花青素类成分广泛存在于植物地上部分花、果、叶、茎中, 均含有C6-C3-C6基本碳架, 隶属于黄酮类成分(图 1), 具有抗氧化、抗炎、抗菌、抗肿瘤、保护神经系统、保护和提高视力等多种生理活性[1-6], 在预防和改善心脑血管疾病和代谢性疾病如冠心病、脑卒中、认知功能衰退、记忆力低下、阿尔茨海默症、脂肪酸和葡萄糖代谢紊乱等方面具有重要价值[7, 8]。阐明花青素类成分的生物合成途径和代谢网络, 将为大幅提高药用植物中花青素的含量、更有效和低成本的获得天然活性成分奠定基础, 已成为药用植物资源及活性产物研究的重要内容和新的发展方向。同其他植物次生代谢物一样, 花青素类成分的生物合成途径步骤繁多, 同时受到昼夜节律钟的严格调控, 形成复杂的次生代谢网络[9], 因此, 分析并阐明昼夜节律钟调控花青素类成分的机制, 对深入了解花青素类成分合成途径的分子调控机制至关重要。但是, 到目前为止, 有关昼夜节律钟调控植物次生代谢途径的研究进展却罕见报道。本文综述了近年来植物昼夜节律系统对次生代谢物花青素类调控的研究进展, 旨在完善花青素类成分生物合成途径, 为加深昼夜节律系统对植物花青素类成分调控的理解以及药用植物品质的定向调控提供新的思路。

|
Figure 1 The basic chemical structure of anthocyanins |
在多数真核生物中, 昼夜节律钟是由多个互相关联的转录和翻译反馈调节环(TTFL)所形成的复杂调节网络, 由正调节和负调节组件构成[10]。理论上可将昼夜节律系统分为3个广泛的结构域:用于感知并传输环境信号以同步中心振荡器的输入路径; 产生与环境变化同步并保持节律的中心振荡器; 连接振荡器与生理和代谢活动的输出路径[11-13]。
1.1 昼夜节律钟的输入系统内部昼夜节律钟与外部昼夜循环保持相对准确且稳定的相位关系, 需要接受外部环境刺激的影响及确认[14]。在菊花、拟南芥、牵牛花中, 可见光对昼夜节律基因CCA1 (circadian clock associated 1)、LHY (late elongated hypocotyl)、PRR9 (pseudo response regulator 9)、GI(gigantea), 时钟相关基因ELF4 (early flowering 4)的翻译有显著影响[15-18]。光敏色素(phytochrome, PHYA~E)作为红光和远红光的感受器, 诱导产生昼夜节律钟同时调节昼夜节律基因, PHYA在黎明通过FHY3 (far-red elongated hypocotyl 3)特异性诱导昼夜节律钟的产生[19], 活化的光敏色素与PIFs (phytochrome interaction factors)结合并诱导其降解, 以此对下游的转录网络产生影响[20, 21], PIFs中的PIF3与TOC1 (time of cab expression 1)相互作用, 且具有部分共同的靶标基因, 由此将光敏通道与昼夜节律钟相关联[22]。PHYA与PHYB对PIF3的作用机制不同, 分别对LHY和CCA1产生调节作用。蓝光感受器隐花色素(CRYs)对节律钟的影响通过CRY2与ZTL (zeitlupe)的相互作用产生[23, 24], 而且CRY2通过COP1和ELF3提高GI的稳定性[25], 同时低强度的UV-B也改变相关时钟基因的表达[14, 26]。
环境温度与光照对于诱导植物产生昼夜节律钟, 以及校准节律钟与外界时间具有同样重要的作用[27, 28]。温度对昼夜节律钟的影响主要通过两个机制:一个是温度补偿的过程, 振荡器通过诱导相关生理活动以抵消环境温度的变化, 这确保了在一个大约24 h周期环境温度变化范围内恒定的振荡模式; 另一机制称为诱导产生的过程, 温度充当重置昼夜节律钟的提示[29, 30]。
已有研究表明, 温度的变化对昼夜节律钟相关的EC夜间阻遏物(ELF4-ELF3-LUX)的影响可控制多种输出基因以改变生理活动[31, 32]。高温降低EC活性, 而低温刺激EC活性。时钟基因PRR7 (PRR9)、GI和LUX是EC夜间阻遏物的靶标基因, 因此PRR7 (PRR9)、GI和LUX的转录水平在黑暗中明显随温度升高而提高[29]。温度补偿发生于温度在一定范围内波动并且不引起大的周期性变化的情况下, 通过由CCA1、LHY、TOC1和GI组成负反馈环, 响应温度变化以维持几乎恒定的循环周期[33, 34], RVE8也参与了这个反馈环的构成[35]。
除温度、光照以外, 研究表明干旱对拟南芥、大豆中昼夜节律相关基因的表达也具有一定的调节作用[36], 同时, 激素以及病原体与昼夜节律钟之间还存在着复杂的相互作用。丁香假单胞菌感染可以缩短昼夜节律周期[37, 38], 而昼夜节律系统影响植物对丁香假单胞菌(Pseudomonas syringae)和海乳白僵菌(Hyaloperonospora arabidopsidis)的防御, 植物先天免疫作为昼夜节律钟输出途径的下游受其调控[39]。在合成类异戊二烯、激素脱落酸、油菜素甾醇、细胞分裂素和赤霉素的前体中起作用的许多基因是时钟控制的[40], 乙烯、生长相关激素吲哚乙酸、赤霉素类、细胞分裂素类和油菜素甾醇表现出昼夜节律调控特点[41]。外源给予ABA、CKs或者BR刺激后, 表现出明显的内源昼夜节律钟相位变化[42, 43]。
1.2 昼夜节律钟的中心振荡器在拟南芥中, 中心振荡器的形成依赖于3个调节回路[44], 菊花[45]、葡萄[46]、大豆[47]、水稻[48]均有相似的调节回路, 药用植物卷柏[49]、铁皮石斛等也已克隆出相关基因, 可能存在相似的分子网络。核心的调节回路是在早上表达量最高的Myb相关基因CCA1和LHY转录因子抑制基因TOC1 (PRR1)的表达, TOC1主要在黄昏出现表达的波峰并且反馈抑制CCA1/LHY的表达水平[10]。在早上CCA1/LHY的表达增强了TOC1同源物PRR7/9基因的转录, 而白天中PRR7/9表现出抑制CCA1/LHY转录的作用, 正午尤其明显[50]。晚上LUX与ELF4、ELF3形成复合物EC, 作为PRR9和LUX的转录抑制物(图 2)。

|
Figure 2 The circadian oscillator diagram. CCA1: Circadian clock associated 1; LHY: Late elongated hypocotyl; TOC1: Time of cab expression 1(PRR1); PRR7/9: Pseudo response regulator 7/9; LUX: Lux arrythmo (PCL1); ELF3/4: Early flowering 3/4; EC: The evening complex; RVE8: REVEILLE8 |
在拟南芥、水稻、玉米、大豆等植物中均显示出昼夜节律钟的输出系统与超过30%的基因转录相关[12, 47, 51]。代表性的输出途径包括植物的生长途径[52]、花期的控制相关途径[53]、植物激素的合成及转导途径[43]、生物和非生物胁迫反应途径[54]、植物代谢相关途径等[55]。
2 昼夜节律调控花青素合成花青素作为一种植物次生代谢产物, 具有利于种子和花粉传播, 并且提高植物对低温、UV胁迫以及病菌攻击的适应能力。同时, 黄酮类成分是许多药用植物的有效成分[56], 花青素也具有抗过敏、抗炎、抗氧化等作用[57, 58]。多种酶介导的苯丙素途径在花青素类生物合成中起着重要的作用[59], 依据这些酶在合成途径中产生作用的位置将编码这些酶的基因分成两类[60]:上游合成基因(EBGs)包括查耳酮合酶基因(CHS)、查耳酮异构酶基因(CHI)、黄酮3-羟化酶基因(F3H)、类黄酮3'-羟化酶基因(F3'H)和黄酮醇合酶基因(FLS); 下游合成基因(LBGs)包括二氢黄酮醇4-还原酶基因(DFR)、无色花色素加氧酶基因(LDOX)和UDP-葡萄糖:类黄酮3-O-葡萄糖基转移酶基因(UF3GT) (图 3)。
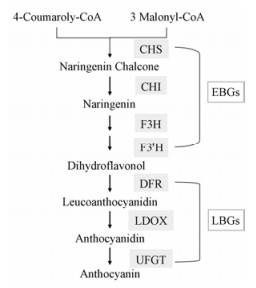
|
Figure 3 Anthocyanin biosynthesis pathways. Enzymes encoded by flavonoid early structural genes (EBGs) and encoded by anthocyanin late structural genes (LBGs) are shown in figure |
近些年对于昼夜节律系统调节花青素类化合物合成的研究, 主要集中在其合成途径结构酶基因的节律性表达上。CHS、CHI和DFR等花青素合成途径的相关基因在光/暗周期下的表达具有昼夜节律性表达的特点[61, 62], 但改变光/暗周期如在连续光照中生长时相关基因mRNA的变化模式与正常昼夜光照变化具有相似的节律, 只是表达时间有一定的移动, 但是在持续黑暗中几乎没有表达。对于这些相关基因的节律性特点是一部分结构酶基因表达在黎明和黄昏时间段表现出双峰的波形(ZT4和ZT12), 在中午表达量明显减少[9]。其中CHS表达水平的波形在早晨出现波峰, 而在UV-B条件下CHS主要在早晨表达, 晚上仅有部分表达[14, 63]。
