扩展功能
文章信息
- 邱德义, 魏晓雅, 吴可量, 岳巧云
- QIU De-yi, WEI Xiao-ya, WU Ke-liang, YUE Qiao-yun
- 德国小蠊及其近缘种的形态和分子鉴定
- Morphological and molecular identification of Blattella germanica and its closely related species
- 中国媒介生物学及控制杂志, 2017, 28(3): 220-225
- Chin J Vector Biol & Control, 2017, 28(3): 220-225
- 10.11853/j.issn.1003.8280.2017.03.006
-
文章历史
- 收稿日期: 2016-12-29
- 网络出版时间: 2017-04-11 19:01
德国小蠊(Blattella germanica)现除南极洲外世界各地均广泛分布,与人类生活密切相关[1]。德国小蠊可携带大肠埃希菌(Escherichia coli)、铜绿假单胞菌(Pseudomonas aeruginosa)等肠道性病原体及肝炎病毒等,诱发某些易感人群哮喘或过敏[2]。德国小蠊体型小,易藏身于狭小的缝隙中,且抗药性强,较难防治[1, 3]。小蠊属除德国小蠊外还存在一些近缘种,如拟德国小蠊(B. lituricollis)、双纹小蠊(B. bisignata)、毛背小蠊(B. sauteri)和单尾小蠊(B. ingularis)等,这些近缘种形态相似,尤其是德国小蠊、拟德国小蠊和双纹小蠊难以区分。雄性外生殖器特征一直被视为昆虫种类鉴定的金标准[4-5],但实际应用中存在一定不便。自2003年Hebert等[6]首次提出DNA条形码概念以来,DNA条形码已广泛应用于多种动物类群的种类鉴定[7-10],是一种非常有效的种类鉴定工具[11-12]。本研究以DNA条形码和形态学鉴定相结合的方式,对德国小蠊及其近缘种进行准确鉴定。
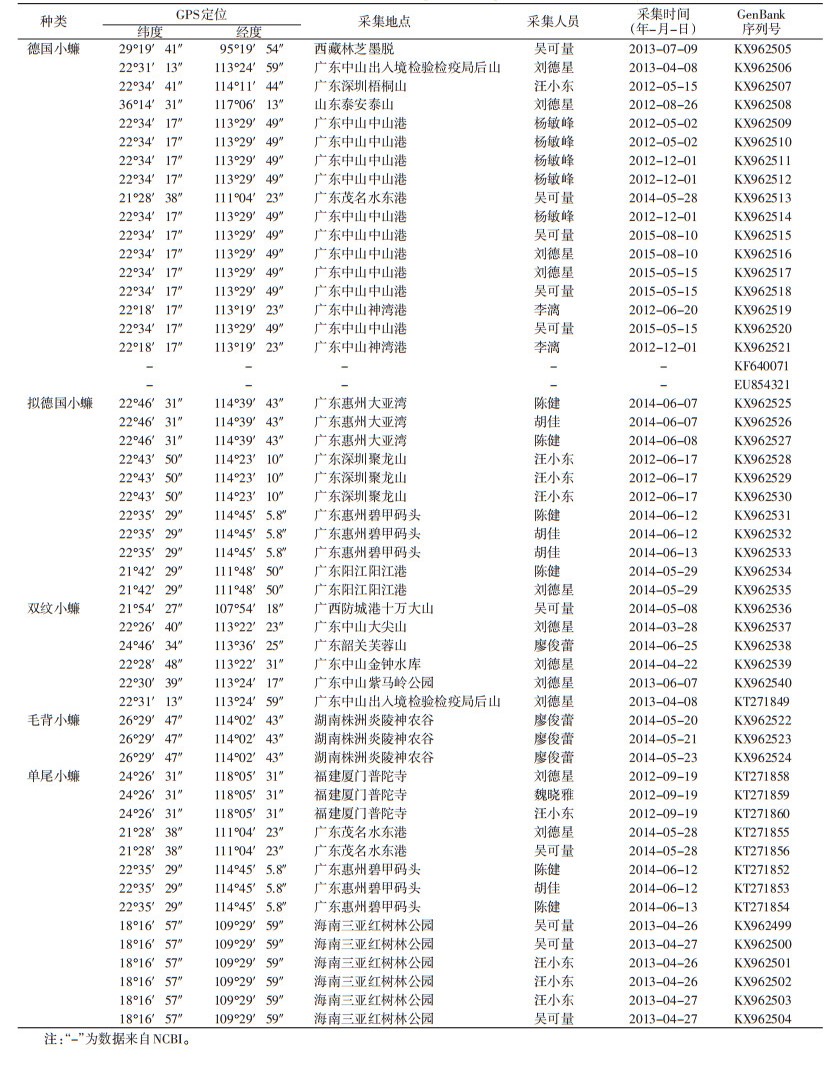
1 材料与方法 1.1 标本的收集2012-2015年共采集不同来源的德国小蠊17只、拟德国小蠊11只、双纹小蠊6只、毛背小蠊3只和单尾小蠊14只,采集时间、地点和序列等信息见表 1。将其制成针插标本存放在国家医学媒介生物监测重点实验室(广东省中山市)。
|
利用蔡司V12体式显微镜观察小蠊外部形态特征,用佳能60D单反相机进行拍照。尾器的解剖需将雄性蜚蠊的生殖节切下并用10%KOH溶液消化30 min后,移入0.9%氯化钠溶液中,仔细剔除阳茎周围的肌肉等组织,在蔡司V12显微镜上安装佳能PowerShot G1X卡片机,观察并拍照,然后用Adobe Illustrator CS6以拍摄的照片为蓝本进行绘图。
1.3 DNA条形码鉴定每只小蠊各取1条后足胫节放入1.5 ml离心管中,用200 μl PBS缓冲液冲洗2次,放入95%乙醇中。拔掉后腿的小蠊标本做成针插的凭证标本,作好唯一性标识,保存在中山出入境检验检疫局媒介生物标本室,备用。
基因组DNA的提取、扩增、PCR产物的纯化及测序方法参照Yue等[13]方法。扩增引物为LCO1490:5′-GGT CAA CAA ATC ATA AAG ATA TTG G-3′;HCO2198:5′-TAA ACT TCA GGG TGA CCA AAA AAT CA-3′[14],由宝生物(大连)有限公司合成。EX-Taq DNA聚合酶、dNTP及其他PCR试剂均购自宝生物(大连)有限公司。
