扩展功能
文章信息
- 刘莹莹, 曹晓梅, 慈颖, 杨燕, 卢云兰, 张晓龙, 方志强
- LIU Ying-ying, CAO Xiao-mei, CI Ying, YANG Yan, LU Yun-lan, ZHANG Xiao-long, FANG Zhi-qiang
- 云南省西南地区4个口岸黄胸鼠种群遗传特性分析
- Genetic characteristics of Rattus tanezumi populations at four ports of entry in southwest Yunnan province, China
- 中国媒介生物学及控制杂志, 2020, 31(4): 451-456
- Chin J Vector Biol & Control, 2020, 31(4): 451-456
- 10.11853/j.issn.1003.8280.2020.04.015
-
文章历史
- 收稿日期: 2020-02-10
2 云南国际旅行卫生保健中心, 云南 昆明 650228
2 Yunnan International Travel Healthcare Center
黄胸鼠(Rattus tanezumi)是我国常见的家栖鼠[1],不仅是鼠疫、肾综合征出血热(HFRS)、钩端螺旋体病、恙虫病等众多传统病媒性疾病的重要宿主[2],还是巴贝西原虫、巴尔通体等新发传染病病原体的宿主,对于鼠传疾病防治具有重要的研究价值[3-5]。
哺乳动物线粒体DNA为母系遗传,共价闭合的环状双链DNA分子,双链分子中外环为重链(H链),内环为轻链(L链)。基因序列组成相对保守,无重组和单拷贝,并易于检测。由于其结构和进化上的特点,线粒体DNA(mtDNA)被作为研究物种进化的重要分子标记[6-8]。Cytb基因在线粒体基因组中进化速度适中,一个较短的片段就可以将种下水平以及科水平的系统发育信息包含在内,适合研究种内到种间甚至科间的系统发育关系[9]。在mtDNA各基因中,因其碱基序列稳定性较高而常用于动物类群的遗传多样性和亲缘关系研究[10]。
动物线粒体上的Cytb基因被认为是探讨近缘种间和种内遗传分化程度的良好指标[11],在物种分类学研究中有较强的适用性,已经在多种动物类群中都得到了有效应用[12]。易金鑫等[13]通过研究Cytb基因序列表明北盘江下游尼罗罗非鱼群体具有丰富的遗传多样性。Luo等[14]通过测定厦门海域和青岛海域的文昌鱼线粒体DNA细胞色素氧化酶Ⅰ基因和Cytb基因的部分序列表明厦门文昌鱼和青岛文昌鱼为不同种。胡群等[15]根据线粒体Cytb基因部分序列,对入境集装箱中截获的鼠样本进行了种类鉴定。
本研究以云南省金水河、清水河、磨憨和姐告4个不同口岸为研究对象,对4个不同地理种群的黄胸鼠线粒体DNA Cytb的序列特征和各种群间的遗传分化和进化关系进行了分析,为进一步深入研究不同地区黄胸鼠种群遗传特性积累了基础资料。此外黄胸鼠还与许多传染性疾病流行有关[2],因此研究黄胸鼠种群遗传特性对于疾病的预防、治疗和鼠害的防控具有重要意义。
1 材料与方法 1.1 标本材料2014-2015年期间4个口岸共捕获黄胸鼠标本55只,金水河、清水河、磨憨和姐告口岸分别为20、10、5和20只。取捕获鼠的肝脏保存于-80 ℃冰箱。
1.2 基因组DNA的提取取出-80 ℃冰箱中保存的黄胸鼠肝脏组织,剪取约25 mg,参照天根生化科技(北京)有限公司的血液/细胞/组织基因组DNA提取试剂盒(DP304)提取基因组DNA。
1.3 Cytb基因的扩增正向引物:5'-GACAGGAAAAATCATCGTTG-3';反向引物:5'-TTCATTACTGGTTTACAAGAC-3'。扩增体系总体积为50 μl:模板DNA 1 μl,正、反向引物各1μl,Premix Taq(TaKaRa Taq Version 2.0 plus dye)25μl,加纯水将体积补至50 μl。PCR反应的循环参数为:95℃预变性5 min;94℃变性1 min,50 ℃退火1 min,72℃延伸90 s,35个循环;72 ℃延伸8 min。PCR扩增产物经1.5 %琼脂糖凝胶电泳后,Goldview染色,紫外凝胶成像仪扫描检测。
