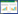文章信息
- 梁欣泉, 李宁, 任勤, 刘继栋
- LIANG Xin-quan, LI Ning, REN Qin, LIU Ji-dong
- 代谢工程改造酿酒酵母生产L-乳酸的研究进展
- Progress in the Metabolic Engineering of Saccharomyces cerevisiae for L-lactic Acid Production
- 中国生物工程杂志, 2016, 36(2): 109-114
- China Biotechnology, 2016, 36(2): 109-114
- http://dx.doi.org/10.13523/j.cb.20160216
-
文章历史
- 收稿日期: 2015-09-11
- 修回日期: 2015-09-24
L-乳酸(L-lactic acid)是一种天然存在的有机酸,广泛应用于食品、医药、纺织、皮革、化妆品及化工领域。同时,L-乳酸还可以用来生产聚乳酸,后者是生产可降解塑料的主要成分[1]。由于L-乳酸在国民经济中所起的重要作用,它已经成为最受关注的化学产物之一。目前,食品安全级的L-乳酸需求量逐年递增,预计在2017年将达到367 000吨,远远超出了目前的供给能力[2]。此外,目前可降解塑料的年产量达到了2亿吨,其原料聚乳酸的年生产量仅为450 000吨[3],这很大程度上是由于乳酸的生产能力欠缺所致。因此,提高L-乳酸市场供应能力及降低L-乳酸的生产成本是当前亟需解决的关键问题之一。
目前,L-乳酸的制备途径主要分为两种,即化学合成法和微生物发酵法[4]。1963年,美国Monsanto公司首次开始采用化学合成法生产乳酸,使用乙醛与剧毒化合物氢氰酸在碱性催化剂作用下经多步反应合成乳酸[5]。然而,该方法除了高能耗及高污染外,其合成路线长、副产物多且无法控制、有害中间产物残留等,严重影响其与人体密切接触的产品使用的安全性。而利用微生物及其酶系发酵生产L-乳酸,具有原料来源广泛、产物安全性高、发酵条件温和、产品单一等特点,正日益得到学者的认可,并成为当前L-乳酸的主要来源。目前,国内发酵法生产L-乳酸常用的菌种为米根霉(Rhizopus oryzae)、乳酸菌、酿酒酵母(Saccharomyces cerevisiae)等。
工业上米根霉的L-乳酸发酵工艺大多采用传统的通风搅拌式发酵。米根霉自身耐酸性较低,L-乳酸积累抑制菌体生长,影响产物浓度进一步提高。加入中和介质控制发酵液pH,增加了乳酸的后续分离成本。而且米根霉发酵对温度、pH值、溶氧量、培养基成分和浓度等条件的控制要求过于严格,发酵成本高,限制了米根霉发酵生产L-乳酸的发展[6]。
与米根霉相比,乳酸菌的理论转化率较高,且没有大量的副产物积累。然而,由于乳酸菌需要严格厌氧发酵且耐酸能力较低,并对pH的调控要求较高,导致发酵控制带来很大不便。此外,乳酸菌复杂的培养基要求、容易感染噬菌体、无法直接利用淀粉等多糖以及难以耐受恶劣环境胁迫等缺陷进一步增加了工业发酵的难度和成本[7]。
在微生物体内,L-乳酸生物合成的前体为丙酮酸(Pyruvate),丙酮酸可以在L-乳酸脱氢酶和其他修饰酶的作用下,生成L-乳酸。近年来,基于合成生物学的微生物细胞工厂构建将外源的L-乳酸合成途径引入到微生物内,辅以加强前体供给、限速步骤调控等策略,使得微生物具备了异源高效生产L-乳酸的能力[8]。由于酿酒酵母自身的特殊优势如食品安全性、遗传背景清晰、环境耐受能力较强等特点,成为目前学者首选的改造宿主[9]。
目前代谢工程改造酿酒酵母生产L-乳酸的研究主要集中在以下几个方面。
1 L-乳酸脱氢酶在酿酒酵母内的表达酿酒酵母自身不含特定的L-乳酸生物合成途径,需要首先引入相应的L-乳酸脱氢酶。1994年,Dequin[10]将源于干酪乳杆菌(Lactobacillus casei)的乳酸脱氢酶基因(Lactate dehydrogenase,LDH)在酿酒酵母内表达,该基因受到乙醇脱氢酶1基因(Alcohol dehydrogenase 1,ADH1)启动子的调控,构建了一株乙醇及乳酸混合发酵的突变株。在以葡萄糖为碳源的培养基中发酵,证实有20%的葡萄糖转化为L-乳酸,L-乳酸产量为10 g/L。1995年,Porro等[11]在酿酒酵母中表达源于牛的乳酸脱氢酶基因(LDHA),L-乳酸产量提高至20 g/L。2003年,Colombie等[12]为提高LDH在菌体中的稳定性,把植物乳酸杆菌(Lactobacellus plantarum)的LDH整合到酿酒酵母基因组中,并在此基础上,用KOH对培养基pH进行控制,发现有27%的葡萄糖转化为L-乳酸,积累50 g/L的L-乳酸。以上结果说明,不同来源的L-乳酸脱氢酶以及其在酿酒酵母内的表达方式对L-乳酸产率有较大影响。此外,仅在酿酒酵母中表达外源LDH得到的L-乳酸积累量都不高,需要通过代谢工程手段进一步提高产率。
2 丙酮酸旁路代谢途径改造酿酒酵母主要有三条丙酮酸代谢途径[13](图 1):(1) 在丙酮酸羧化酶途径中,丙酮酸在丙酮酸羧化酶(pyruvate carboxylase,PYC)的催化下,转化为草酰乙酸(oxaloacetic acid,OAA),进入三羧酸循环(TCA);(2) 在丙酮酸脱氢酶途径中,丙酮酸在线粒体蛋白酶丙酮酸脱氢酶(pyruvate dehydrogenase,PDH)的催化下,转化为线粒体乙酰辅酶A(acetyl-CoA),进入TCA;(3)在丙酮酸脱羧酶途径中,丙酮酸在丙酮酸脱羧酶(pyruvate decarboxylase,PDC)的作用下转化为乙醛,后者在乙醇脱氢酶(alcohol dehydrogenase,ADH)的进一步作用下生成乙醇(ethanol)或者在乙醛脱氢酶(acetaldehyde dehydrogenase,ALD)的进一步作用下生成乙酸(acetate)。L-乳酸是丙酮酸在乳酸脱氢酶(lactate dehydrogenase,LDH)的作用下生成的,因此,要提高L-乳酸的产率,多位学者采用了降低丙酮酸旁路代谢通量的策略。