有研究发现外部刺激感受器对花青素合成通道中的某些相关基因及转录因子(如: HY5、PIFs等)有直接调控作用[64, 65], 此外, 拟南芥中花青素仅在光照环境中积累, 并且多种非生物和生物胁迫条件(例如寒冷、干旱和病原体攻击等)对其积累具有促进作用。有趣的是, 这些对花青素积累具有刺激作用的条件对昼夜节律系统相关基因均有调节作用。
由此可推测, 或许外部刺激通过两个途径共同调控花青素类化合物的合成(图 4)。有大量研究关于光与温度对花青素合成的影响, 不同强度、类型的光对花青素生物合成具有不同的影响, 低温会增强花青素的积累。
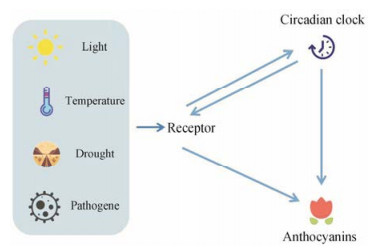
|
Figure 4 Exogenous stimuli and circadian clocks regulate the synthesis of anthocyanins |
花青素类成分只在光照下积累, 而且不同强度及类型的光对花青素的生物合成具有不同的效果[66]。隐花色素和光敏色素是植物中已确认的蓝光和红光的光感受器, 光敏色素也接受远红光的刺激[67]。
此外, 紫外光也是许多研究的关注点, UV-B所占比例不足到达地球表面的总光能的0.5%, 但它具有日光光谱最高的能量并且对生物圈具有实质性的影响[68, 69]。已有研究显示UV-B虽然具有较低的注量率, 但作为重要的环境信号控制发育, 促进光形态发生并促进合成黄酮类成分所需的基因的表达[70-73]。为了抵抗高强度UV-B对植物造成的损伤, UV-B诱导黄酮类成分的积累保护植物体, 在自然条件下这种植物的自我保护性是常见的[73]。
2.1.1 光诱导花青素类成分合成的分子机制 2.1.1.1 WD40-bHLH-MYB复合物及HY5介导光诱导花青素类成分合成大量研究结论已证实WD40-bHLH-MYB复合物使调控花青素合成通道中的酶基因尤其是下游合成基因DFR、LDOX、UF3GT的表达增加, 该复合物包括R2R3 MYB TFs (MYB75、MYB90、MYB113、MYB114)、bHLH TFs (TT8、GL3、EGL3)和WD40蛋白TTG1[74-76]。其中MYB家族的MYB75转录因子PAP1, 在花青素合成途径中具有主要的调节作用, MYB75/PAP1的转录和翻译后水平都强烈依赖于光的存在, 除此之外糖和细胞分裂素对其有正向调节作用, 乙烯具有负调节作用[77-79]。具有光依赖性的PAP1转录由亮氨酸拉链转录因子HY5调节, HY5与PAP1启动子区域(G-和ACE-boxes)位点结合调节其表达[80]。HY5不仅通过PAP1调节花青素合成途径功能酶基因, 某些EGBs和LGBs中也存在着G-box和ACE元件, 有研究表明, CHS、DFR、LDOX、UF3GT的转录水平受到HY5的正向调节[81, 82]。然而, 比较在LBG转录激活中的有效性, PAP1似乎比HY5起更重要的作用。HY5已被证实作为植物光敏色素(PHY)、隐花色素(CRY)和UV-B (UVR8)光感受器的下游组分[69, 83]。CRY1、CRY2、PHYA、PHYB均能与COP1发生强烈的相互作用, 抑制COP1对底物HY5的降解作用, 提高HY5蛋白含量[84-87]。在低强度的UV-B下, HY5和HY5 HOMOLOG (HYH)是在黄酮类化合物生物合成途径的相关基因水平升高中起关键作用的转录因子[70, 73, 88]。COP1与UVR8之间的相互作用是UV-B的受体信号转导的必要条件, 而UVR8-COP1主要靶标因子是转录因子HY5和HYH[89]。
对于MBW复合物的调控除了上述对MYB37的特征性的一系列调控外, 有研究发现HY5通过MYB-LIKE 2 (MYBL2)和MYBD通道改变MBW的稳定性。已有报道的几种负调节剂, 例如MYBL2和SPL9干扰各种条件下MBW复合物的形成[90, 91]。拟南芥中由MYBL2基因编码的R3 MYB-related蛋白与bHLH蛋白直接相互作用, 破坏MBW复合物的形成, 导致花青素积累减少[90, 92], MYBD通过直接结合MYBL2启动子区域抑制MYBL2表达, 间接调控MBW复合物对花青素合成有正向调节作用。而HY5能够结合MYBD启动子上的G-box区域, 诱导MYBD在光响应中的表达, 最近的研究发现HY5也结合MYBL2的启动子, 这种转录抑制与结合位点的特异性组蛋白修饰有关[93, 94]。
2.1.1.2 PIF4、PIF5介导光诱导花青素类成分合成PIF4和PIF5对某些LGBs和EGBs的表达有抑制作用, PIF4和PIF5降低了F3'H和DFR的活性[64], PIF4与HY5调节光敏基因通过共同的G-box区域[95], 抑制PAP1的转录水平, 也由此影响花青素的合成, 且PIF4和PIF5均为PHYB的靶标基因, PHYB靶向抑制其基因表达。
PIF4和HY5在光通道合成花青素中某些方面具有相反的作用, 两者均受到COP1的调控。COP1可促进HY5蛋白的降解[96], 增加PIF4活性[97]。在光条件下, COP1的总细胞量不变, 但是COP1在细胞核中的量在改变。在黑暗中, COP1主要存在于细胞核中, 通过降解核转录因子如HY5而作为光形态发生的负调节子[98]。此外, COP1在黑暗中介导MYB75的降解, MYB75转录物受COP1/SPA复合物的影响[66]。在光照条件下, COP1/SPA复合物和MAP KINASE4 (MPK4)对花青素的合成具有双重调节作用[66, 99]。MPK4对MYB75磷酸化阻止了COP1对MYB75识别, 这种方法可增强MYB75的稳定性。对于紫外光而言, 如上所述COP1与UVR8相互作用, COP1作为紫外线诱导光形态发生反应的正调节器, 这与其在可见光中的作用相反[89]。
2.1.2 昼夜节律钟在光诱导花青素合成中的影响光作为昼夜节律钟的门控系统已有大量相关研究, 与此同时, 昼夜节律钟对光依赖的花青素合成途径具有调控作用。PIFs作为花青素合成的负调节子, CCA1和LHY在早上促进PIF4和PIF5的表达, 而在夜晚TOC1降低其蛋白活性, PIF3也有类似的受到昼夜节律基因调节的现象。对于隐花色素, ZTL对HY5表达有抑制作用。RVE8作为TOC1的类似物通过与LNKs结合, 调节PAP1的蛋白活性。在催眠睡茄中UVR8通过HY5影响花青素合成酶基因的转录和表达[100]。通过以上方法, 昼夜节律基因调控了花青素的合成途径。
结合上述研究结果分析, 对于光诱导花青素的生物合成具有两种途径:一种是经由昼夜节律钟调控的花青素的预见性合成, 另一种是花青素作为防护强光对植物伤害的应激性合成。与调控花青素合成的昼夜节律基因表达结合, CHS等基因在上午表达量增高, 花青素的合成也增多, 与自然环境中阳光到达地球表面的光强度及种类相应的是中午紫外线的强度增加, 花青素合成规律与此相应似乎预见性防止紫外线对植物的损伤(图 5)。
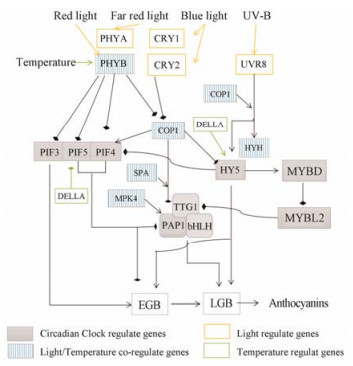
|
Figure 5 Effects of light and temperature on regulation of anthocyanin synthesis in circadian rhythm system. PHYA/B: Phytochrome A/B; CRY1/2: Cryptochrome 1/2; UVR8: Ultraviolet-B receptor UVR8; COP1: E3 ubiquitin-protein ligase COP1; PIFs: Phytochrome interacting factor; HY5: Basic-leucine zipper (bZIP) transcription factor family protein; HYH: HY5-homolog; MYBL2: MYB-like 2; MYBD: MYB domain-containing protein; PAP1: Production of anthocyanin pigment 1; TTG1: Transducin/WD40 repeat-like superfamily protein; bHLH: Basic helix-loop-helix family; MPK4: Putative mitogen activated protein kinase 4 |
生物内部昼夜节律钟结合外部温度变化对花青素的合成进行调节, 当温度过低或过高都将对生物造成损伤, 和光照强度与类型对生物的影响相似, 过低的温度促进花青素类成分的合成是植物对低温的应激反馈[101]。关于温度信号与昼夜节律相互作用对花青素合成造成影响的研究目前还很缺乏, 但是, 目前已有的一些研究证实低温通过上调花青素合成基因PAL、CHS、CHI、FLS促进花青素类生物合成[64], 由此入手, 试图探索昼夜节律在其中的调节作用。
通常日间温度高于夜间, 而在白天, 早晚温度低, 正午时分的温度高, 这样温度的波形情况与黄酮合成途径相关的一些基因的表达节律符合。在拟南芥中昼夜节律钟通过调节转录因子参与低温适应的过程[32, 102], 低温诱导花青素的生物合成依赖于光的存在, 在光和温度信号通道上有许多同样的转录因子作为整合的节点[103]。
