扩展功能
文章信息
- 邱德义, 廖俊蕾, 魏晓雅, 岳巧云
- QIU De-yi, LIAO Jun-lei, WEI Xiao-ya, YUE Qiao-yun
- 基于DNA条形码丽蝇科49种常见种的分类鉴定
- Identification of 49 common fly species in Calliphoridae based on DNA barcodes
- 中国媒介生物学及控制杂志, 2017, 28(4): 322-331
- Chin J Vector Biol & Control, 2017, 28(4): 322-331
- 10.11853/j.issn.1003.8280.2017.04.005
-
文章历史
- 收稿日期: 2017-04-17
- 网络出版时间: 2017-06-12 16:24
2 中山大学生命科学学院, 广东 广州 510275
2 Life College of Sun Yat-sen University
丽蝇科(Calliphoridae)为双翅目(Diptera)下较大的类群,已知47属,约1 100种,其中中国有45属,约270种[1-2]。丽蝇科绝大多数种类是居住区蝇种,与人、畜关系非常密切[3-4]并传播疾病[5],至少有80种丽蝇能引起蝇蛆症,在医学研究中有重要意义,其嗜尸性在法医昆虫学研究中有重要地位[4, 6]。目前,丽蝇科属的鉴定主要是根据成虫的外部形态[7],而种的鉴定大多依据雄性外生殖器形态,特别是阳体形态[1]。在检验检疫或法医学等研究中,残体、幼虫、卵、蛹或雌性个体等蝇类无法根据形态特征进行鉴定[8]。大多数研究表明,DNA条形码技术(DNA barcoding)可对该类个体进行鉴定[9-12],是目前最有效的分子鉴定方法[13-17],基本原理是以线粒体细胞色素C氧化酶亚基Ⅰ(COⅠ)基因片段作为大多数物种的通用分子标记[13, 18],当COⅠ不足以区分亲缘关系较近的某些物种时,一般以ITS2作为辅助基因,国际专用DNA条形码数据库中仅接收以COⅠ和ITS2作为标准的动物种类鉴定DNA条形码序列(http://www.boldsystems.org/),而进化速率较快的cytb等基因一般用于亚种、姊妹种或不同地理种群间的分子溯源[19-20]。
DNA条形码技术被广泛应用于不同生物类群鉴定中,如节肢动物[16, 21-24]、丽蝇[9, 25-27]、麻蝇[28]和寄蝇[29]等双翅目昆虫,包括蝇蛹皮的鉴定[30-31]。