|
| 图 1 酿酒酵母改造菌株的丙酮酸代谢及L-乳酸合成途径 Fig. 1 The metabolism of pyruvate and the biosynthetic pathway of L-Lactic acid in the engineered Saccharomyces cerevisiae strain A: Glycolysis; B: Pyruvate decarboxylase; C: Alcohol dehydrogenase; D: Acetaldehy dehydrogenase; E: Acetyl-CoA synthetase; F: Pyruvate carboxylase; G: Pyruvate dehydrogenase; H: Lactate dehydrogenase |
1998年,Adachi等[14]为降低副产物乙醇的积累量,在酿酒酵母中导入外源LDH的基础上敲除PDC1,使乳酸对葡萄糖的产率从0.155 g/g提高到0.2 g/g,而副产物乙醇对葡萄糖的产率则从0.35 g/g降为0.2 g/g。2005年,为进一步提高L-乳酸的转化率,Ishida等[15]通过同源重组的方式用牛的LDHA替换掉酿酒酵母的PDC1,前者受到PDC1启动子的控制,构建了LDH双拷贝并缺失PDC1的突变株,结果发现62.2%的葡萄糖转化为L-乳酸,乳酸积累量达到55.6 g/L,副产物乙醇为16.9 g/L。用外源D-LDH进行转化也出现类似的结果[16]。随后,该研究小组还分别构建了4个拷贝LDH和6个拷贝LDH的基因工程菌[17, 18]。结果表明,L-乳酸的积累量与LDH拷贝数呈正相关,其中6个拷贝改造菌株的L-乳酸积累量可达到122 g/L,但副产物乙醇的产量也超过40 g/L。此时,乙醇成为了制约L-乳酸积累量进一步提高的关键因素,若将乙醇合成途径完全切断,乙醇将不再积累,但此时丙酮酸是否会顺利流向目的产物L-乳酸尚需验证。该小组在前期构建的双拷贝LDH并缺失PDC1的菌株的基础上敲除了PDC5,副产物乙醇积累量降低到5 g/L以下,L-乳酸的积累量达到82.3 g/L,L-乳酸对葡萄糖的转化率为81.5%,然而其发酵周期为192 h[19]。通过微氧条件下的代谢流分析[20]发现,改造菌株的生长受抑制可能是由于戊糖磷酸途径代谢的减弱,用于组氨酸合成的核糖-5-磷酸供应不足。因此,大幅度弱化代谢中的某一条途径,而不补偿生长所必需的中间代谢产物,势必会导致菌体生长受到抑制。
2.2 乙醇脱氢酶(ADH)途径的改造乙醇脱氢酶1(ADH1)是酿酒酵母乙醇发酵中最主要的同工酶[21]。2003年,Skory[22]在酿酒酵母中导入真菌LDH的基础上敲除了ADH1,但无氧条件下改造菌株无法生长,有氧条件下的L-乳酸积累为20 g/L,同时有近40 g/L的甘油生成,这对提高L-乳酸的产率是不利的。较之表达外源LDH并敲除ADH1的菌株,仅表达LDH菌株的L-乳酸积累量更高,L-乳酸为38 g/L,对葡萄糖的产率为0.44 g/g。这结果可能是因为前者受到过量积累的乙醛的抑制所致,类似的结果也在酿酒酵母生产脂肪酸中发现[23]。2009年,在Ishida等[15]构建的菌株的基础上,Tokuhiro等[24]通过同源重组方式用LDHA替换掉ADH1,构建了4拷贝LDH并缺失PDC1和ADH1的突变株,乳酸积累量达到74.1 g/L,L-乳酸对葡萄糖的产率为0.69 g/g,副产物乙醇约7 g/L。因此,在弱化某条途径的代谢通量时,必须要保证上游中间代谢产物的积累不会对菌体的生长产生抑制。
2.3 丙酮酸脱氢酶(PDH)途径的改造酿酒酵母丙酮酸脱氢酶复合物(PDH)能够催化丙酮酸转化形成线粒体乙酰辅酶A。目前尚未见到关于弱化酿酒酵母内PDH活性的相关报道,不过,Bianchi等[25]在克鲁维酵母(Kluyeromyces lactis)中导入外源LDH的基础上敲除PDH的E1α亚基基因,构建含有LDH并缺失PDH活性的突变株,L-乳酸对葡萄糖产率达到0.85 g/g,比含有LDH活性并缺失PDC活性的突变株高,后者为0.58 g/g。但是关于构建缺失PDH活性的酿酒酵母突变株是否会使碳源顺利流向目的产物L-乳酸仍需要验证。
3 辅因子工程利用酿酒酵母生产L-乳酸所面临的主要问题是如何能获得较高的L-乳酸产量和较高的产率,并缩短菌体的发酵周期。通过改变菌体代谢通量达到目的产物积累最大化时,势必也会造成某些中间代谢产物合成的不足以及目的产物的过量积累而造成菌体生长受抑制。因此,增加目的产物积累量的同时必须补偿必需中间代谢产物的供给以及减轻对目的产物的抑制。
3.1 胞质乙酰辅酶A的补偿胞质乙酰辅酶A(acetyl-CoA)是酿酒酵母中众多代谢产物的前体,弱化菌体的PDC酶活会影响胞质Acetyl-CoA的供应,从而影响菌体的生长[26]。在含有乙酸或乙醇时,缺失PDC的突变株呈现出野生型一样生长能力的事实则更进一步支持了这个推测[27]。目前,尚未发现关于弱化了PDC的酿酒酵母突变株的胞质Acetyl-CoA补偿策略相关报道。Shiba等[28]过表达乙醛脱氢酶基因6(ALD6)和乙酰辅酶A合成酶基因1(ACS1),构建过表达ALD6和ACS1酿酒酵母突变株,提高了胞质Acetyl-CoA含量。Lian等[29]失活酿酒酵母ADH1、ADH4以及用于甘油合成的甘油-3-磷酸脱氢酶基因,同时还引入Acetyl-CoA外源合成途径,构建了突变株,大大提高胞质Acetyl-CoA含量。这些研究有助于在酿酒酵母工程菌中应用外源合成途径,提高菌体生长率。