2.2.1 温度调节花青素合成的分子机制与光变化类似, 温度的变化可以将D2 (Pfr同型二聚体)变为phyB, 表明phyB也可以被定义为温度变化受体[30]。在细胞核中, PIFs与phyA、phyB各自的APA或APB相互作用位点结合然后被降解[104, 105]。PIF蛋白属于拟南芥bHLH超家族的第15个亚家族, 它们在调节光和温度的昼夜节律中起着不同的作用。bHLH转录因子PIF4已被证明是拟南芥中对光和温度多重反应的枢纽[30]。PIF的转录受温度调节的机制与温度调节EC的靶标时钟基因PRR7 (PRR9)、GI和LUX的分子机制相似[106]。此外, 温度可以调节PIF4结合其靶标的能力[30]。研究表明PIF4和PIF5通过抑制编码花青素生物合成途径基因和MBW复合体各组分的转录抑制花青素的生物合成。PIF4/PIF5具有抑制由红光诱导产生的F3'H和DFR的启动子活性的功能[64]。而且PIF1/PIF4和HY5通过直接结合到共同的靶标基因的相同G-box顺式元件区域产生相反的作用, PIF4介导热的温度信号传导, HY5在冷的温度传导中发挥作用[107]。通过低温诱导表达增加的EBGs基因(如CHS、CHI、F3H)较大程度上依赖于HY5/HYH的作用。HY5蛋白在低温中的积累可能是由于DELLA可以在低温中提高HY5蛋白水平[108]。而DELLA蛋白通过未知的E3连接酶促进泛素-蛋白酶体系中的PIF降解[109], 建立PIF4和HY5之间的关系的另一个桥梁是COP1, COP1参与降解诸如HY5的26S蛋白酶体并且它的活性影响了PIF4的稳定性[110]这与光通道相同[111]。而COP1基因的表达随着温度升高而增加, COP1蛋白的稳定性随温度升高而降低[112], 也有研究表明低温降低了细胞核中COP1丰度[107]。
2.2.2 昼夜节律钟在低温诱导花青素合成中的作用昼夜节律系统的内部振荡器对温度补偿的调节对花青素生物合成也产生了一定的影响。昼夜节律钟中促进PRR5表达的RVE8, 在温度补偿和温度信号传导中起着重要的作用[30]。RVE8在内部振荡器中具有类似于CCA1和LHY的作用, 通过与其他昼夜节律基因的相互作用设置与外界环境相吻合的节律[30]。在花青素合成途径中, RVE8具有正调节作用, 在中午与负调节子LNK蛋白相互作用明显, 与此相应花青素的合成在早上达到峰值, 中午明显下降。
2.3 光和温度的相互作用影响花青素合成途径照射到达地球表面的阳光类型和强度发生着周期性变化, 同时温度也在一定波动范围内每天周期性地循环。阳光辐射不仅增加空气温度, 同时植物吸收的光除了用于进行光合作用转化为化学能其余转化为热能提高了植物温度[97, 110]。植物组织温度除了受太阳辐射和空气温度的影响, 风和湿度等也是影响因素。因此, 光和温度之间的关系, 以及它们调节昼夜节律钟以影响花青素生物合成的昼夜振荡是非常复杂的。
有研究发现在远红光脉冲之后, 在黑暗环境使光敏受体活性最小化, phyB突变体的幼苗仍然对温度有反应[113]。这意味着并不是所有的温度传感器都涉及光敏通道, 感光器phyB被认为是温度传感器, 但是对单独的温度传感器的了解依旧很少。总之, 红光激活phyB, 远红光和高温使其失活[113]。UVR8二聚体/单体的稳定性受温度的影响, 较低的温度(8~10 ℃)降低了单体到二聚体的回复率[114], ZTL介导的反应过程也受到温度影响[115]。除此之外, PIFs及HY5等影响因子也同时在光敏通道及温敏通道调节花青素合成中发挥着重要作用。
3 讨论花青素类成分作为一类重要的植物次生代谢产物, 具有抗氧化、抗炎、抗菌、等多种生理活性, 在预防和改善心血脑管疾病和代谢性疾病等方面具有广阔的应用前景。植物昼夜节律系统不仅影响植物许多生理和分子过程, 而且严格调控着植物次生代谢成分花青素类的生物合成。阐明昼夜节律钟调控花青素类成分的分子机制, 将为后续开展以昼夜节律系统关键基因为遗传操作靶标的精准代谢工程、改良药用植物种质资源提供充分科学依据。
近年来, 关于药用植物花青素类成分生物合成及调控机制的研究已经取得了较大进展, 如淫羊藿、枸杞、葡萄、桑椹、大豆、花生等[116-120]。目前已基本阐明花青素生物合成途径的多种结构基因(CHS、CHI、F3H、FLS、DFR、LDOX和UF3GT)。同时, 这些结构基因的表达还受到转录因子的严格调控。但是迄今为止, 对于结构基因与转录因子之间复杂的相互作用网络以及转录因子对花青素合成途径相关基因调控方式的了解仍十分有限。而且花青素类成分的生物合成受多种外源性刺激的影响, 而昼夜节律钟通过整合外源刺激信号接收网络、转录因子网络整体调节花青素代谢通道次生产物的生物合成。因此, 研究昼夜节律钟对花青素类成分合成途径的调节机制对于完善整个花青素合成调控网络至关重要。
稳定而敏锐的昼夜节律钟的存在使植物体内的生命活动与外界环境相协调, 在促进植物生长及提高适应性上起着重要作用。低温和高强度的光照尤其是高强度紫外线都不利于植物体的生长, 甚至对植物体造成损伤。大量的研究数据表明, 花青素生物合成上游大部分结构合成酶、具有正调节作用的转录因子等的表达量在黎明与正午之间以及黄昏出现波峰, 这与正午光照尤其是紫外强度最高, 以及夜晚温度较低相呼应。温度与光照作为主要的授时因子, 改变并校准着昼夜节律中心振荡器, 而受昼夜节律钟调控的相关基因, 如HY5、PIFs、WBM复合物等与花青素合成途径相关酶基因的启动子结合影响其转录水平, 进而调控该途径中化合物的形成。
除此之外, 低温和强光刺激直接通过其感受器影响WBM复合物的转录及翻译水平从而对花青素类成分的合成造成影响。也就是说, 光照和温度通过两个途径影响花青素的合成, 一是作为授时因子通过昼夜节律系统调控花青素合成以预见性适应昼夜节律的变化; 二是作为外源刺激, 诱导花青素化合物的补偿合成以响应环境胁迫。光照、温度的改变影响着昼夜节律钟的相位变化, 昼夜节律系统对感受器也具有调控作用, 而探明其中具体的分子机制及对花青素类化合物的调控网络还有待进一步研究。
| [1] | He J, Giusti MM. Anthocyanins:natural colorants with health-promoting properties[J]. Annu Rev Food Sci Technol, 2010, 1: 163–187. DOI:10.1146/annurev.food.080708.100754 |
| [2] | Seeram NP, Momin RA, Nair MG, et al. Cyclooxygenase inhibitory and antioxidant cyanidin glycosides in cherries and berries[J]. Phytomedicine, 2001, 8: 362–369. DOI:10.1078/0944-7113-00053 |
| [3] | Hou DX, Yanagita T, Uto T, et al. Anthocyanidins inhibit cyclooxygenase-2 expression in LPS-evoked macrophages:structure-activity relationship and molecular mechanisms involved[J]. Biochem Pharmacol, 2005, 70: 417–425. DOI:10.1016/j.bcp.2005.05.003 |
| [4] | Zhang H, Wang L, Deroles S, et al. New insight into the structures and formation of anthocyanic vacuolar inclusions in flower petals[J]. BMC Plant Biol, 2006, 6: 29. DOI:10.1186/1471-2229-6-29 |
| [5] | Cisowska A, Wojnicz D, Hendrich AB. Anthocyanins as antimicrobial agents of natural plant origin[J]. Nat Prod Commun, 2011, 6: 149–156. |
| [6] | Naz S, Siddiqi R, Ahmad S, et al. Antibacterial activity directed isolation of compounds from Punica granatum[J]. J Food Sci, 2007, 72: M341–M345. DOI:10.1111/jfds.2007.72.issue-9 |
| [7] | Wallace TC, Slavin M, Frankenfeld CL. Systematic review of anthocyanins and markers of cardiovascular disease[J]. Nutrients, 2016, 8: 32–45. DOI:10.3390/nu8010032 |
| [8] | Wang X, Ouyang YY, Liu J, et al. Flavonoid intake and risk of CVD:a systematic review and meta-analysis of prospective cohort studies[J]. Br J Nutr, 2014, 111: 1–22. |