国内有些应用对DNA条形码概念理解不准确,与其他分子鉴定标记概念相混淆,主要是应用cytb、COⅡ、16S rDNA及核基因等进化速率过快或过慢的基因片段进行种类鉴定,及利用DNA条形码鉴定物种的差异度标准应用于其他基因,忽视了不同基因片段进化速率不同的问题,且在利用DNA条形码对物种进行比对时过多地依赖美国国立生物技术信息中心(NCBI)的GenBank数据库,对DNA条形码专业数据库BOLD的认识和使用不足,均会降低物种鉴定的准确度,因此,需统一观念和鉴定标准。大多数研究证明,DNA条形码技术突破了传统物种鉴定方法对经验的过度依赖,实现了利用标准基因片段对物种进行快速准确地鉴定,是传统分类学的有力补充[32]。
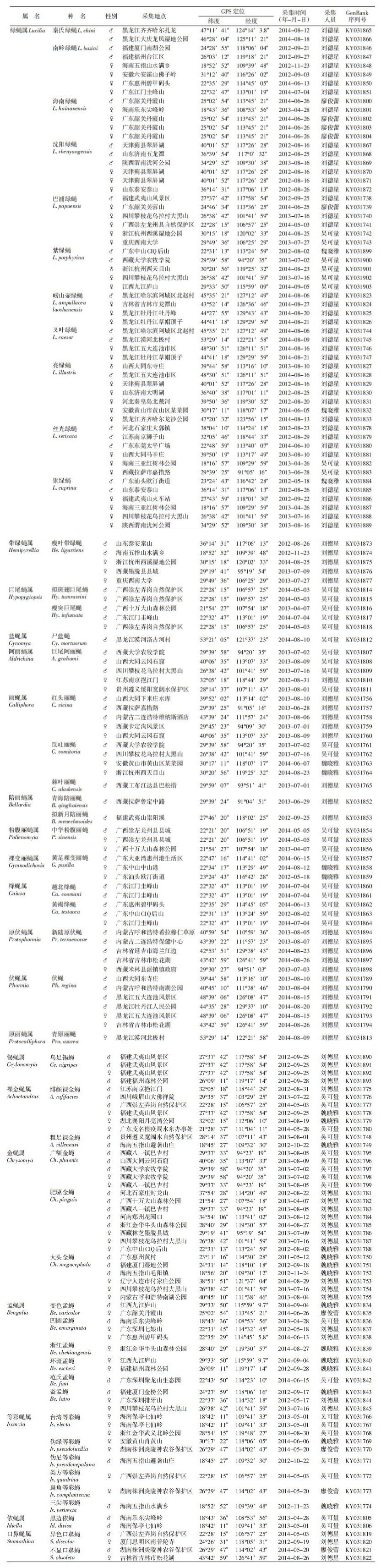
1 材料与方法 1.1 标本来源① 于2012-2015年采自全国各地,成蝇以网捕为主,同时辅以笼诱。网捕的成蝇立即用毒瓶熏死,转移至另一洁净离心管中短暂保存;笼诱法捕获的成蝇放入冰箱冷冻30 min后取出。标本一般于当天处理,制作成针插标本并记录,现保存于中山出入境检验检疫局媒介生物标本室;② 中山大学生物博物馆馆藏标本。
1.2 形态学鉴定参照文献[1, 33-34],利用蔡司V12体视显微镜对成蝇的外部形态进行观察并鉴定。1.3 DNA条形码序列的获得每只蝇各取2条后足置1.5 ml离心管中,用200 μl PBS缓冲液洗涤2次,放入95%乙醇中。拔掉后腿的蝇标本制作成针插凭证标本,作好唯一性标识,保存于中山出入境检验检疫局媒介生物标本室,备用。
1.4 PCR基因组DNA的提取、扩增、PCR产物纯化及测序方法见文献[35]。扩增引物由宝生物工程(大连)有限公司合成,为多细胞无脊椎动物COⅠ的通用引物:LOC1490 5′ -GGT CAA CAA ATC ATA AAG ATA TTG G-3′;HCO2198 5′ -TAA ACT TCA GGG TGA CCA AAA AAT CA-3′ [36]。Ex⁃Taq DNA聚合酶购自宝生物工程(大连)有限公司;dNTP、Loading Buffer、DNA Marker Ⅱ和SYBR GreenⅠ等试剂均购自天根生化科技(北京)有限公司。PCR产物送上海立菲生物技术有限公司进行双向测序。将拼接后的序列去掉引物部分,用蛋白质翻译等工具验证编码阅读框的正确性及校对序列的有效性。
1.5 序列的上传及分析将166条序列上传至GenBank,每条序列及其凭证标本详细信息见表 1,并在GenBank中引入相关21种丽蝇的49条序列共同分析(表 2)。利用Mega 6.06软件[37],以Kimura 2-parameter模型计算碱基差异度,以邻接法(Neighbor-joining method,NJ)构建系统树。

|