1.4 序列测定及数据分析将PCR产物进行正反链双向测序。测序由英潍捷基(上海)贸易有限公司完成。测序结果与选自GenBank的同源序列使用Clustal X 2.0软件进行多重序列比对[16],并辅以人工校对。按照Kimura双参数法(kimura-2-parameter),用MEGA 7.0软件分析每条序列的碱基、氨基酸组成和密码子使用频率,并选用Kimura-2-Parameter模型计算物种间DNA序列间遗传距离差[17]。选用小家鼠(Mus musculus)为外群。并用MEGA 7.0软件包以距离矩阵邻接法(Neighbor-Joining,NJ)构建系统进化树[18]。在NJ树中分枝处的置信度采用1 000次自引导法重复检测。
2 结 果 2.1 PCR扩增Cytb基因扩增后产物经琼脂糖凝胶电泳可见清晰、明亮的约1 140 bp的预期目标条带,见图 1。

|
| 注:M为DL2 000 Marker,CK为阴性对照。4个口岸共有55个黄胸鼠样本,图中仅列出32个,1~20为金水河口岸样本,21~30为清水河口岸样本,31~32为磨憨口岸样本(部分)。 图 1 云南省4个口岸的黄胸鼠Cytb基因电泳检测(部分) Figure 1 Electrophoresis detection of the cytochrome b gene of Rattus tanezumi at four ports in Yunnan province (partial samples) |
| |
对Cytb基因序列采用双向测序,并且重复测定样品。
2.3 数据处理 2.3.1 Cytb基因序列的校正4个口岸地区黄胸鼠种群个体的Cytb基因序列用软件Clustal X 2.0和DNAMAN v8.0加以排序比较,可以得到1 125 bp比较完整的序列片段。为了验证Cytb基因片断的正确性,将转换成FASTA格式后的序列在NCBI数据库中(http://www.ncbi.nih.gov./BLAST/)进行BLASTn同源相似性搜索。结果表明,试验所测得的序列均为Cytb基因序列。
2.3.2 4个口岸地区黄胸鼠Cytb基因序列测定了金水河、清水、磨憨以及姐告4个口岸地区黄胸鼠的线粒体DNA基因约1 140 bp片段序列,经比对后,每个口岸选取1条Cytb基因序列,片段长度约为1 125 bp,其中金水河和姐告口岸黄胸鼠Cytb基因序列已录入GeneBank数据库。
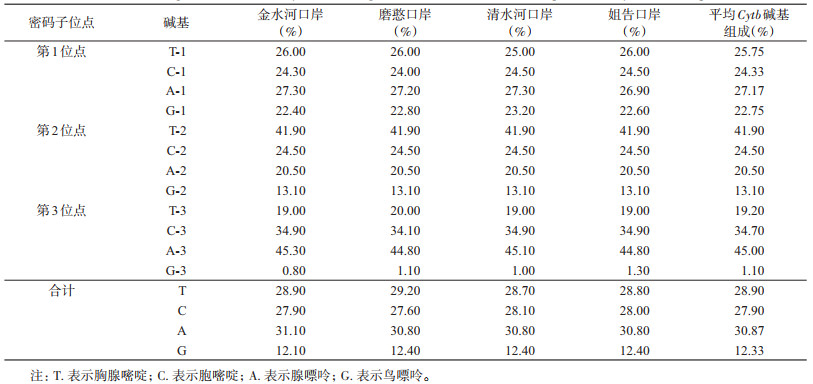
2.3.3 黄胸鼠Cytb部分序列的组成用MEGA 7. 0软件对获得的Cytb基因进行比对分析,见表 1。所测定的Cytb基因部分序列中,T、C、A、G平均含量分别为28.90%、27.90%、30.87%和12.33%。其中(A+T)含量占59.77%,(C+G)含量占40.23%。密码子第1~3位点碱基组成存在差异,第1位点富含A,平均含量为27.17%,第2位点富含T,平均含量为41.90%,第3位点富含A,平均含量为45.00%;同一碱基在密码子不同位点存在差异,T在第2位点含量最高,为41.90%,C在第3位点含量最高,为34.70%,A在第3位点含量最高,为45.00%,G在第1位的含量最高,为22.75%。
|
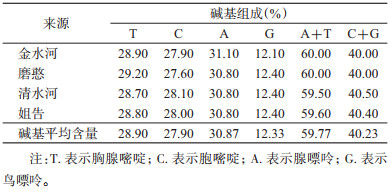
Cytb基因序列碱基组成如表 2所示。Cytb基因的碱基T、C、A、G的平均含量分别为28.90%,27.90%,30.87%和12.33%,清水河口岸的(A + T)含量最低为59.50%,金水河与磨憨口岸的(A + T)含量最高,均为60.00%。总体看来,(A + T)含量高于(C+G)。
|
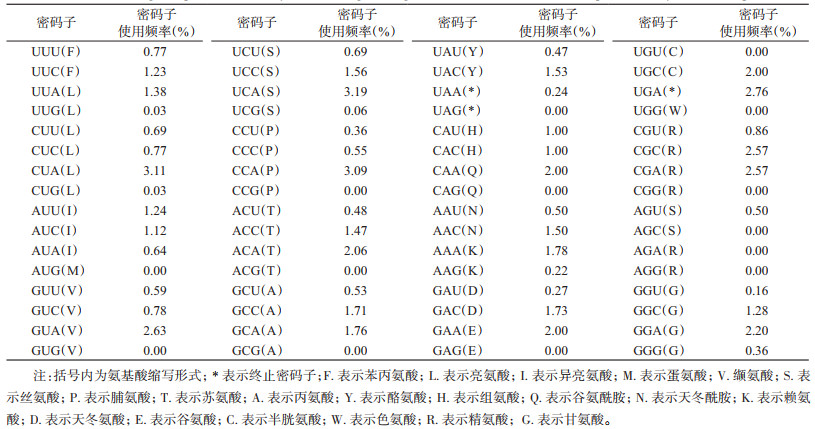
通过MEGA v7.0软件分析,统计4个不同口岸地区的黄胸鼠的密码子使用频率,从而推断Cytb编码区的密码子使用频率和氨基酸组成。从表 3、4可知,密码子UCA、CUA、CCA的使用频率较高,分别是3.19%、3.11%、3.09%。也有一些氨基酸并未出现,包括AUG、GUG、CCG、ACG、GCG等。氨基酸的组成显示,亮氨酸、异亮氨酸、苯丙氨酸使用频率较高,分别为15.13%、10.44%、8.17%。
|

|
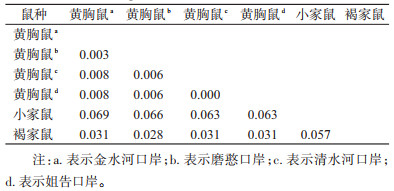
遗传距离最近的是姐告和清水河口岸黄胸鼠,为0.000;遗传距离最远的分别是金水河与清水河,金水河与姐告口岸黄胸鼠,均为0.008。将4个口岸黄胸鼠的Cytb基因序列与美国国立生物技术信息中心(NCBI)网站的小家鼠和褐家鼠(R. norvegicus)进行比对,结果见表 5。