采用PCR产物直接进行双向测序,将拼接后的序列去掉引物部分,用蛋白质翻译等工具验证编码阅读框正确性及校对序列的有效性。以2条美洲大蠊(Periplaneta americana)的细胞色素C氧化酶亚基Ⅰ(COⅠ)序列为外源,5种小蠊的51条COⅠ序列和2条来自GenBank的德国小蠊序列,利用Mega 6.0软件构建Neighbor-Joining(NJ)树,重复步数为5 000。
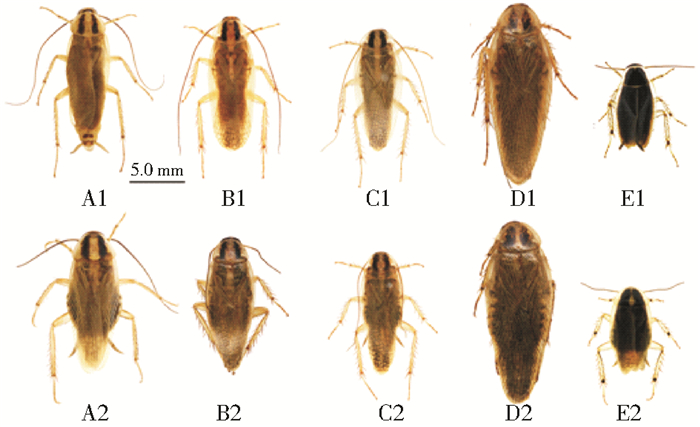
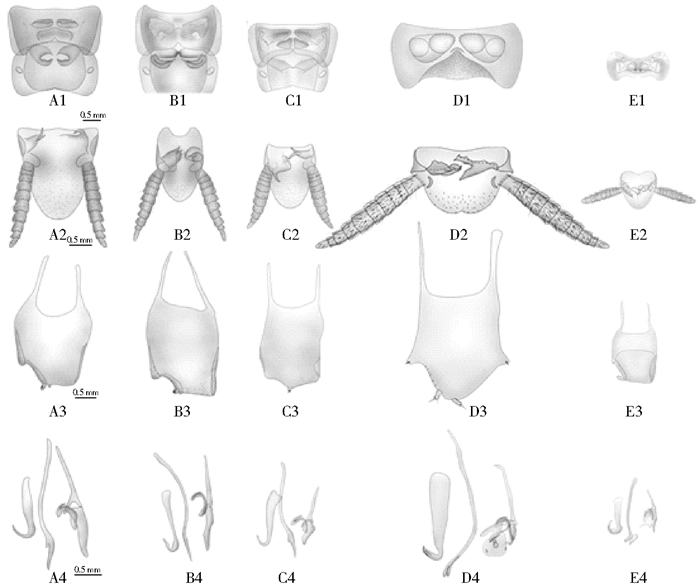
2 结果 2.1 形态特征德国小蠊及其近缘种的外部形态见图 1,其近缘种的雄性尾器见图 2,雄性尾器特征描述见表 2。
|
| 注:A.德国小蠊;B.拟德国小蠊;C.双纹小蠊;D.毛背小蠊;E.单尾小蠊。1.雄虫;2.雌虫。 图 1 德国小蠊及其近缘种的背面观 Figure 1 Dorsal view of B. germanica and its closely related species |
| |
|
| 注:A.德国小蠊;B.拟德国小蠊;C.双纹小蠊;D.毛背小蠊;E.单尾小蠊。1.腹部第7、8背板背面观;2.肛上板腹面观;3.下生殖板背面观;4.左中右阳茎背面观。 图 2 德国小蠊及其近缘种的雄性尾器 Figure 2 Morphology of the male genitalia among B. germanica and its closely related species |
| |
对5种小蠊的51条COⅠ扩增产物进行序列测序分析,与2条来自GenBank的德国小蠊序列共53条序列凭证标本的采集信息及GenBank序列号见表 1。GenBank和DNA条形码BOLD数据库中无拟德国小蠊、双纹小蠊、毛背小蠊和单尾小蠊的DNA条形码序列,因此,该4种小蠊的序列全部为原始获得。
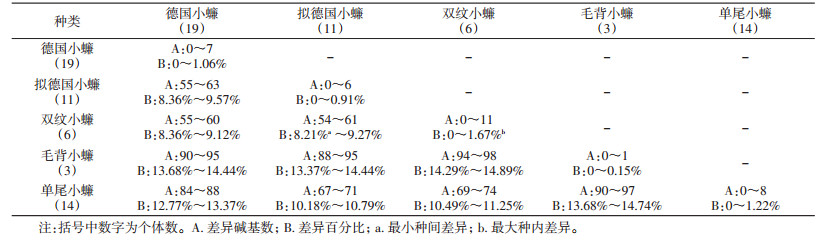
经序列比对分析发现,5种小蠊最大种内差异为1.67%,小于种类鉴定界限(2%);最小种间差异为8.21%,高于限值(2%),种内个体间与种间有明显的差异间隔。5种小蠊的种间及种内碱基差异见表 3。
|
德国小蠊及其近缘种均聚在同一支内,种间无交叉,见图 3,说明DNA条形码用于德国小蠊及其近缘种的鉴定非常准确。

|
| 图 3 德国小蠊及其近缘种的NJ进化树 Figure 3 NJ tree of B. germanica and its closely related species |
| |
德国小蠊及其近缘种外部形态非常相似,很难准确鉴定到种。前胸背板两条纵条纹的相对宽窄及曲直以及面部的斑纹常作为种类鉴定的依据,但这些表征在不同个体间不稳定,常发生变异。雄性外生殖器的形态特征一直是德国小蠊及其近缘种种类鉴定的金标准,但因其需解剖,在实际应用中存在一定限制。近年来,DNA条形码技术逐渐兴起,是物种鉴定的有力工具,为形态学鉴定的有力补充。德国小蠊及其近缘种DNA条形码的种内最大差异小于限值(2%),而最小种间差异为8.21%,明显高于限值(2%),与种间差异>2%的报道一致[15]。
形态学鉴定在许多生物学研究中仍被普遍使用,其地位在短时期内无法被其他学科所取代[16]。多数研究及事实证明,DNA条形码与形态特征相结合的分类学方法可为传统的鉴定方法带来光明的发展前景[17-19]。
| [1] | 冯平章, 郭予元, 吴福桢. 中国蟑螂种类及防治[M]. 北京: 中国科学技术出版社, 1997: 4-15. |