共获得丽蝇科20属49种长度为658 bp的COⅠ序列166条,其中雄虫82条,雌虫84条,秦氏绿蝇等28个种的DNA条形码序列在NCBI的GenBank或BOLD中未找到相应序列,为本研究首次获得。经分析,所获得序列均无缺失和插入,可编码219个氨基酸,并从GenBank数据库中引用21种丽蝇的49条序列,共214条DNA条形码序列用于分析。
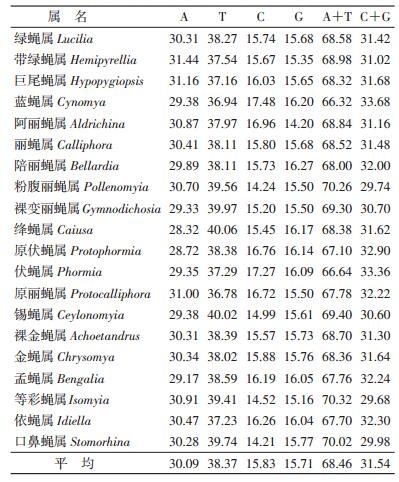
2.1.1 碱基组成分析丽蝇科平均碱基组成为A(30.09%)、T(38.37%)、G(15.71%)、C(15.83%),(A+T)(68.46%)远高于(G+C)(31.54%),见表 3。与关于丽蝇科[38]和其他双翅目昆虫[13, 39]线粒体序列的研究结果基本一致。
|
丽蝇科种内碱基差异度平均值为0.34%,49种丽蝇的种内差异度几乎均<1%,部分种类如南岭绿蝇(0~2.02%)、巴浦绿蝇(0~1.23%)和盗孟蝇(0.15%~1.54%)均在1.00%~ 2.00%之间,与DNA条形码鉴定物种差异度<2%的原则基本相符。南岭绿蝇种内碱基差异度为2.02%,略>2%,经分析发现,与GenBank中的序列KC249692和KC249693有关,可能因上传数据种类鉴定的准确度较低所致。同种丽蝇雌雄个体间的种内碱基差异度与同性个体间的碱基差异度相当,无明显差别,均在种内差异度范围内。
丽蝇科属间、种间碱基差异度如表 4所示,同属种间碱基差异度平均值几乎均>2%,如绿蝇属(4.24%)、巨尾蝇属(8.03%)、丽蝇属(4.26%)、裸金蝇属(4.17%)、孟蝇属(8.13%)和等彩蝇属(6.97%)。绿蝇属的某些种间差异度相对较低(0.44%~7.93%),与Sonet等[40]用511 bp的COⅠ序列计算的数值(1.40%~8.80%)相近,与两者有相似的形态学特征相对应。绿蝇属亚属的紫绿蝇与壶绿蝇(0.77%~1.08%,平均值0.83%)、叉叶绿蝇与亮绿蝇(0.77%~1.86%,平均值1.18%)、丝光绿蝇与铜绿蝇(0.46%~0.92%,平均值0.62%),金蝇属的广额金蝇与肥躯金蝇(0.15%~0.61%),绛蝇属的越北绛蝇与黄褐绛蝇(0.92%~1.23%),其种间碱基差异度与各自的种内碱基差异度相近或出现重合,说明绿蝇属、金蝇属和绛蝇属不同种间分化年代较近,甚至不能称之为不同种,可能是姊妹种或不同地理亚种。形态差异较大的广额金蝇与肥躯金蝇种间碱基差异度的平均值仅为0.29%,与各自的种内碱基差异度相当,且在658 bp的COⅠ序列中仅在碱基位点上出现固定的核苷酸差异,说明应用该段序列不能用于两物种DNA条形码的有效鉴定,可能需要增加其他基因片段,如核基因ITS2或cytb等。该部分的研究结果将在后续的研究中报道。

|
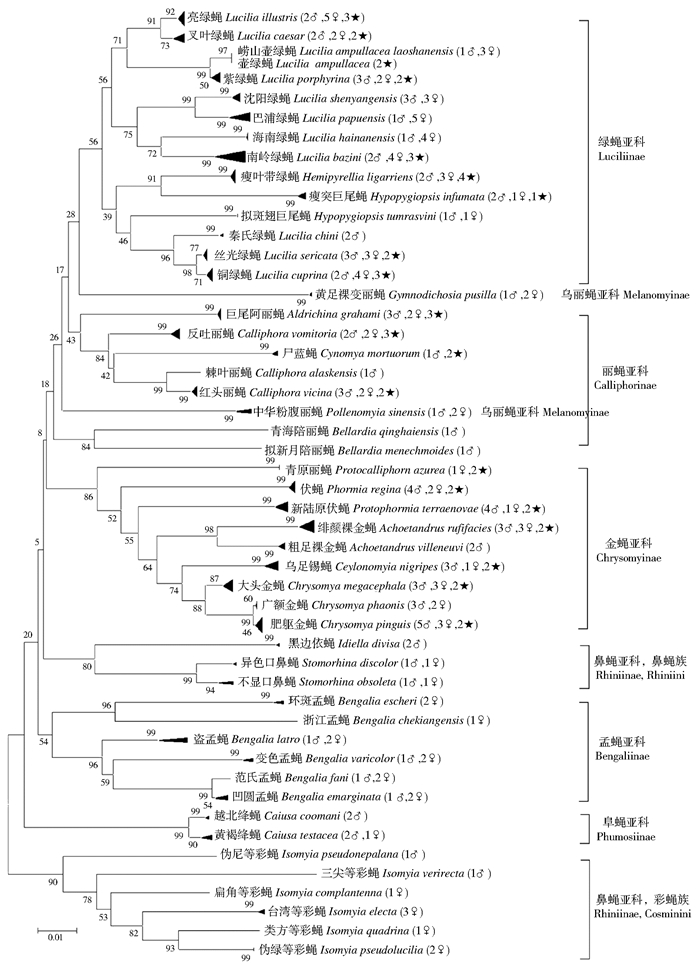