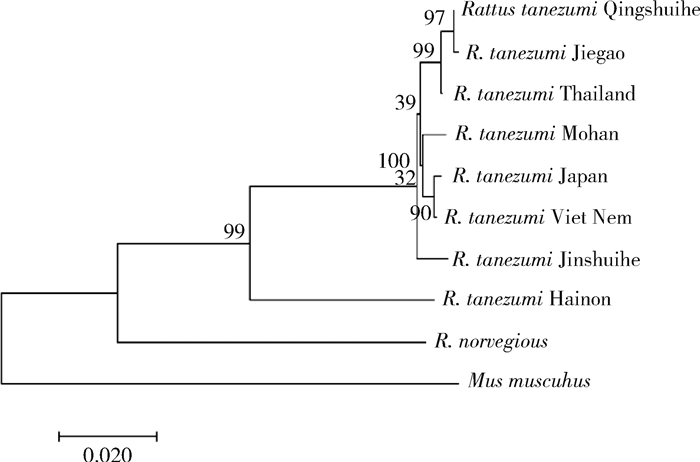
以同属于啮齿目的小家鼠(NC_005089.1)和褐家鼠(NC_001665.2)为外群,采用NJ法、“Bootstrap”1 000循环检验构建分子系统进化树(图 2)。根据进化树可知:总体来看分为3个大聚类簇,R. tanezumi Jinshuihe独立1个分支,R. tanezumi Mohan与R. tanezumi Viet Nem、R. tanezumi Japan聚在一起,R. tanezumi Qingshuihe与R. tanezumi Jiegao、R. tanezumi Thailand聚在一起,两个分支构成第1个聚类簇。对外群分析发现,两个外群各自聚为1支,与4个口岸地区黄胸鼠分开,各自构成1个聚类簇,且外群亲缘关系基本符合其阶元划分,从而可知外群选择的正确性。
|
| 图 2 利用邻接法构建云南省4个口岸黄胸鼠Cytb基因序列的进化树 Figure 2 Phylogenetic tree of cytochrome b gene sequences of Rattus tanezumi from four ports in Yunnan province based on the neighbor-joining method |
| |
Cytb基因在线粒体基因组中的进化速度适中[19],较短的1个DNA片段就能包含从种水平到属水平乃至纲水平的系统发育信息。而且在一定的进化尺度内,Cytb基因不受饱和效应的严重影响,所提供的系统发育信息和遗传分化水平更适于分析种间和属间差异[20],因此本研究通过测定黄胸鼠Cytb基因序列来研究4个不同口岸地区黄胸鼠的种群特性。测定的4个不同口岸黄胸鼠mtDNA Cytb基因部分序列的碱基组成中,T、C、A 3种碱基的含量差异较小,G含量最低。其中(A+T)含量明显高于(C+G)含量,推测黄胸鼠mtDNA可能存在(A+T)偏倚性。4个不同口岸地区黄胸鼠间的4种碱基组成相似,具有相同的变化规律。Cytb基因序列中A含量最高,其次是T,然后是C,最后是G含量最低,这与啮齿类动物mtDNA基因组中碱基含量A>T>C>G的研究结果一致[21]。
4个不同口岸黄胸鼠mtDNA Cytb基因部分序列中,不同位点碱基含量不同,均为密码子的第1位点和第3位点富含碱基A,第2位点富含碱基T。其中C在第3位点最高,G在第2位点含量最高。(A+T)含量在第3位点最高,而(C+G)含量在第3位点最低,可见在不同位点黄胸鼠间密码子的碱基组成存在较大偏倚。比较其密码子的使用频率,同一密码子在黄胸鼠间的使用频率不平衡,表明密码子的使用频率存在偏倚性。对4个不同口岸地区的黄胸鼠密码子使用频率进行分析发现,CUA、AUU、UUC等密码子使用频率较高,而GUG、ACG、GCG等密码子未出现。通过分析Cytb基因片段氨基酸组成发现亮氨酸、异亮氨酸、苯丙氨酸等氨基酸使用频率较高。
黄胸鼠分布与城市化进程中环境的变迁,交通工具的发达,气候的改变,以及人为因素的影响有关[22-24]。遗传距离分析发现,姐告与清水河口岸黄胸鼠Cytb基因差异程度很小,两个口岸同属于南亚热带季风性湿润气候,且两个口岸交通运输发达、人口密集,可能与黄胸鼠种群特性有关。Brouat等[25]在塞内加尔东南部的多乳鼠研究结果发现种群存在明显的距离隔离,表明地理障碍对于种群基因流的阻碍作用。当在局限空间内分散的物种达到突变-迁移-漂变平衡时,遗传分化是随着地理距离的增加而增加的[26]。本研究中金水河与清水河、姐告口岸的Cytb基因差异程度较大,可能与地理距离有关。
根据Musser和Carleton[27]的研究,黄胸鼠是印度次大陆北部,华南和中部以及印度支那大陆(云南省为中部地区)的本地物种。云南省位于中国西南和东南亚的边界地区,由于复杂的地质和气候历史,物种的种内遗传多样性较高[28-29]。临沧高速公路在建,清水河口岸地区交通便利,有利于黄胸鼠迁移扩散。NJ进化树分析可知,清水河口岸黄胸鼠与瑞丽姐告口岸黄胸鼠亲缘关系较近,且与泰国地区黄胸鼠亲缘关系也较近,口岸地区贸易往来密切,黄胸鼠可能通过商品贸易货车等交通工具运输发生迁移。磨憨口岸黄胸鼠与日本、越南地区黄胸鼠亲缘关系较近,可能是通过汽车或国际航行船舶等交通工具进行扩散。黄胸鼠在云南地区的传播受到地理障碍的限制,不适合IBD(Isolation by distance)模式,但可以利用人类运输实现远距离扩散[30],金水河口岸黄胸鼠与其他3个口岸黄胸鼠的亲缘关系相对较远,可能与红河金水河口岸地理位置有关。黄胸鼠作为1个家栖鼠种,其活动和繁殖都会受到人为因素的影响,地理距离较远的黄胸鼠随着火车和汽车等交通工具迀移而发生基因交流,后续可以通过分析黄胸鼠种群的遗传分化系数FST与地理距离的关系来确定这种基因流与地理距离的相关性。
