扩展功能
文章信息
- 岳巧云, 邱德义, 单振菊, 汪小东, 胡佳, 张文庆
- YUE Qiao-yun, QIU De-yi, SHAN Zhen-ju, WANG Xiao-dong, HU Jia, ZHANG Wen-qing
- 基于DNA条形码和子宫骨片形态特征的8种雌性麻蝇种类鉴定
- Identification of 8 female Sarcopha flies based on mitochondrial DNA and signum morphological characteristics
- 中国媒介生物学及控制杂志, 2016, 27(5): 436-442
- Chin J Vector Biol & Control, 2016, 27(5): 436-442
- 10.11853/j.issn.1003.8280.2016.05.004
-
文章历史
- 收稿日期: 2016-06-16
- 网络出版时间: 2016-08-11
2 中山出入境检验检疫局检验检疫技术中心, 广东 中山 528403
2 Zhongshan Entry-Exit Inspection and Quarantine Technology Center
麻蝇是一种潜在的病原生物的传播媒介[1-2]。传统麻蝇的鉴定主要根据其形态特征,特别是雄性个体的外生殖器,被视为种类鉴定的金标准[3-6]。而雌性麻蝇的形态鉴定非常复杂,不同种类雌性麻蝇个体间外部形态极为相似,仅根据其外部形态很难准确地鉴定[7]。
DNA条形码技术利用线粒体细胞色素C氧化酶亚基Ⅰ(COⅠ),为多细胞动物种类鉴定的参考标志[8]。自2003年Hebert等[9]提出DNA条形码概念以来,COⅠ基因已广泛地应用于多种动物的种类鉴定[10-13],且非常有效[14-15]。DNA条形码正在实施一个全球性的iBOL计划[16]。相同物种雌雄个体DNA条形码序列变异并未超过种内变异范围,通过比对雌雄性个体的DNA序列,可以准确地鉴定雌性个体。尽管大多研究者建议完全取消形态学鉴定,但其依然被使用且在短时期内无法被取代[17]。“整合分类学”[18-19]或DNA条形码数据与形态特征相结合的分类学方法,将促进传统分类学的研究发展[20]。
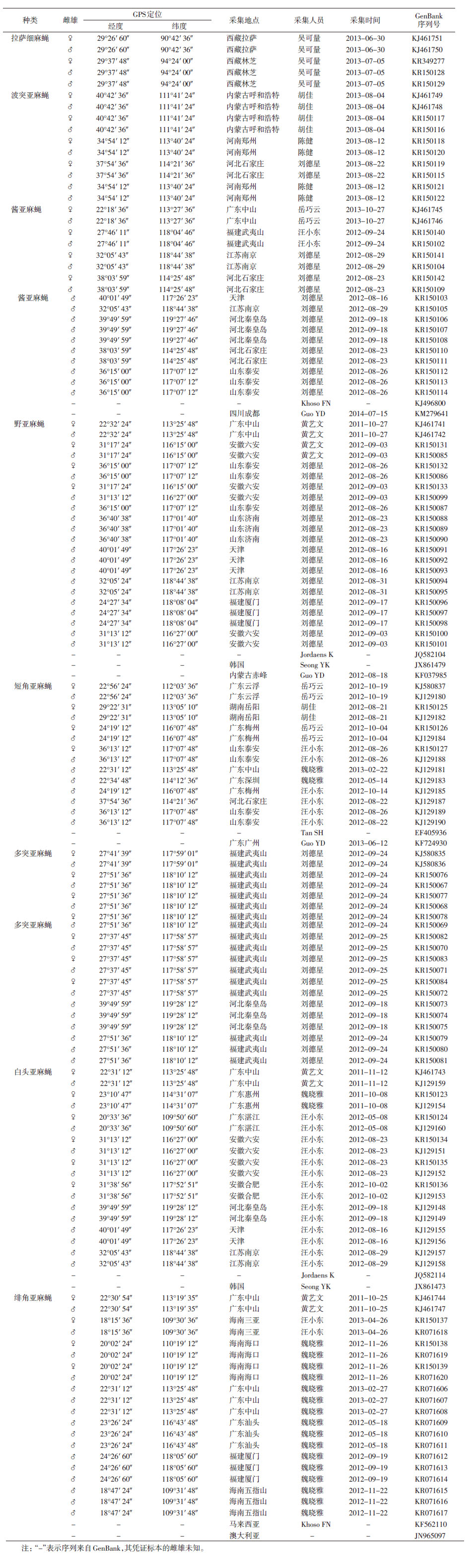
1 材料与方法 1.1 蝇的收集2011年10月25日至2013年10月27日,以腐鱼作诱饵用诱捕笼或者昆虫网共采集8种麻蝇127只,包括35对正在交配的蝇类〔其中6对白头亚麻蝇(Parasarcophaga albiceps),2对拉萨细麻蝇(Pierretia lhasae),4对波突亚麻蝇(P. jaroschevskyi),4对绯角亚麻蝇(P. ruficornis),7对多突亚麻蝇(P. polystylata),4对野亚麻蝇(P. similis),4对酱亚麻蝇(P. dux)和4对短角亚麻蝇(P. brevicornis)〕。将麻蝇置95%乙醇中或制成针插标本,标本存放在国家质检总局医学媒介生物监测重点实验室(中山)。
1.2 DNA条形码鉴定每只蝇各取1条后足放入1.5 ml离心管中,用200 μl PBS缓冲液洗2次,置入95%乙醇中。拔掉后腿的蝇标本做成针插的凭证标本,做好唯一性标识,保存于国家质检总局医学媒介生物监测重点实验室(中山),备用。基因组DNA的提取、扩增、PCR产物纯化及测序按文献[21]操作。扩增引物:LCO1490:5′-GGT CAA CAA ATC ATA AAG ATA TTG G-3′;HCO2198:5′-TAA ACT TCA GGG TGA CCA AAA AAT CA-3′[22]。由宝生物工程(大连)有限公司合成。EX-Taq DNA 聚合酶、dNTP 及其他PCR 试剂均购自宝生物工程(大连)有限公司。
8种雌性麻蝇的35对配对个体70条COⅠ序列,57条雄性个体的COⅠ序列和11条来自GenBank的性别未知个体的序列,共138条构建Neighbor-Joining(NJ)树。138条序列的凭证标本采集信息、雌雄及GenBank序列号见表 1。GenBank中无拉萨细麻蝇和波突亚麻蝇的序列,均为原始获得。利用Mega 6.0软件构建 NJ树,重复步数为5 000。