系统树结果如图 1所示,相同物种的不同个体均聚在同一分支下,且相同亚科不同属也聚在同一大支,鼻蝇亚科的彩蝇族和鼻蝇族除外,但两个族分别聚在同一分支,支持度分别为90%和80%,每个支序均为同族丽蝇。在绿蝇亚科中,带绿蝇属和巨尾蝇属与绿蝇属混在不同的支序内,但因支持度不高(<50%),无法说明其亲缘关系,符合关于COⅠ基因不适合进行系统进化关系分析的结论,对上述两属并入绿蝇属的现象,与Wells等[27]研究结果相似,并提出带绿蝇属是否应从绿蝇属种区分开的质疑。在丽蝇亚科中,除陪丽蝇属的5种丽蝇外均聚在同一支序下,其中尸蓝蝇较反吐丽蝇先与丽蝇属的红头丽蝇和棘叶丽蝇聚成一支(支持度为42%),说明其可能与该两种蝇的亲缘关系较反吐丽蝇近。而陪丽蝇属的青海陪丽蝇和拟新月陪丽蝇在系统发育树中介于丽蝇亚科与金蝇亚科之间且种类数量不足,无法确切地判断其进化位置。乌丽蝇亚科的2属2种分别处于NJ树的不同分支,支持度均<30%,无法判断其亲缘关系,与Weils和Sperling[41]研究结果相互验证。在金蝇亚科中,9种丽蝇均为单系群,同处于一个支序下。金蝇属的大头金蝇、广额金蝇和肥躯金蝇最先聚为一支(支持度为88%),其中广额金蝇与肥躯金蝇碱基差异度最小(支持度为99%);然后乌足锡蝇与绯颜裸金蝇和粗足裸金蝇依次聚在一起,再与新陆原伏蝇、伏蝇和青原丽蝇聚在一起。在孟蝇亚科中,6种孟蝇分为两支,赭孟蝇亚属的环斑孟蝇与浙江孟蝇聚在一起,而优孟蝇亚属的盗孟蝇、变色孟蝇、范氏孟蝇和凹圆孟蝇则聚于另一分支,凹圆孟蝇与范氏孟蝇的碱基差异度很小(支持度为99%)。6种孟蝇分别处在两个相邻分支上,说明浙江孟蝇与环斑孟蝇的碱基差异度较小,而盗孟蝇、变色孟蝇、范氏孟蝇和环斑孟蝇的碱基差异度较小,与形态学研究结果相一致。
|
| 注:♂.雄性丽蝇序列;♀.雌性丽蝇序列;★. GenBank引用的序列。进化树三角形的大小代表每个物种含有个体数目的多少 图 1 基于DNA条形码构建的NJ系统进化树(Kimura 2-parameter模型) Figure 1 Neighbor-joining tree based on 658 bp DNA barcodes(Kimura 2-parameter model) |
| |
对获得的序列进行比对发现,同种蝇类的相似度均>99%,而不同种间相似度随形态学差异的增加而降低,说明DNA条形码技术适合种类的鉴定。所得序列的种间距离(>2%)大于种内距离(<1%),与Reibe等[42]研究结果一致。
由NJ系统发育树可知,丽蝇科中的绿蝇亚科、丽蝇亚科、金蝇亚科、孟蝇亚科和阜蝇亚科均为单系类群(monophyletic group),与Renaud等[39]对德国常见法医丽蝇种类的研究结果一致。本研究获得的DNA条形码和GenBank序列中同种蝇均聚在一起,说明COⅠ序列不适用于样本的分子溯源,与Harvey等[9]研究结论一致。