3.2 胞质NAD+的补偿NADH/NAD+在细胞中的比例影响着酿酒酵母的代谢途径方向[30],NADH积累,甘油合成也会相应提高[21]。2006年,Heux等[31]在酿酒酵母中表达乳酸乳球菌(Lactococcus lactis)NADH氧化酶基因,NADH浓度降低5倍,NADH/NAD+降低6倍,乙醇、甘油、琥珀酸、羟戊二酸产率明显降低,乙醛、乙酸、羟基丁酮产率则增加。2007年,Vemuri等[32]超表达酿酒酵母NADH氧化酶,甘油产率降低。2012年,Wang等[33]将来源于乳酸菌的NADH氧化酶基因(NOXE)和大肠杆菌可溶解的吡啶核苷酸转氢酶基因(UDHA)表达于酿酒酵母中,丙酮酸产量提高21%。2014年,Hou等[34]在酿酒酵母中表达L. lactis NADH氧化酶基因,构建一株乙醇发酵突变株,在以木糖为碳源的培养基中发酵,木糖醇、甘油产率分别降低60%和83%,同时提高了乙醇产率。因此,降低NADH/NAD+在胞质中的比例将有利于葡萄糖向L-乳酸方向转化。
4 胞外运输由于代谢生成的乳酸会在工程菌中积累,降低胞内pH并增加酿酒酵母菌体负荷,LDH酶活受到抑制,L-乳酸合成降低[35]。1999年,Casal等[36]将JEN1表达于乳酸-质子同向转运缺陷的酿酒酵母菌株中,菌株恢复乳酸转运。van Maris等[37]的研究表明酿酒酵母的乳酸胞外运输消耗ATP,而消耗方式还需进一步探究。Pacheco等[38]在酿酒酵母工程菌内表达JEN1及ADY2后,乳酸产量提高15%,但是Wakamatsu等[39]的研究仅证实JEN1参与乳酸的转胞内运输。当前,对于酿酒酵母自身转运蛋白的研究一直停留在JEN1、ADY2这两个蛋白上,其他菌种乳酸转运蛋白也有相关研究[40]。关于乳酸胞外运输机制和动力学特征有待进一步研究。
5 展 望利用代谢工程技术对细胞代谢进行修饰与改造,定向改变细胞特性,是一种生产各类化学物质的重要方法[41]。在改造酿酒酵母生产L-乳酸方面已实现L-乳酸的积累,产率的提高,同时副产物明显降低。但是改造菌株生长能力降低的问题还需解决,而且酿酒酵母工程菌株的L-乳酸产量也未能达到乳酸菌菌株的L-乳酸产量水平[3],因而还无法替代乳酸菌作为L-乳酸的微生物生产的主要菌株。
当前,利用酿酒酵母生产L-乳酸还需要在以下三个方面进一步研究:(1)围绕丙酮酸碳源流向的特点,适当调节各个基因的表达强度,精确控制细胞内的代谢流,促使代谢流流向L-乳酸,减少分支代谢流量,减少副产物积累;(2)虽然通过代谢工程改造增强丙酮酸流向L-乳酸相关基因的表达,能提高L-乳酸产量,但是其他生理特性的改变也促使甘油等副产物含量上升、胞质乙酰辅酶A含量降低、辅因子供应不平衡,造成菌体生长受到抑制,需要从全局代谢网络和调控网络出发,寻找更为有效的代谢工程改造位点;(3)目的产物的不断积累,抑制菌体的生长,而且产物的后续分离也比较困难,这些都是待解决的问题。
通过在上述三方面研究的不断深入,有望提高L-乳酸产量,减少细胞内代谢副产物,增加代谢改造工程菌生长率,降低后续分离成本,实现工业大规模生产。这些研究的重要性不仅体现在建立经济的L-乳酸生产上,还将对设计微生物细胞工厂有着重要的意义。
致谢 本课题得到广西大学科研基金(XJZ140293)的资助,特此致谢。
| [1] | Nduko J M,Matsumoto K,Ooi T,et al. Enhanced production of poly(lactate-co-3-hydroxybutyrate) from xylose in engineered Escherichia coli overexpressing a galactitol transporter. Applied Microbiology and Biotechnology,2014,98(6):2453-2460. |
| [2] | Taskila S,Ojamo H. The current status and future expectations in industrial production of lactic acid by lactic acid bacteria. In:Kongo M,ed. Lactic Acid Bacteria-R&D for Food,Health and Livestock Purposes. Rijeka,Croatia:Tech Open Access Publisher,2013.615-632. |
| [3] | Abdel-Rahman M A,Tashiro Y,Sonomoto K. Recent advances in lactic acid production by microbial fermentation processes. Biotechnology Advances,2013,31(6):877-902. |
| [4] | Maki-Arvela P,Simakova I L,Salmi T,et al. Production of lactic acid/lactates from biomass and their catalytic transformations to commodities. Chemical Reviews,2014,114(3):1909-1971. |
| [5] | Martinez F,Balciunas E M. Lactic acid properties,applications and production:a review. Trends in Food Science & Technology,2013,30(1):70-83. |
| [6] | 赵亮亮. 代谢工程改造酿酒酵母生产L-乳酸. 无锡:江南大学,2011. Zhao L L. The reconstruction of Saccharomyces cerevisiae by metabolic engineering to product L-lactic acid. Wuxi:Jiangnan University,2011. |