雄性个体的种类鉴定依据《中国蝇类》检索表进行鉴定,并经过专家确认。雌性个体的种类通过与之交配的雄性个体进行确认。分类术语依照文献[3, 5, 23]。
2 结果 2.1 8种雌性麻蝇的DNA条形码鉴定所有个体COⅠ片段均为658 bp,无终止密码子、插入或缺失片段,可无间断地翻译成219个氨基酸,各碱基的平均含量为A(30.0%)、T(38.0%)、G(15.9%)、C(16.1%),(A+T)的平均含量为68.0%,高于(G+C)的含量(32.0%),与其他线粒体序列的碱基含量基本一致[24-26]。
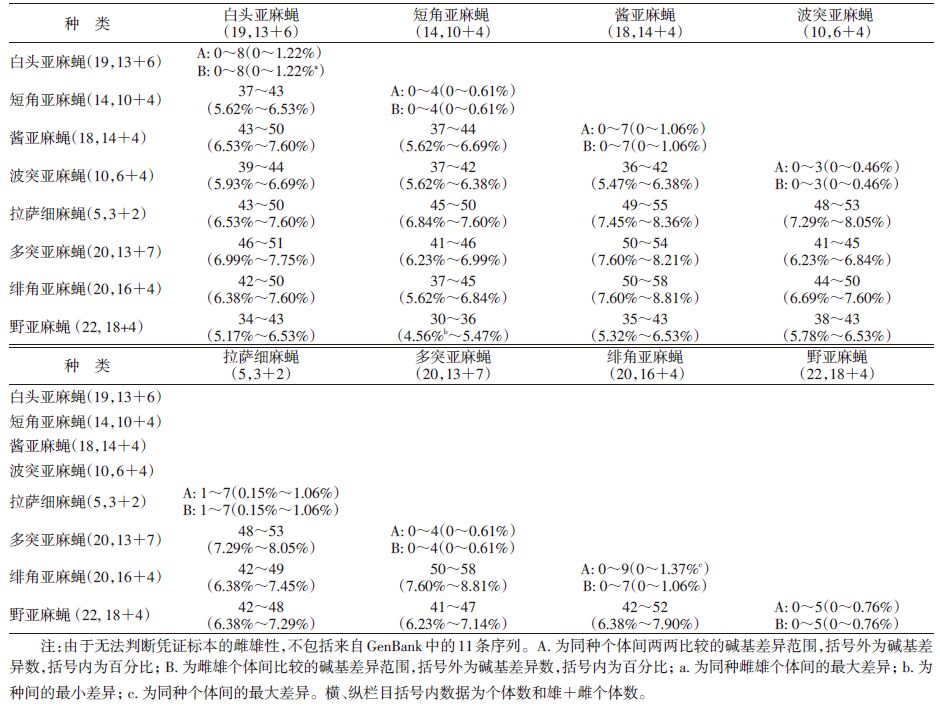
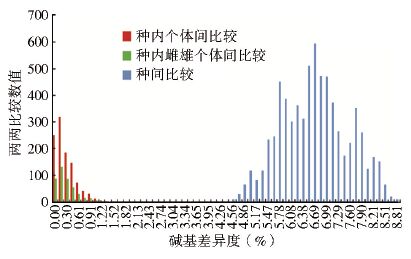
8种麻蝇中配对的雌性和雄性个体间最大的种内差异为1.37%,<2%,同种雌雄性个体间的碱基差异范围与同种间个体间的差异范围一致,不同种间的变异范围为4.56%~8.81%,高于种类鉴定界限(2%),种内个体间与种间有明显的差异间隔。8个种的同种两性间和种间的碱基对差异见表 2和图 1。
|
|
| 图 1 种内个体间、雌雄个体间及种间DNA条形码序列碱基差异度比较 Figure 1 Intra-species,inter-sexes and inter-species differences of the DNA barcodes |
| |
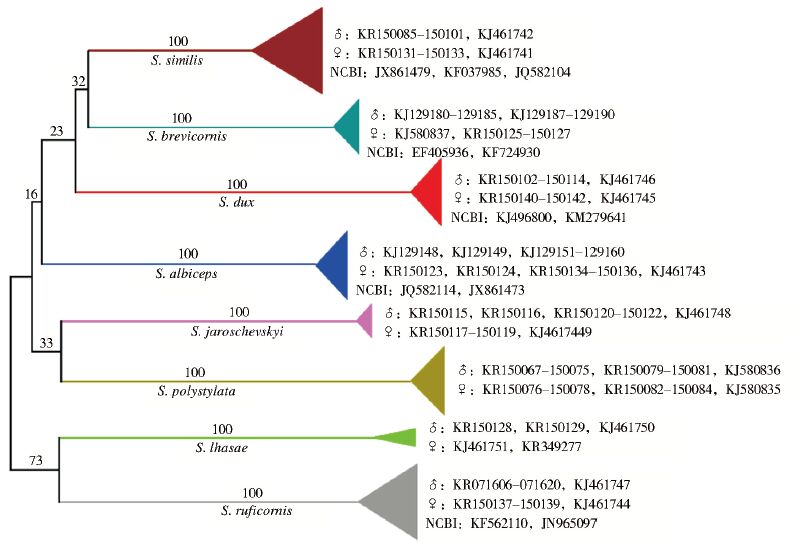
所有雌雄性个体和同种内未知雌雄的个体聚在同一个分支,置信度为100%,结果与形态学分类完全一致,见图 2。
|
| 图 2 8种麻蝇138只个体的DNA条形码序列构建的NJ树 Figure 2 Neighbor-Joining based on the DNA barcodes of the 138 individuals of 8 species |
| |
1. 子宫骨片强烈骨化,结构空间立体感强,外形呈两熊合抱状,中部具1个膜状小窝(图 3a)

|
| 注: a. 拉萨细麻蝇〔S (P). lhasae〕; b. 绯角亚麻蝇〔S (P). ruficornis〕; c. 多突亚麻蝇〔S (P). polystylata〕; d. 野亚麻蝇〔S (P). similis〕; e. 白头亚麻蝇 〔S (P). albiceps〕; f. 短角亚麻蝇〔S (P). brevicornis〕; g. 波突亚麻蝇〔S (P). jaroschevskyi〕; h. 酱亚麻蝇〔S (P). dux〕。 图 3 8种麻蝇的子宫骨片形态特征 Figure 3 Signum morphological characteristics of 8 species |
| |
…………………………拉萨细麻蝇S(P). lhasae子宫骨片扁平或具轻微的立体感,绝不呈现上述外形………………………………………………2
2. 子宫骨片骨化较弱,较柔软色浅………………3
子宫骨片强烈骨化,深色或浅色………………4
3. 子宫骨片三角状,前端部分内卷(图 3b)
……………………绯角亚麻蝇S (P). ruficornis子宫骨片梯形,后缘中部具有1个突起(图 3c)
……………………多突亚麻蝇S (P). polystylata
4. 子宫骨片浅色,瓶状,微拱起(图 3d)
……………………………野亚麻蝇S (P). similis
子宫骨片深褐色或者黑色,交替出现浅色骨化带……5
5. 子宫骨片具数条清晰的浅色骨化带……………6
子宫骨片黑褐色,不具浅色骨化带,2片(图 3e)
………………………白头亚麻蝇S (P). albiceps
6. 子宫骨片长明显长于宽,基部骤窄,具3个骨化条带和1个板状的基部宽带,第1条骨化带最宽,基部的板状带骨化强烈(图 3f)
……………………短角亚麻蝇S (P). brevicornis
子宫骨片宽度大于长度,具6条骨化带,其中1条或者3条断裂(图 3g,3h)………………………7
7. 子宫骨片具3条完整的骨化带和3条断裂的骨化带,最后1条骨化带呈“V”形,子宫骨片前缘的凹陷宽浅(图 3g)……波突亚麻蝇S (P). jaroschevskyi
子宫骨片具5条完整骨化带和1条断裂骨化带,前缘具有1个乳状突起(图 3h)