应用DNA条形码对丽蝇科常见种类的鉴定结果与形态学分类结果一致,证明了以COⅠ基因5′一段长度为658 bp的基因片段作为标准片段的DNA条形码可准确地鉴定大部分丽蝇科种类。DNA条形码是传统分类学的有力补充,特别针对昆虫残体或幼虫等非成虫态的成体及雌性个体均可进行鉴定,并可提高物种鉴定的准确度,对国境口岸准确快速地鉴定媒介生物种类有积极意义,省去实验室孵化培育的过程,从而减少了媒介生物及其携带病原体的扩散机会,对公共卫生安全有重大意义。
| [1] |
范滋德, 陈之梓, 方建明, 等. 中国动物志.昆虫纲.第6卷.双翅目:丽蝇科[M]. 北京: 科学出版社, 1997, 120-125.
|
| [2] |
吴薇, 夏德峰, 郑炜, 等. 浙江省丽蝇科区系研究[J]. 广东农业科学, 2014, 41(9): 88-90. |
| [3] |
Crosskey RW, Lane RP. House-flies, blow-flies and their allies (Calyptrate:Diptera)[M]//Lane RP, Crosskey RW. Medical Insects and Arachnids. London:Chapman and Hall, 1993:403-428.
|
| [4] |
Hall M, Wall R. Myiasis of humans and domestic animals[J]. Adv Parasitol, 1995, 35: 257-334. DOI:10.1016/S0065-308X(08)60073-1 |
| [5] |
Sherman RA, Hall MJR, Thomas S. Medicinal maggots:an ancient remedy for some contemporary afflictions[J]. Annu Rev Entomol, 2000, 45(1): 55-81. DOI:10.1146/annurev.ento.45.1.55 |
| [6] |
Amendt J, Krettek R, Zehner R. Forensic entomology[J]. Naturwissenschaften, 2004, 91(2): 51-65. DOI:10.1007/s00114-003-0493-5 |
| [7] |
Rognes K. Blowflies (Diptera:Calliphoridae) of Fennoscandia and Denmark[M]. New York: Brill Academic Publishers, 1991, 1-272.