| [9] | Perez-Garcia P, Ma Y, Yanovsky MJ, et al. Time-dependent sequestration of RVE8 by LNK proteins shapes the diurnal oscillation of anthocyanin biosynthesis[J]. Proc Natl Acad Sci USA, 2015, 112: 5249–5253. DOI:10.1073/pnas.1420792112 |
| [10] | Nagel DH, Kay SA. Complexity in the wiring and regulation of plant circadian networks[J]. Curr Biol, 2012, 22: R648–657. DOI:10.1016/j.cub.2012.07.025 |
| [11] | Covington MF, Maloof JN, Straume M, et al. Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development[J]. Genome Biol, 2008, 9: R130. DOI:10.1186/gb-2008-9-8-r130 |
| [12] | Filichkin SA, Breton G, Priest HD, et al. Global profiling of rice and poplar transcriptomes highlights key conserved circadian-controlled pathways and cis-regulatory modules[J]. PLoS One, 2011, 6: e16907. DOI:10.1371/journal.pone.0016907 |
| [13] | Khan S, Rowe SC, Harmon FG. Coordination of the maize transcriptome by a conserved circadian clock[J]. BMC Plant Biol, 2010, 10: 126. DOI:10.1186/1471-2229-10-126 |
| [14] | Feher B, Kozmabognar L, Kevei E, et al. Functional interaction of the circadian clock and UV RESISTANCE LOCUS 8-controlled UV-B signaling pathways in Arabidopsis thaliana[J]. Plant J, 2011, 67: 37–48. DOI:10.1111/tpj.2011.67.issue-1 |
| [15] | Wang ZY, Kenigsbuch D, Sun L, et al. A Myb-related transcription factor is involved in the phytochrome regulation of an Arabidopsis Lhcb gene[J]. Plant Cell, 1997, 9: 491–507. DOI:10.1105/tpc.9.4.491 |
| [16] | Locke JC, Millar AJ, Turner MS. Modelling genetic networks with noisy and varied experimental data:the circadian clock in Arabidopsis thaliana[J]. J Theor Biol, 2005, 234: 383–393. DOI:10.1016/j.jtbi.2004.11.038 |
| [17] | Kikis EA, Khanna R, Quail PH. ELF4 is a phytochrome-regulated component of a negative-feedback loop involving the central oscillator components CCA1 and LHY[J]. Plant J, 2005, 44: 300–313. DOI:10.1111/j.1365-313X.2005.02531.x |
| [18] | Higuchi Y, Sumitomo K, Oda A, et al. Day light quality affects the night-break response in the short-day plant chrysanthemum, suggesting differential phytochrome-mediated regulation of flowering[J]. J Plant Physiol, 2012, 169: 1789–1796. DOI:10.1016/j.jplph.2012.07.003 |
| [19] | Allen T, Koustenis A, Theodorou G, et al. Arabidopsis FHY3 specifically gates phytochrome signaling to the circadian clock[J]. Plant Cell, 2006, 18: 2506–2516. DOI:10.1105/tpc.105.037358 |
| [20] | Louise N, Peter K, Paulina S, et al. Circadian and plastid signaling pathways are integrated to ensure correct expression of the CBF and COR genes during photoperiodic growth[J]. Plant Physiol, 2016, 171: 1392–1406. |
| [21] | Leivar P, Monte E. PIFs:systems integrators in plant development[J]. Plant Cell, 2014, 26: 56–78. DOI:10.1105/tpc.113.120857 |
| [22] | Soy J, Leivar P, Gonzalez-Schain N, et al. Molecular convergence of clock and photosensory pathways through PIF3-TOC1 interaction and co-occupancy of target promoters[J]. Proc Natl Acad Sci USA, 2016, 113: 4870–4875. DOI:10.1073/pnas.1603745113 |
| [23] | Liu H, Wang Q, Liu Y, et al. Arabidopsis CRY2 and ZTL mediate blue-light regulation of the transcription factor CIB1 by distinct mechanisms[J]. Proc Natl Acad Sci USA, 2013, 110: 17582–17587. DOI:10.1073/pnas.1308987110 |
| [24] | Jarillo JA, Capel J, Tang RH, et al. An Arabidopsis circadian clock component interacts with both CRY1 and phyB[J]. Nature, 2001, 410: 487–490. DOI:10.1038/35068589 |
| [25] | Baudry A, Ito S, Song YH, et al. F-box proteins FKF1 and LKP2 act in concert with ZEITLUPE to control Arabidopsis clock progression[J]. Plant Cell, 2010, 22: 606–622. DOI:10.1105/tpc.109.072843 |
| [26] | McWatters HG, Bastow RM, Hall A, et al. The ELF3 zeitnehmer regulates light signalling to the circadian clock[J]. Nature, 2000, 408: 716–720. DOI:10.1038/35047079 |
| [27] | McClung CR, Davis SJ. Ambient thermometers in plants:from physiological outputs towards mechanisms of thermal sensing[J]. Curr Biol, 2010, 20: R1086–1092. DOI:10.1016/j.cub.2010.10.035 |