…………………………………酱亚麻蝇S (P). dux
3 讨论雄性外生殖器的形态特征是麻蝇种类鉴定的金标准,但雌性麻蝇的形态鉴定存在困难,Povoln--和Verves[23]首次指出不同种雌性麻蝇间的子宫骨片形态特征差异较大,为根据子宫骨片的特征鉴定雌性麻蝇提供了可能。目前仅少数研究提及子宫骨片的特征但并无明确的表述[5, 27]。
8种麻蝇DNA条形码的种内最大差异远低于2%的限值,而种间最小差异高达4.56%,明显高于2%的限值,与麻蝇种间差异高于3%的报道一致[28]。麻蝇分布广泛且种类繁多,该文限于在中国收集的8种麻蝇,将子宫骨片普遍作为雌性麻蝇种类鉴定的依据,应加强研究其他种类雌性麻蝇子宫骨片的结构。
| [1] | Greenberg B. Flies and disease: vol. 1,ecology, classification and biotic associations[M]. New Jersey,USA: Princeton University Press, 1971 : 1 -5. |
| [2] | Sukontason KL, Bunchu N, Methanitikorn R, et al. Ultrastructure of adhesive device in fly in the families Calliphoridae, Muscidae and Sarcophagidae, and their implication as mechanical carriers of pathogens[J]. Parasitol Res, 2006, 98 (5) : 477–481 .DOI:10.1007/s00436-005-0100-0. |
| [3] | Pape T. The Sarcophagidae (Diptera) of fennoscandia and denmark: fauna entomologica scandinavica[M]. Leiden, Holland: Dutch Academic Publishing House Brill, 1987 : 1 -203. |
| [4] | Pape TH. Catalogue of the Sarcophagidae of the world (Insecta:Diptera): memoirs on entomology, international, Volume: 8[M]. Gainesville,Florida,USA: American Entomological Institute, 1996 : 3 -7. |
| [5] | 范滋德. 中国常见蝇类检索表[M]. 2版. 北京: 科学出版社, 1992 : 580 -718. |
| [6] | 薛万琦, 赵建铭. 中国蝇类[M]. 沈阳: 辽宁科学技术出版社, 1996 : 1518 -1656. |
| [7] | Byrd JH, Castner JL. Forensic entomology: the utility of arthropods in legal investigations[M]. 2nd ed. London, England: CRC Press Taylor & Francis Group, 2010 : 1 -10. |
| [8] | Hebert PDN, Ratnasingham S, de Waard JR. Barcoding animal life: cytochrome c oxidase subunitⅠ divergences among closely related species[J]. Proc Roy Soc B Biol Sci, 2003, 270 (Suppl 1) : S96–99 . |
| [9] | Hebert PDN, Cywinska A, Ball SL. Biological identifications through DNA barcodes[J]. Proc Roy Soc B Biol Sci, 2003, 270 (1512) : 313–321 .DOI:10.1098/rspb.2002.2218. |