|
| [8] |
Stevens J, Wall R. Species, sub-species and hybrid populations of the blowflies Lucilia cuprina and Lucilia sericata (Diptera:Calliphoridae)[J]. Proc Roy Biol Soc B, 1996, 263(1375): 1335-1341. DOI:10.1098/rspb.1996.0196 |
| [9] |
Harvey ML, Dadour IR, Gaudieri S. Mitochondrial DNA cytochrome oxidase I gene:potential for distinction between immature stages of some forensically important fly species (Diptera) in western Australia[J]. Forensic Sci Int, 2003, 131(2/3): 134-139. |
| [10] |
Chen WY, Hung TH, Shiao SF. Molecular identification of forensically important blow fly species (Diptera:Calliphoridae) in Taiwan[J]. J Med Entomol, 2004, 41(1): 47-57. DOI:10.1603/0022-2585-41.1.47 |
| [11] |
Saigusa K, Takamiya M, Aoki Y. Species identification of the forensically important flies in Iwate prefecture, Japan based on mitochondrial cytochrome oxidase gene subunitⅠ (COⅠ) sequences[J]. Legal Med, 2005, 7(3): 175-178. DOI:10.1016/j.legalmed.2005.01.004 |
| [12] |
Wells JD, Stevens JR. Application of DNA-based methods in forensic entomology[J]. Annu Rev Entomol, 2008, 53(1): 103-120. DOI:10.1146/annurev.ento.52.110405.091423 |
| [13] |
Hebert PDN, Cywinska A, Ball SL, et al. Biological identifications through DNA barcodes[J]. Proc Biol Sci, 2003, 270(1512): 313-321. DOI:10.1098/rspb.2002.2218 |
| [14] |
Hebert PDN, Ratnasingham S, deWaard JR. Barcoding animal life:cytochrome c oxidase subunit 1 divergences among closely related species[J]. Pro Biol Sci, 2003, 270(Suppl 1): S96-99. |
| [15] |
Hebert PDN, Penton EH, Burns JM, et al. Ten species in one:DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astraptes fulgerator[J]. Proc Natl Acad Sci USA, 2004, 101(41): 14812-14817. DOI:10.1073/pnas.0406166101 |
| [16] |
Hajibabaei M, Janzen DH, Burn JM, et al. DNA barcodes distinguish species of tropical Lepidoptera[J]. Proc Natl Acad Sci USA, 2006, 103(4): 968-971. DOI:10.1073/pnas.0510466103 |
| [17] |
Ratnasingham S, Hebert PDN. BOLD:the barcode of life data system (http://www.barcodinglife.org)[J]. Mol Ecol Res, 2007, 7(3):355-364.
|
| [18] |
Stoeckle M. Taxonomy, DNA, and the barcode of life[J]. Bio Sci, 2003, 53(9): 796-797. |
| [19] |
GilArriortua M, Bordas MIS, Cainé LM, et al. Cytochrome b as a useful tool for the identification of blowflies of forensic interest (Diptera:Calliphoridae)[J]. Forensic Sci Int, 2013, 228(1/3): 132-136. |
| [20] |
Rolo EA, Oliveira AR, Dourado CG, et al. Identification of sarcosaprophagous Diptera species through DNA barcoding in wildlife forensics[J]. Forensic Sci Int, 2013, 228(1/3): 160-164. |
| [21] |
Will KW, Rubinoff D. Myth of the molecule:DNA barcodes for species cannot replace morphology for identification and classification[J]. Cladistics, 2004, 20(1): 47-55. DOI:10.1111/cla.2004.20.issue-1 |
| [22] |
Monaghan MT, Balke M, Gregory TR, et al. DNA-based species delineation in tropical beetles using mitochondrial and nuclear markers[J]. Philos Trans R Soc Lond B Biol Sci, 2005, 360(1462): 1925-1933. DOI:10.1098/rstb.2005.1724 |
| [23] |
Foley DH, Wilkerson RC, Cooper RD, et al. A molecular phylogeny of Anopheles annulipes (Diptera:Culicidae) sensu lato:the most species-rich anopheline complex[J]. Mol Phylogenet Evol, 2007, 43(1): 287-297. |
| [24] |
Blacket MJ, Semeraro L, Malipatil MB. Barcoding Queensland fruit flies (Bactrocera tryoni):impediments and improvements[J]. Mol Ecol Res, 2012, 12(3): 428-436. DOI:10.1111/men.2012.12.issue-3 |
| [25] |