| [1] |
胡秋波, 吴太平, 蒋洪. 黄胸鼠生态及防治研究进展[J]. 中华卫生杀虫药械, 2014, 20(2): 180-185. Hu QB, Wu TP, Jiang H. A review of ecology and control of Rattus tanezumi[J]. Chin J Hyg Insect Equip, 2014, 20(2): 180-185. DOI:10.19821/j.1671-2781.2014.02.028 |
| [2] |
谌苏燕. 2015年南昌县鼠类生物监测与疾病相关性分析报告[J]. 世界最新医学信息文摘, 2017, 17(21): 152-153. Shen SY. Correlation analysis report of rodents biological surveillance and disease in 2015 from Nanchang[J]. World Latest Med Inf (Electron Version), 2017, 17(21): 152-153. DOI:10.3969/j.issn.1671-3141.2017.21.138 |
| [3] |
年庆功, 邓兵, 王征, 等. 内蒙古某部训练基地鼠类中检测到巴贝西原虫DNA片段[J]. 医学动物防制, 2018, 34(12): 1149-1151. Nian QG, Deng B, Wang Z, et al. DNA fragment of Babesia protozoa detected in rodents in an army base in Inner Mongolia[J]. J Med Pest Control, 2018, 34(12): 1149-1151. DOI:10.7629/yxdwfz201812008 |
| [4] |
陈波, 宗素进, 朱志宏, 等. 义乌市鼠类携带耶尔森菌情况调查[J]. 上海预防医学, 2015, 27(12): 787-788. Chen B, Zong SJ, Zhu ZH, et al. Investigation on Yersinia carry in rats in Yiwu[J]. Shanghai J Pre Med, 2015, 27(12): 787-788. DOI:10.19428/j.cnki.sjpm.2015.12.018 |
| [5] |
万道正, 杨会敏, 梁亮, 等. 中越边境口岸鼠类及其自然感染病原体调查[J]. 中国国境卫生检疫杂志, 2017, 40(5): 341-344. Wan DZ, Yang HM, Liang L, et al. Distribution and pathogens of natural infection on rodents in China-Vietnam border ports[J]. Chin Front Health Quar, 2017, 40(5): 341-344. DOI:10.16408/j.1004-9770.2017.05.011 |
| [6] |
鞠妍, 刘华淼, 邢秀梅. 鹿科动物线粒体DNA序列差异和系统进化关系[J]. 吉林农业大学学报, 2017, 39(1): 94-99. Ju Y, Liu HM, Xing XM. Relationship between sequence divergence and phyletic evolution of deer (Cervidae) mtDNA[J]. J Jilin Agric Univ, 2017, 39(1): 94-99. DOI:10.13327/j.jjlau.2016.2973 |
| [7] |