| [10] | Blaxter M, Mann J, Chapman T, et al. Defining operational taxonomic units using DNA barcode data[J]. Philos Trans Roy Soc B Biol Sci, 2005, 360 (1462) : 1935–1943 .DOI:10.1098/rstb.2005.1725. |
| [11] | Evans KM, Wortley AH, Mann DG. An assessment of potential diatom “barcode” genes (cox1, rbcL, 18S and ITS rDNA) and their effectiveness in determining relationships in Sellaphora (Bacillariophyta)[J]. Protist, 2007, 158 (3) : 349–364 .DOI:10.1016/j.protis.2007.04.001. |
| [12] | Hebert PDN, Penton EH, Burns JM, et al. Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astraptes fulgerator[J]. Proc Natl Acad Sci USA, 2004, 101 (41) : 14812–14817 .DOI:10.1073/pnas.0406166101. |
| [13] | Hajibabaei M, Singer GAC, Clare EL, et al. Design and applicability of DNA arrays and DNA barcodes in biodiversity monitoring[J]. BMC Biol, 2007, 5 (1) : 24.DOI:10.1186/1741-7007-5-24. |
| [14] | Yang ZH, Rannala B. Bayesian species delimitation using multilocus sequence data[J]. Proc Natl Acad Sci USA, 2010, 107 (20) : 9264–9269 .DOI:10.1073/pnas.0913022107. |
| [15] | Bergsten J, Bilton DT, Fujisawa T, et al. The effect of geographical scale of sampling on DNA barcoding[J]. Syst Biol, 2012, 61 (5) : 851–869 .DOI:10.1093/sysbio/sys037. |
| [16] | Vernooy R, Haribabu E, Muller MR, et al. Barcoding life to conserve biological diversity: beyond the taxonomic imperative[J]. PLoS Biol, 2010, 8 (7) : e1000417.DOI:10.1371/journal.pbio.1000417. |
| [17] | Tautz D, Arctander P, Minelli A, et al. A plea for DNA taxonomy[J]. Trends Ecol Evol, 2003, 18 (2) : 70–74 .DOI:10.1016/S0169-5347(02)00041-1. |
| [18] | Dayrat B. Towards integrative taxonomy[J]. Biol J Linn Soc, 2005, 85 (3) : 407–415 .DOI:10.1111/bij.2005.85.issue-3. |
| [19] | Padial JM, Miralles A, De la Riva I, et al. Review: the integrative future of taxonomy[J]. Front Zool, 2010, 7 (1) : 16.DOI:10.1186/1742-9994-7-16. |