| [28] | Boikoglou E, Ma Z, von Korff M, et al. Environmental memory from a circadian oscillator:the Arabidopsis thaliana clock differentially integrates perception of photic vs. thermal entrainment[J]. Genetics, 2011, 189: 655–664. DOI:10.1534/genetics.111.131417 |
| [29] | Mizuno T, Nomoto Y, Oka H, et al. Ambient temperature signal feeds into the circadian clock transcriptional circuitry through the EC night-time repressor in Arabidopsis thaliana[J]. Plant Cell Physiol, 2014, 55: 958–976. DOI:10.1093/pcp/pcu030 |
| [30] | Wigge PA. Ambient temperature signalling in plants[J]. Curr Opin Plant Biol, 2013, 16: 661–666. DOI:10.1016/j.pbi.2013.08.004 |
| [31] | Helfer A, Nusinow DA, Chow BY, et al. LUX ARRHYTHMO encodes a nighttime repressor of circadian gene expression in the Arabidopsis core clock[J]. Curr Biol, 2011, 21: 126–133. DOI:10.1016/j.cub.2010.12.021 |
| [32] | Liu T, Carlsson J, Takeuchi T, et al. Direct regulation of abiotic responses by the Arabidopsis circadian clock component PRR7[J]. Plant J, 2013, 76: 101–114. |
| [33] | Gould PD, Locke JC, Larue C, et al. The molecular basis of temperature compensation in the Arabidopsis circadian clock[J]. Plant Cell, 2006, 18: 1177–1187. DOI:10.1105/tpc.105.039990 |
| [34] | Salome PA, Weigel D, McClung CR. The role of the Arabidopsis morning loop components CCA1, LHY, PRR7, and PRR9 in temperature compensation[J]. Plant Cell, 2010, 22: 3650–3661. DOI:10.1105/tpc.110.079087 |
| [35] | Rawat R, Takahashi N, Hsu PY, et al. Reveille 8 and pseudo-reponse regulator 5 form a negative feedback loop within the Arabidopsis circadian clock[J]. PLoS Gen, 2011, 7: e1001350. DOI:10.1371/journal.pgen.1001350 |
| [36] | Dubois M, Claeys H, Van den Broeck L, et al. Time of day determines Arabidopsis transcriptome and growth dynamics under mild drought[J]. Plant Cell Environ, 2017, 40: 180–189. DOI:10.1111/pce.v40.2 |
| [37] | Boller T, Felix G. A renaissance of elicitors:perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptor[J]. Annu Rev Plant Biol, 2009, 60: 379–406. DOI:10.1146/annurev.arplant.57.032905.105346 |
| [38] | Felix G, Duran JD, Volko S, et al. Plants have a sensitive perception system for the most conserved domain of bacterial flagellin[J]. Plant J, 1999, 18: 265–276. DOI:10.1046/j.1365-313X.1999.00265.x |
| [39] | Zhang C, Xie Q, Anderson RG, et al. Crosstalk between the circadian clock and innate immunity in Arabidopsis[J]. PLoS Pathogens, 2013, 9: e1003370. DOI:10.1371/journal.ppat.1003370 |
| [40] | Vranova E, Coman D, Gruissem W. Network analysis of the MVA and MEP pathways for isoprenoid synthesis[J]. Annu Rev Plant Biol, 2013, 64: 665–700. DOI:10.1146/annurev-arplant-050312-120116 |
| [41] | Novakova M, Motyka V, Dobrev PI, et al. Diurnal variation of cytokinin, auxin and abscisic acid levels in tobacco leaves[J]. J Exp Bot, 2005, 56: 2877–2883. DOI:10.1093/jxb/eri282 |
| [42] | Hanano S, Domagalska MA, Nagy F, et al. Multiple phytohormones influence distinct parameters of the plant circadian clock[J]. Genes Cells, 2006, 11: 1381–1392. DOI:10.1111/gtc.2006.11.issue-12 |
| [43] | Atamian HS, Harmer SL. Circadian regulation of hormone signaling and plant physiology[J]. Plant Mol Biol, 2016, 91: 691–702. DOI:10.1007/s11103-016-0477-4 |
| [44] | Locke JC, Kozma-Bognar L, Gould PD, et al. Experimental validation of a predicted feedback loop in the multi-oscillator clock of Arabidopsis thaliana[J]. Mol Syst Biol, 2006, 2: 59. |
| [45] | Fu J, Yang L, Dai S. Conservation of Arabidopsis thaliana circadian clock genes in Chrysanthemum lavandulifolium[J]. Plant Physiol Biochem, 2014, 80: 337–347. DOI:10.1016/j.plaphy.2014.04.001 |
| [46] | Carbonell-Bejerano P, Rodriguez V, Royo C, et al. Circadian oscillatory transcriptional programs in grapevine ripening fruits[J]. BMC Plant Biol, 2014, 14: 78. DOI:10.1186/1471-2229-14-78 |
| [47] | Marcolino-Gomes J, Rodrigues FA, Fuganti-Pagliarini R, et al. Diurnal oscillations of soybean circadian clock and drought responsive genes[J]. PLoS One, 2014, 9: e86402. DOI:10.1371/journal.pone.0086402 |