周凡莉, 黄卫平, 金素钰, 等. 建昌马mtDNA D-loop区遗传多样性及系统进化分析[J]. 中国畜牧兽医, 2020, 47(1): 63-73. Zhou FL, Huang WP, Jin SY, et al. Genetic diversity and evolutionary analysis of mtDNA D-loop region in Jianchang horse[J]. China Anim Husb Vet Med, 2020, 47(1): 63-73. DOI:10.16431/j.cnki.1671-7236.2020.01.008 |
| [8] |
Motitz C, Dowling TE, Brown WM. Evolution of animal mitochondrial DNA:relevance for population biology and systematics[J]. Ann Rev Ecol Syst, 1987, 18: 269-292. DOI:10.1146/annurev.es.18.110187.001413 |
| [9] |
Meyer A, Wilson AC. Origin of tetrapods inferred from their mitochondrial DNA affiliation to lungfish[J]. J Mol Evol, 1990, 31(5): 359-364. DOI:10.1007/BF02106050 |
| [10] |
任艳玲, 沈志强, 马泽彬, 等. 渤海黑牛mtDNA Cytb基因遗传多样性及亲缘关系分析[J]. 山东畜牧兽医, 2017, 38(5): 1-2. Ren YL, Shen ZQ, Ma ZB, et al. Genetic diversity and relationship analysis of mtDNA Cytb gene in Bohai black cattle[J]. Shandong J Anim Sci Vet Med, 2017, 38(5): 1-2. DOI:10.3969/j.issn.1007-1733.2017.05.001 |
| [11] |
Irwin DM, Kocher TD, Wilson AC. Evolution of the cytochrome b gene of mammals[J]. J Mol Evol, 1991, 32(2): 128-144. DOI:10.1007/BF02515385 |
| [12] |
Hay JM, Ruvinsky I, Hedges SB, et al. Phylogenetic relationships of amphibian families inferred from DNA sequences of mitochondrial 12S and 16S ribosomal RNA genes[J]. Mol Biosci Evol, 1995, 12(5): 928-937. DOI:10.1093/oxfordjournals.molbev.a040270 |
| [13] |
易金鑫, 代应贵, 孙际佳, 等. 北盘江下游尼罗罗非鱼群体Cytb基因多态性[J]. 水产科学, 2019, 38(5): 716-720. Yi JX, Dai YG, Sun JJ, et al. Polymorphism of Cytb gene in population of Nile Tilapia Oreochromis niloticus from lower Beipan River[J]. Fish Sci, 2019, 38(5): 716-720. DOI:10.16378/j.cnki.1003-1111.2019.05.019 |
| [14] |
Luo JC, Xiao YS, Song L, et al. Phylogenetic separation of lancelets in China revealed by mitochondrial COI and Cytb gene analysis[J]. South China Fish Sci, 2007, 3(2): 8-14. DOI:10.3969/j.issn.2095-0780.2007.02.002 |