| [20] | Bergsten J, Brilmyer G, Crampton-Platt A, et al. Sympatry and colour variation disguised well-differentiated sister species: Suphrodytes revised with integrative taxonomy including 5 kbp of housekeeping genes (Coleoptera:Dytiscidae)[J]. DNA Barcodes, 2012 : 1–18 .DOI:10.2478/dna-2012-0001. |
| [21] | Yue QY, Wu KL, Qiu DY, et al. A formal re-description of the cockroach Hebardina concinna anchored on DNA barcodes confirms wing polymorphism and identifies morphological characters for field identification[J]. PLoS One, 2014, 9 (9) : e106789.DOI:10.1371/journal.pone.0106789. |
| [22] | Folmer O, Black M, Hoeh W, et al. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates[J]. Mol Mar Biol Biotechnol, 1994, 3 (5) : 294–299 . |
| [23] | Povolný D, Verves Y. The flesh-flies of central Europe (Insecta, Diptera, Sarcophagidae)[J]. Spixiana, 1997 : 1–260 . |
| [24] | Renaud AK, Savage J, Adamowicz SJ. DNA barcoding of northern Nearctic Muscidae (Diptera) reveals high correspondence between morphological and molecular species limits[J]. BMC Ecol, 2012 : 24.DOI:10.1186/1472-6785-12-24. |
| [25] | Rivera J, Currie DC. Identification of Nearctic black flies using DNA barcodes (Diptera:Simuliidae)[J]. Mol Ecol Resour, 2009, 9 (Suppl 1) : S224–236 . |
| [26] | e Schuehli GS, de Carvalho CJB, Wiegmann BM. Molecular phylogenetics of the Muscidae (Diptera: Calyptratae): new ideas in a congruence context[J]. Invertebr Syst, 2007, 21 (3) : 263–278 .DOI:10.1071/IS06026. |
| [27] | 叶宗茂. 麻蝇族3种雌性麻蝇的发现(双翅目:麻蝇科)[J]. 动物分类学报,1985, (1) :107–110. |
| [28] | Wells JD, Pape T, Sperling FA. DNA-based identification and molecular systematics of forensically important Sarcophagidae (Diptera)[J]. J Forensic Sci, 2001, 46 (5) : 1098–1102 . |
 2016, Vol. 27
2016, Vol. 27