| [48] | Matsuzaki J, Kawahara Y, Izawa T. Punctual transcriptional regulation by the rice circadian clock under fluctuating field conditions[J]. Plant Cell, 2015, 27: 633–648. DOI:10.1105/tpc.114.135582 |
| [49] | Banks JA, Nishiyama T, Hasebe M, et al. The Selaginella genome identifies genetic changes associated with the evolution of vascular plants[J]. Science, 2011, 332: 960–963. DOI:10.1126/science.1203810 |
| [50] | Farre EM, Harmer SL, Harmon FG, et al. Overlapping and distinct roles of PRR7 and PRR9 in the Arabidopsis circadian clock[J]. Curr Biol, 2005, 15: 47–54. DOI:10.1016/j.cub.2004.12.067 |
| [51] | Hayes KR, Beatty M, Meng X, et al. Maize global transcriptomics reveals pervasive leaf diurnal rhythms but rhythms in developing ears are largely limited to the core oscillator[J]. PLoS One, 2010, 5: e12887. DOI:10.1371/journal.pone.0012887 |
| [52] | Nozue K, Fau CM, Fau DP, et al. Rhythmic growth explained by coincidence between internal and external cues[J]. Nature, 2007, 448: 358–361. DOI:10.1038/nature05946 |
| [53] | Yu JW, Rubio V, Lee NY, et al. COP1 and ELF3 control circadian function and photoperiodic flowering by regulating GI stability[J]. Mol Cell, 2008, 32: 617–630. DOI:10.1016/j.molcel.2008.09.026 |
| [54] | Grundy J, Stoker C, Carre IA. Circadian regulation of abiotic stress tolerance in plants[J]. Front Plant Sci, 2015, 6: 648. |
| [55] | Choudhary MK, Nomura Y, Wang L, et al. Quantitative circadian phosphoproteomic analysis of Arabidopsis reveals extensive clock control of key components in physiological, metabolic, and signaling pathways[J]. Mol Cell Proteomics, 2015, 14: 2243–2260. DOI:10.1074/mcp.M114.047183 |
| [56] | Hichri I, Barrieu F, Bogs J, et al. Recent advances in the transcriptional regulation of the flavonoid biosynthetic pathway[J]. J Exp Bot, 2011, 62: 2465–2483. DOI:10.1093/jxb/erq442 |
| [57] | Chen CF, Li YD, Xu Z. Chemical principles and bioactivities of blueberry[J]. Acta Pharm Sin (药学学报), 2010, 45: 422–429. |
| [58] | Jin JR, Hong H, Jin GY, et al. Anthocyanidin inhibits immunoglobulin E-mediated allergic response in mast cells[J]. Acta Pharm Sin (药学学报), 2012, 47: 34–38. |
| [59] | Koes R, Verweij W, Quattrocchio F. Flavonoids:a colorful model for the regulation and evolution of biochemical pathways[J]. Trends Plant Sci, 2005, 10: 236–242. DOI:10.1016/j.tplants.2005.03.002 |
| [60] | Tanaka Y, Ohmiya A. Seeing is believing:engineering anthocyanin and carotenoid biosynthetic pathways[J]. Curr Opin Biotechnol, 2008, 19: 190–197. DOI:10.1016/j.copbio.2008.02.015 |
| [61] | Deikman J, Hammer PE. Induction of anthocyanin accumulation by cytokinins in Arabidopsis thaliana[J]. Plant Physiol, 1995, 108: 47–57. DOI:10.1104/pp.108.1.47 |
| [62] | Kim YB, Park SY, Thwe AA, et al. Metabolomic analysis and differential expression of anthocyanin biosynthetic genes in white-and red-flowered buckwheat cultivars (Fagopyrum esculentum)[J]. J Agric Food Chem, 2013, 61: 10525–10533. DOI:10.1021/jf402258f |
| [63] | Takeuchi T, Newton L, Burkhardt A, et al. Light and the circadian clock mediate time-specific changes in sensitivity to UV-B stress under light/dark cycles[J]. J Exp Bot, 2014, 65: 6003–6012. DOI:10.1093/jxb/eru339 |
| [64] | Liu Z, Zhang Y, Wang J, et al. Phytochrome-interacting factors PIF4 and PIF5 negatively regulate anthocyanin biosynthesis under red light in Arabidopsis seedlings[J]. Plant Sci, 2015, 238: 64–72. DOI:10.1016/j.plantsci.2015.06.001 |
| [65] | Loyola R, Herrera D, Mas A, et al. The photomorphogenic factors UV-B RECEPTOR 1, ELONGATED HYPOCOTYL 5, and HY5 HOMOLOGUE are part of the UV-B signalling pathway in grapevine and mediate flavonol accumulation in response to the environment[J]. J Exp Bot, 2016, 67: 5429–5445. DOI:10.1093/jxb/erw307 |
| [66] | Maier A, Schrader A, Kokkelink L, et al. Light and the E3 ubiquitin ligase COP1/SPA control the protein stability of the MYB transcription factors PAP1 and PAP2 involved in anthocyanin accumulation in Arabidopsis[J]. Plant J, 2013, 74: 638–651. DOI:10.1111/tpj.2013.74.issue-4 |