| [15] |
胡群, 马思杰, 邹春颖, 等. 基于Cytb基因序列对入境集装箱中截获的疑似鼠样本的种类鉴定[J]. 中国国境卫生检疫杂志, 2014, 37(1): 37-39. Hu Q, Ma SJ, Zou CY, et al. Species identification of suspected samples of rats found in imported containers based on Cytb gene[J]. Chin Front Health Quar, 2014, 37(1): 37-39. DOI:10.16408/j.1004-9770.2014.01.008 |
| [16] |
Thompson JD, Gibson TJ, Plewniak F, et al. The Clustal_X windows interface:flexible strategies for multiple sequence alignment aided by quality analysis tools[J]. Nucleic Acids Research, 1997, 25(24): 4876-4882. DOI:10.1093/nar/25.24.4876 |
| [17] |
Stecher G, Tamura K, Kumar S. Molecular evolutionary genetics analysis (MEGA) for macOS[J]. Mol Biol Evol, 2020, 37(4): 1237-1239. DOI:10.1093/molbev/msz312 |
| [18] |
赵庆, 吴彪, 刘志鸿, 等. 利用DNA条形码COI基因分析我国重要贝类系统进化关系[J]. 中国水产科学, 2018, 25(4): 847-857. Zhao Q, Wu B, Liu ZH, et al. Analysis of evolutionary relationships of important shellfish in China based on the DNA barcoding COI gene[J]. J Fish Sci China, 2018, 25(4): 847-857. DOI:10.3724/SP.J.1118.2018.17363 |
| [19] |
Ladoukakis ED, Zouros E. Evolution and inheritance of animal mitochondrial DNA:rules and exceptions[J]. J Biol Res(Thessalon), 2017, 24: 2. DOI:10.1186/s40709-017-0060-4 |
| [20] |
Meyer A. Shortcomings of the cytochrome b gene as a molecular marker[J]. Trends Ecol Evol, 1994, 9(8): 278-280. DOI:10.1016/0169-5347(94)90028-0 |
| [21] |
Gadaleta G, Pepe G, De Candia G, et al. Complete nucleotide sequence of the Rattus norvegicus mitochondrial genome:cryptic signals revealed by comparative analysis between vertebrates[J]. J Mol Evol, 1989, 28(6): 497-516. DOI:10.1007/BF02602930 |
| [22] |
Mora MS, Mapelli FJ, Gaggiotti OE, et al. Dispersal and population structure at different spatial scales in the subterranean rodent Ctenomys australis[J]. BMC Genet, 2010, 11: 9. DOI:10.1186/1471-2156-11-9 |
| [23] |