| [7] | Upadhyaya B P,DeVeaux L C,Christopher L P. Metabolic engineering as a tool for enhanced lactic acid production. Trends in Biotechnology,2014,32(12):637-644. |
| [8] | Su J,Wang T,Wang Y,et al. The use of lactic acid-producing,malic acid-producing,or malic acid-degrading yeast strains for acidity adjustment in the wine industry. Applied Microbiology and Biotechnology,2014,98(6):2395-2413. |
| [9] | Kavšcek M,Stražar M,Curk T,et al. Yeast as a cell factory:current state and perspectives. Microbial Cell Factories,2015,14(1):1-10. |
| [10] | Dequin S. Mixed lactic acid-alcoholic fermentation by Saccharomyces cerevisiae expressing the Lactobacillus casei L(+)-LDH. Biotechnology,1994,12:173-177. |
| [11] | Porro D,Brambilla L,Ranzi B M,et al. Development of metabolically engineered Saccharomyces cerevisiae cells for the production of lactic acid. Biotechnology Progress,1995,11(3):294-298. |
| [12] | Colombie S,Dequin S,Sablayrolles J M. Control of lactate production by Saccharomyces cerevisiae expressing a bacterial LDH gene. Enzyme and Microbial Technology,2003,33(1):38-46. |
| [13] | Koivistoinen O M,Kuivanen J,Barth D,et al. Glycolic acid production in the engineered yeasts Saccharomyces cerevisiae and Kluyveromyces lactis. Microbial Cell Factories,2013,12(6):839-850. |
| [14] | Adachi E,Torigoe M,Sugiyama M,et al. Modification of metabolic pathways of Saccharomyces cerevisiae by the expression of lactate dehydrogenase and deletion of pyruvate decarboxylase genes for the lactic acid fermentation at low pH value. Journal of Fermentation and Bioengineering,1998,86(3):284-289. |
| [15] | Ishida N,Saitoh S,Tokuhiro K,et al. Efficient production of L-lactic acid by metabolically engineered Saccharomyces cerevisiae with a genome-integrated L-lactate dehydrogenase gene. Applied and Environmental Microbiology,2005,71(4):1964-1970. |
| [16] | Ishida N,Suzuki T,Tokuhiro K,et al. D-lactic acid production by metabolically engineered Saccharomyces cerevisiae. Journal of Bioscience and Bioengineering,2006,101(2):172-177. |
| [17] | Saitoh S,Ishida N,Onishi T,et al. Genetically engineered wine yeast produces a high concentration of L-lactic acid of extremely high optical purity. Applied and Environmental Microbiology,2005,71(5):2789-2792. |
| [18] | Ishida N,Saitoh S,Ohnishi T,et al. Metabolic engineering of Saccharomyces cerevisiae for efficient production of pure L-(+)-lactic acid. Applied Biochemistry and Biotechnology,2006,131(1-3):795-807. |