| [2] | 胡修元. 蜚蠊可能引起/传播的疾病[J]. 中国媒介生物学及控制杂志, 1999, 10(3): 168–169. |
| [3] | 吴福桢. 中国常见蜚蠊种类及其为害、利用与防治的调查研究[J]. 昆虫学报, 1987, 30(4): 430–438. |
| [4] | 范滋德. 中国常见蝇类检索表[M]. 2版. 北京: 科学出版社, 1992: 580-719. |
| [5] | 薛万琦, 赵建铭. 中国蝇类[M]. 沈阳: 辽宁科学技术出版社, 1996: 1367-1656. |
| [6] | Hebert PDN, Cywinska A, Ball SL, et al. Biological identifications through DNA barcodes[J]. Proc Roy Soc B Biol Sci, 2003, 270(1512) : 313–321 .DOI:10.1098/rspb.2002.2218. |
| [7] | Blaxter M, Mann J, Chapman T, et al. Defining operational taxonomic units using DNA barcode data[J]. Philos Trans Roy Soc B Biol Sci, 2005, 360(1462) : 1935–1943 .DOI:10.1098/rstb.2005.1725. |
| [8] | Evans KM, Wortley AH, Mann DG. An assessment of potential diatom "barcode" genes(cox1, rbcL, 18S and ITS rDNA)and their effectiveness in determining relationships in Sellaphora (Bacillariophyta)[J]. Protist, 2007, 158(3) : 349–364 .DOI:10.1016/j.protis.2007.04.001. |
| [9] | Hebert PDN, Penton EH, Burns JM, et al. Ten species in one:DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astraptes fulgerator[J]. Proc Natl Acad Sci USA, 2004, 101(41) : 14812–14817 .DOI:10.1073/pnas.0406166101. |
| [10] | Hajibabaei M, Singer GAC, Clare EL, et al. Design and applicability of DNA arrays and DNA barcodes in biodiversity monitoring[J]. BMC Biol, 2007, 5(1) : 24.DOI:10.1186/1741-7007-5-24. |
| [11] | Yang ZH, Rannala B. Bayesian species delimitation using multilocus sequence data[J]. Proc Natl Acad Sci USA, 2010, 107(20) : 9264–9269 .DOI:10.1073/pnas.0913022107. |