Nelson LA, Wallman JF, Dowton M. Using COⅠ barcodes to identify forensically and medically important blowflies[J]. Med Vet Entomol, 2007, 21(1): 44-52. DOI:10.1111/mve.2007.21.issue-1 |
| [26] |
Wallman JF, Donnellan SC. The utility of mitochondrial DNA sequences for the identification of forensically important blowflies (Diptera:Calliphoridae) in southeastern Australia[J]. Forensic Sci Int, 2001, 120(1/2): 60-67. |
| [27] |
Wells JD, Wall R, Stevens JR. Phylogenetic analysis of forensically important Lucilia flies based on cytochrome oxidaseⅠ sequence:a cautionary tale for forensic species determination[J]. Int J Legal Med, 2007, 121(3): 229-233. DOI:10.1007/s00414-006-0147-1 |
| [28] |
Meiklejohn KA, Wallman JF, Dowton M. DNA-based identification of forensically important Australian Sarcophagidae (Diptera)[J]. Int J Legal Med, 2011, 125(1): 27-32. DOI:10.1007/s00414-009-0395-y |
| [29] |
Smith MA, Woodley NE, Janzen DH, et al. DNA barcodes reveal cryptic host-specificity within the presumed polyphagous members of a genus of parasitoid flies (Diptera:Tachinidae)[J]. Proc Natl Acad Sci USA, 2006, 103(10): 3657-3662. DOI:10.1073/pnas.0511318103 |
| [30] |
岳巧云, 邱德义, 黄艺文, 等. DNA条形码技术在未知昆虫幼虫种类鉴定中的应用[J]. 中国卫生检验杂志, 2011, 21(3): 615-617. |
| [31] |
刘德星, 聂维忠, 邱德义, 等. 应用DNA条形码技术快速鉴定入境船舶上截获的昆虫蛹[J]. 检验检疫学刊, 2014, 24(5): 53-57. |
| [32] |
Besansky NJ, Severson DW, Ferdig MT. DNA barcoding of parasites and invertebrate disease vectors:what you don't know can hurt you[J]. Trends Parasitol, 2003, 19(12): 545-546. DOI:10.1016/j.pt.2003.09.015 |
| [33] |
范滋德. 中国常见蝇类检索表[M]. 2版. 北京: 科技出版社, 1992, 457-580.
|
| [34] |
薛万琦, 赵建铭. 中国蝇类[M]. 沈阳: 辽宁科学技术出版社, 1998, 1366-1506.
|
| [35] |
Yue QY, Wu KL, Qiu DY, et al. A formal re-description of the cockroach Hebardina concinna anchored on DNA barcodes confirms wing polymorphism and identifies morphological characters for field identification[J]. PLoS One, 2014, 9(9): e106789. DOI:10.1371/journal.pone.0106789 |
| [36] |
Folmer O, Black M, Hoeh W, et al. DNA primers for amplification of mitochondrial cytochrome c oxidase subunitⅠ from diverse metazoan invertebrates[J]. Mol Mar Biol Biotechnol, 1994, 3(5): 294-299. |
| [37] |
Kumar S, Nei M, Dudley J, et al. MEGA:a biologist-centric software for evolutionary analysis of DNA and protein sequences[J]. Brief Bioinform, 2008, 9(4): 299-306. DOI:10.1093/bib/bbn017 |
| [38] |
GilArriortua M, Bordas MIS, K-hnemann S, et al. Molecular differentiation of central European blowfly species (Diptera:Calliphoridae) using mitochondrial and nuclear genetic markers[J]. Forensic Sci Int, 2014, 242: 274-282. DOI:10.1016/j.forsciint.2014.07.018 |
| [39] |
Renaud AK, Savage J, Adamowicz SJ. DNA barcoding of northern Nearctic Muscidae (Diptera) reveals high correspondence between morphological and molecular species limits[J]. BMC Ecol, 2012, 12: 24. DOI:10.1186/1472-6785-12-24 |
| [40] |
Sonet G, Jordaens K, Braet Y, et al. Why is the molecular identification of the forensically important blowfly species Lucilia caesar and L. illustris (family Calliphoridae) so problematic?[J]. Forensic Sci Int, 2012, 223(1/3): 153-159. |
| [41] |
Wells JD, Sperling FAH. DNA-based identification of forensically important Chrysomyinae (Diptera:Calliphoridae)[J]. Forensic Sci Int, 2001, 120(1/2): 110-115. |
| [42] |
Reibe S, Schmitz J, Madea B. Molecular identification of forensically important blowfly species (Diptera:Calliphoridae) from Germany[J]. Parasitol Res, 2009, 106(1): 257-261. DOI:10.1007/s00436-009-1657-9 |
 2017, Vol. 28
2017, Vol. 28