| [19] | Ishida N,Saitoh S,Onishi T,et al. The effect of pyruvate decarboxylase gene knockout in Saccharomyces cerevisiae on L-lactic acid production. Bioscience Biotechnology and Biochemistry,2006,70(5):1148-1153. |
| [20] | Nagamori E,Shimizu K,Fujita H,et al. Metabolic flux analysis of genetically engineered Saccharomyces cerevisiae that produces lactate under micro-aerobic conditions. Bioprocess and Biosystems Engineering,2013,36(9):1261-1265. |
| [21] | Ida Y,Furusawa C,Hirasawa T,et al. Stable disruption of ethanol production by deletion of the genes encoding alcohol dehydrogenase isozymes in Saccharomyces cerevisiae. Journal of Bioscience and Bioengineering,2012,113(2):192-195. |
| [22] | Skory C D. Lactic acid production by Saccharomyces cerevisiae expressing a Rhizopus oryzae lactate dehydrogenase gene. Journal of Industrial Microbiology & Biotechnology,2003,30(1):22-27. |
| [23] | Li X W,Guo D Y,Cheng Y B,et al. Overproduction of fatty acids in engineered Saccharomyces cerevisiae. Biotechnology and Bioengineering,2014,111(9):1841-1852. |
| [24] | Tokuhiro K,Ishida N,Nagamori E,et al. Double mutation of the pdc1 and adh1 genes improves lactate production in the yeast Saccharomyces cerevisiae expressing the bovine lactate dehydrogenase gene. Applied Microbiology and Biotechnology,2009,82(5):883-890. |
| [25] | Bianchi M M,Brambilla L,Protani F,et al. Efficient homolactic fermentation by Kluyveromyces lactis strains defective in pyruvate utilization and transformed with the heterologous ldh gene. Applied and Environmental Microbiology,2001,67(12):5621-5625. |
| [26] | Kozak B U,van Rossum H M,Benjamin K R,et al. Replacement of the Saccharomyces cerevisiae acetyl-CoA synthetases by alternative pathways for cytosolic acetyl-CoA synthesis. Metabolic Engineering,2014,21:46-59. |
| [27] | Flikweert M T,vanDijken J P,Pronk J T. Metabolic responses of pyruvate decarboxylase-negative Saccharomyces cerevisiae to glucose excess. Applied and Environmental Microbiology,1997,63(9):3399-3404. |
| [28] | Shiba Y,Paradise E M,Kirby J,et al. Engineering of the pyruvate dehydrogenase bypass in Saccharomyces cerevisiae for high-level production of isoprenoids. Metabolic Engineering,2007,9(2):160-168. |
| [29] | Lian J Z,Si T,Nair N U,et al. Design and construction of acetyl-CoA overproducing Saccharomyces cerevisiae strains. Metabolic Engineering,2014,24:139-149. |
