-cell lung cancer cell. PLoS One , 2013, 5 (8) : 1–12. |
| [156] | Thai P, Statt S, Chen C H, et al. Characterization of a novel long noncoding RNA, SCAL1, induced by cigarette smoke and elevated in lung cancer cell lines. American Journal of Respiratory Cell and Molecular Biology , 2013, 49 (2) : 204–211. DOI:10.1165/rcmb.2013-0159RC |
| [157] | Shi X, Sun M, Liu H, et al. A critical role for the long non-coding RNA GAS5 in proliferation and apoptosis in non-small-cell lung cancer. Molecular Carcinogenesis , 2013, 54 (1) : 1–12. |
| [158] | Qiu M, Xu Y, Yang X, et al. CCAT2 is a lung adenocarcinoma-specific long non-coding RNA and promotes invasion of non-small cell lung cancer. Tumor Biology , 2014, 35 (6) : 5375–5380. DOI:10.1007/s13277-014-1700-z |
| [159] | Fan M, Li X, Jiang W, et al. A long non-coding RNA, PTCSC3, as a tumor suppressor and a target of miRNAs in thyroid cancer cells. Experimental and Therapeutic Medicine , 2013, 5 (4) : 1143–1146. |
| [160] | Yang F, Bi J, Xue X, et al. Up-regulated long non-coding RNA H19 contributes to proliferation of gastric cancer cells. FEBS Journal , 2012, 279 (17) : 3159–3165. DOI:10.1111/j.1742-4658.2012.08694.x |
| [161] | Li H, Yu B, Li J, et al. Overexpression of lncRNA H19 enhances carcinogenesis and metastasis of gastric cancer. Oncotarget , 2014, 5 (8) : 2318–2329. DOI:10.18632/oncotarget |
| [162] | Sun W, Wu Y, Yu X, et al. Decreased expression of long noncoding RNA AC096655. 1-002 in gastric cancer and its clinical significance. Tumor Biology , 2013, 34 (5) : 2697–2701. DOI:10.1007/s13277-013-0821-0 |
| [163] | Sun M, Xia R, Jin F, et al. Downregulated long noncoding RNA MEG3 is associated with poor prognosis and promotes cell proliferation in gastric cancer. Tumor Biology , 2014, 35 (2) : 1065–1073. DOI:10.1007/s13277-013-1142-z |
| [164] | Zhao Y, Guo Q, Chen J, et al. Role of long non-coding RNA HULC in cell proliferation, apoptosis and tumor metastasis of gastric cancer:a clinical and in vitro investigation. Oncology Reports , 2014, 31 (1) : 358–364. |
| [165] | Kim K, Jutooru I, Chadalapaka G, et al. HOTAIR is a negative prognostic factor and exhibits pro-oncogenic activity in pancreatic cancer. Oncogene , 2013, 32 (13) : 1616–1625. DOI:10.1038/onc.2012.193 |
| [166] | Ma C, Nong K, Zhu H, et al. H19 promotes pancreatic cancer metastasis by derepressing let-7's suppression on its target HMGA2-mediated EMT. Tumor Biology , 2014, 35 (9) : 9163–9169. DOI:10.1007/s13277-014-2185-5 |
| [167] | Wang Y, He L, Du Y, et al. The Long Noncoding RNA lncTCF7 Promotes Self-Renewal of Human Liver Cancer Stem Cells through Activation of Wnt Signaling. Cell Stem Cell , 2015, 16 (4) : 413–425. DOI:10.1016/j.stem.2015.03.003 |
| [168] | Tsang F H, Au S L, Wei L, et al. HOTTIP, an oncogenic long non-coding RNA, is frequently up-regulated in hepatocellular carcinoma and is negatively regulated by tumor suppressive microRNA miR-125b. Cancer Research , 2015, 75 (15 Supplement) : 145–145. DOI:10.1158/1538-7445.AM2015-145 |
 2016, Vol. 36
2016, Vol. 36