| [67] | Fankhauser C. The phytochromes, a family of red/far-red absorbing photoreceptors[J]. J Biol Chem, 2001, 276: 11453–11456. DOI:10.1074/jbc.R100006200 |
| [68] | Jenkins GI. Signal transduction in responses to UV-B radiation[J]. Annu Rev Plant Biol, 2009, 60: 407–431. DOI:10.1146/annurev.arplant.59.032607.092953 |
| [69] | Heijde M, Ulm R. UV-B photoreceptor-mediated signalling in plants[J]. Trends Plant Sci, 2012, 17: 230–237. DOI:10.1016/j.tplants.2012.01.007 |
| [70] | Ulm R, Baumann A, Oravecz A, et al. Genome-wide analysis of gene expression reveals function of the bZIP transcription factor HY5 in the UV-B response of Arabidopsis[J]. Proc Natl Acad Sci USA, 2004, 101: 1397–1402. DOI:10.1073/pnas.0308044100 |
| [71] | Brown BA, Cloix C, Jiang GH, et al. A UV-B-specific signaling component orchestrates plant UV protection[J]. Proc Natl Acad Sci USA, 2005, 102: 18225–18230. DOI:10.1073/pnas.0507187102 |
| [72] | Oravecz A, Baumann A, Mate Z, et al. Constitutively photomorphogenic1 is required for the UV-B response in Arabidopsis[J]. Plant Cell, 2006, 18: 1975–1990. DOI:10.1105/tpc.105.040097 |
| [73] | Stracke R, Favory JJ, Gruber H, et al. The Arabidopsis bZIP transcription factor HY5 regulates expression of the PFG1/MYB12 gene in response to light and ultraviolet-B radiation[J]. Plant Cell Environ, 2010, 33: 88–103. |
| [74] | Dooner HK, Robbins TP, Jorgensen RA. Genetic and developmental control of anthocyanin biosynthesis[J]. Annu Rev Genet, 1991, 25: 173–199. DOI:10.1146/annurev.ge.25.120191.001133 |
| [75] | Gonzalez A, Zhao M, Leavitt JM, et al. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings[J]. Plant J, 2008, 53: 814–827. DOI:10.1111/tpj.2008.53.issue-5 |
| [76] | Shi MZ, Xie DY. Biosynthesis and metabolic engineering of anthocyanins in Arabidopsis thaliana[J]. Recent Pat Biotechnol, 2014, 8: 47–60. DOI:10.2174/1872208307666131218123538 |
| [77] | Pan Y, Michael TP, Hudson ME, et al. Cytochrome P450 monooxygenases as reporters for circadian-regulated pathways[J]. Plant Physiol, 2009, 150: 858–878. DOI:10.1104/pp.108.130757 |
| [78] | Das PK, Shin DH, Choi SB, et al. Sugar-hormone cross-talk in anthocyanin biosynthesis[J]. Mol Cells, 2012, 34: 501–507. DOI:10.1007/s10059-012-0151-x |
| [79] | Cominelli E, Gusmaroli G, Allegra D, et al. Expression analysis of anthocyanin regulatory genes in response to different light qualities in Arabidopsis thaliana[J]. J Plant Physiol, 2008, 165: 886–894. DOI:10.1016/j.jplph.2007.06.010 |
| [80] | Shin DH, Choi M, Kim K, et al. HY5 regulates anthocyanin biosynthesis by inducing the transcriptional activation of the MYB75/PAP1 transcription factor in Arabidopsis[J]. FEBS Lett, 2013, 587: 1543–1547. DOI:10.1016/j.febslet.2013.03.037 |
| [81] | Lee J, He K, Stolc V, et al. Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development[J]. Plant Cell, 2007, 19: 731–749. DOI:10.1105/tpc.106.047688 |
| [82] | Zhang H, He H, Wang X, et al. Genome-wide mapping of the HY5-mediated gene networks in Arabidopsis that involve both transcriptional and post-transcriptional regulation[J]. Plant J, 2011, 65: 346–358. DOI:10.1111/tpj.2011.65.issue-3 |
| [83] | Chattopadhyay S, Ang LH, Puente P, et al. Arabidopsis bZIP protein HY5 directly interacts with light-responsive promoters in mediating light control of gene expression[J]. Plant Cell, 1998, 10: 673–683. DOI:10.1105/tpc.10.5.673 |
| [84] | Wang H, Ma LG, Li JM, et al. Direct interaction of Arabidopsis cryptochromes with COP1 in light control development[J]. Science, 2001, 294: 154–158. DOI:10.1126/science.1063630 |
| [85] | Yang HQ, Tang RH, Cashmore AR. The signaling mechanism of Arabidopsis CRY1 involves direct interaction with COP1[J]. Plant Cell, 2001, 13: 2573–2587. DOI:10.1105/tpc.13.12.2573 |
| [86] | Seo HS, Watanabe E, Tokutomi S, et al. Photoreceptor ubiquitination by COP1 E3 ligase desensitizes phytochrome A signaling[J]. Genes Dev, 2004, 18: 617–622. DOI:10.1101/gad.1187804 |