McKinney ML. Urbanization as a major cause of biotic homogenization[J]. Biol Conser, 2006, 127(3): 247-260. DOI:10.1016/j.biocon.2005.09.005 |
| [24] |
Midgley GF, Hughes GO, Thuiller W, et al. Migration rate limitations on climate change-induced range shifts in Cape Proteaceae[J]. Divers Distrib, 2006, 12(5): 555-562. DOI:10.1111/j.1366-9516.2006.00273.x |
| [25] |
Brouat C, Loiseau A, Kane M, et al. Population genetic structure of two ecologically distinct multimammate rats:the commensal Mastomys natalensis and the wild Mastomys erythroleucus in southeastern Senegal[J]. Mol Ecol, 2007, 16(14): 2985-2997. DOI:10.1111/j.1365-294X.2007.03353.x |
| [26] |
Hutchison DW, Templeton AR. Correlation of pairwise genetic and geographic distance measures:inferring the relative influences of gene flow and drift on the distribution of genetic variability[J]. Evol, 1999, 53(6): 1898-1914. DOI:10.1111/j.1558-5646.1999.tb04571.x |
| [27] |
Musser GG, Carleton MD. Superfamily muroidea[M]//Wilson DE, Reeder DM. Mammal species of the world: a taxonomic and geographic reference. Baltimore, Maryland: Johns Hopkins University Press, 2005: 894-1531.
|
| [28] |
Chen WC, Li Y, Liu Y, et al. Complex topographic configuration in the Hengduan Mountains shaped the phylogeographic structure of Chinese white-bellied rats[J]. J Zool, 2011, 284(3): 215-223. DOI:10.1111/j.1469-7998.2011.00797.x |
| [29] |
Ge DY, Lu L, Cheng JL, et al. An endemic rat species complex is evidence of moderate environmental changes in the terrestrial biodiversity Centre of China through the late Quaternary[J]. Sci Rep, 2017, 7: 46127. DOI:10.1038/srep46127 |
| [30] |
Guo S, Li GC, Liu JL, et al. Dispersal route of the Asian house rat (Rattus tanezumi) on mainland China:insights from microsatellite and mitochondrial DNA[J]. BMC Genet, 2019, 20: 11. DOI:10.1186/s12863-019-0714-3 |
 2020, Vol. 31
2020, Vol. 31