| [30] | Ida Y,Hirasawa T,Furusawa C,et al. Utilization of Saccharomyces cerevisiae recombinant strain incapable of both ethanol and glycerol biosynthesis for anaerobic bioproduction. Applied Microbiology and Biotechnology,2013,97(11):4811-4819. |
| [31] | Heux S,Cachon R,Dequin S. Cofactor engineering in Saccharomyces cerevisiae:expression of a H2O-forming NADH oxidase and impact on redox metabolism. Metabolic Engineering,2006,8(4):303-314. |
| [32] | Vemuri G N,Eiteman M A,McEwen J E,et al. Increasing NADH oxidation reduces overflow metabolism in Saccharomyces cerevisiae. Proceedings of the National Academy of Sciences of the United States of America,2007,104(7):2402-2407. |
| [33] | Wang Z K,Gao C J,Wang Q,et al. Production of pyruvate in Saccharomyces cerevisiae through adaptive evolution and rational cofactor metabolic engineering. Biochemical Engineering Journal,2012,67:126-131. |
| [34] | Hou J,Suo F,Wang C Q,et al. Fine-tuning of NADH oxidase decreases byproduct accumulation in respiration deficient xylose metabolic Saccharomyces cerevisiae. BMC Biotechnology,2014,14(4):452-457. |
| [35] | Branduardi P,Sauer M,De Gioia L,et al. Lactate production yield from engineered yeasts is dependent from the host background,the lactate dehydrogenase source and the lactate export. Microbial Cell Factories,2006,5(1):107-109. |
| [36] | Casal M,Paiva S,Andrade R P,et al. The lactate-proton symport of Saccharomyces cerevisiae is encoded by jen1. Journal of Bacteriology,1999,181(8):2620-2623. |
| [37] | van Maris A J A,Winkler A A,Porro D,et al. Homofermentative lactate production cannot sustain anaerobic growth of engineered Saccharomyces cerevisiae:possible consequence of energy-dependent lactate export. Applied and Environmental Microbiology,2004,70(5):2898-2905. |
| [38] | Pacheco A,Talaia G,Sa-Pessoa J,et al. Lactic acid production in Saccharomyces cerevisiae is modulated by expression of the monocarboxylate transporters Jen1 and Ady2. FEMS Yeast Research,2012,12(3):375-381. |
| [39] | Wakamatsu M,Tomitaka M,Tani T,et al. Improvement of ethanol production from D-lactic acid by constitutive expression of lactate transporter Jen1p in Saccharomyces cerevisiae. Bioscience Biotechnology and Biochemistry,2013,77(5):1114-1116. |
| [40] | Skory C D,Hector R E,Gorsich S W,et al. Analysis of a functional lactate permease in the fungus Rhizopus. Enzyme and Microbial Technology,2010,46(1):43-50. |
| [41] | Pfleger B F,Gossing M,Nielsen J. Metabolic engineering strategies for microbial synthesis of oleochemicals. Metabolic Engineering,2015,29:1-11. |
 2016, Vol. 36
2016, Vol. 36