| [12] | Bergsten J, Bilton DT, Fujisawa T, et al. The effect of geographical scale of sampling on DNA barcoding[J]. Syst Biol, 2012, 61(5) : 851–869 .DOI:10.1093/sysbio/sys037. |
| [13] | Yue QY, Wu KL, Qiu DY, et al. A formal re-description of the cockroach Hebardina concinna anchored on DNA barcodes confirms wing polymorphism and identifies morphological characters for field identification[J]. PLoS One, 2014, 9(9) : e106789.DOI:10.1371/journal.pone.0106789. |
| [14] | Folmer O, Black M, Hoeh W, et al. DNA primers for amplification of mitochondrial cytochromec oxidase subunit Ⅰ from diverse metazoan invertebrates[J]. Mol Mar Biol Biotechnol, 1994, 3(5) : 294–299 . |
| [15] | Wells JD, Pape T, Sperling FA. DNA-based identification and molecular systematics of forensically important Sarcophagidae (Diptera)[J]. J Forensic Sci, 2001, 46(5) : 1098–1102 . |
| [16] | Tautz D, Arctander P, Minelli A, et al. A plea for DNA taxonomy[J]. Trend Ecol Evol, 2003, 18(2) : 70–74 .DOI:10.1016/S0169-5347(02)00041-1. |
| [17] | Dayrat B. Towards integrative taxonomy[J]. Biol J Linn Soc, 2005, 85(3) : 407–415 .DOI:10.1111/bij.2005.85.issue-3. |
| [18] | Padial JM, Miralles A, De la Riva I, et al. Review:the integrative future of taxonomy[J]. Front Zool, 2010, 7(1) : 16.DOI:10.1186/1742-9994-7-16. |
| [19] | Bergsten J, Brilmyer G, Crampton-Platt A, et al. Sympatry and colour variation disguised well-differentiated sister species:Suphrodytes revised with integrative taxonomy including 5 kbp of housekeeping genes (Coleoptera:Dytiscidae)[J]. DNA Barcodes, 2012, 1(1) : 1–18 . |
 2017, Vol. 28
2017, Vol. 28