| [87] | Jang S, Marchal V, Panigrahi KC, et al. Arabidopsis COP1 shapes the temporal pattern of CO accumulation conferring a photoperiodic flowering response[J]. EMBO J, 2008, 27: 1277–1288. DOI:10.1038/emboj.2008.68 |
| [88] | Brown BA, Jenkins GI. UV-B signaling pathways with different fluence-rate response profiles are distinguished in mature Arabidopsis leaf tissue by requirement for UVR8, HY5, and HYH[J]. Plant Physiol, 2008, 146: 576–588. |
| [89] | Favory JJ, Stec A, Gruber H, et al. Interaction of COP1 and UVR8 regulates UV-B-induced photomorphogenesis and stress acclimation in Arabidopsis[J]. EMBO J, 2009, 28: 591–601. DOI:10.1038/emboj.2009.4 |
| [90] | Dubos C, Le Gourrierec J, Baudry A, et al. MYBL2 is a new regulator of flavonoid biosynthesis in Arabidopsis thaliana[J]. Plant J, 2008, 55: 940–953. DOI:10.1111/tpj.2008.55.issue-6 |
| [91] | Gou JY, Felippes FF, Liu CJ, et al. Negative regulation of anthocyanin biosynthesis in Arabidopsis by a miR156-targeted SPL transcription factor[J]. Plant Cell, 2011, 23: 1512–1522. DOI:10.1105/tpc.111.084525 |
| [92] | Xu W, Dubos C, Lepiniec L. Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes[J]. Trends Plant Sci, 2015, 20: 176–185. DOI:10.1016/j.tplants.2014.12.001 |
| [93] | Wang YL, Wang YQ, Song ZQ, et al. Repression of MYBL2 by both microRNA858a and HY5 leads to the activation of anthocyanin biosynthetic pathway in Arabidopsis[J]. Mol Plant, 2016, 9: 1395–1405. DOI:10.1016/j.molp.2016.07.003 |
| [94] | Nguyen NH, Jeong CY, Kang GH, et al. MYBD employed by HY5 increases anthocyanin accumulation via repression of MYBL2 in Arabidopsis[J]. Plant J, 2015, 84: 1192–1205. DOI:10.1111/tpj.13077 |
| [95] | Toledo-Ortiz G, Johansson H, Lee KP, et al. The HY5-PIF regulatory module coordinates light and temperature control of photosynthetic gene transcription[J]. PLoS Gen, 2014, 10: e1004416. DOI:10.1371/journal.pgen.1004416 |
| [96] | Osterlund MT, Hardtke CS, Wei N, et al. Targeted destabilization of HY5 during light-regulated development of Arabidopsis[J]. Nature, 2000, 405: 462–466. DOI:10.1038/35013076 |
| [97] | Legris M, Nieto C, Sellaro R, et al. Perception and signalling of light and temperature cues in plants[J]. Plant J, 2017, 90: 683–697. DOI:10.1111/tpj.2017.90.issue-4 |
| [98] | Hardtke CS, Gohda KF, Osterlund MF, et al. HY5 stability and activity in Arabidopsis is regulated by phosphorylation in its COP1 binding domain[J]. EMBO J, 2000, 19: 4997–5006. DOI:10.1093/emboj/19.18.4997 |
| [99] | Li S, Wang W, Gao J, et al. MYB75 phosphorylation by MPK4 is required for light-induced anthocyanin accumulation in Arabidopsis[J]. Plant Cell, 2016, 28: 2866–2883. DOI:10.1105/tpc.16.00130 |
| [100] | Takshak S, Agrawal SB. Secondary metabolites and phenylpropanoid pathway enzymes as influenced under supplemental ultraviolet-B radiation in Withania somnifera Dunal, an indigenous medicinal plant[J]. J Photochem Photobiol B-Biol, 2014, 140: 332–343. DOI:10.1016/j.jphotobiol.2014.08.011 |
| [101] | Lokhande SD, Ogawa K, Tanaka A, et al. Effect of temperature on ascorbate peroxidase activity and flowering of Arabidopsis thaliana ecotypes under different light conditions[J]. J Plant Physiol, 2003, 160: 57–64. DOI:10.1078/0176-1617-00990 |
| [102] | Nakamichi N, Kusano M, Fukushima A, et al. Transcript profiling of an Arabidopsis pseudo response regulator arrhythmic triple mutant reveals a role for the circadian clock in cold stress response[J]. Plant Cell Physiol, 2009, 50: 447–462. DOI:10.1093/pcp/pcp004 |
| [103] | Zhang YQ, Zheng S, Liu ZJ, et al. Both HY5 and HYH are necessary regulators for low temperature-induced anthocyanin accumulation in Arabidopsis seedlings[J]. J Plant Physiol, 2011, 168: 367–374. DOI:10.1016/j.jplph.2010.07.025 |
| [104] | Leivar P, Quail PH. PIFs:pivotal components in a cellular signaling hub[J]. Trends Plant Sci, 2011, 16: 19–28. |
| [105] | Lorrain S, Allen T, Duek PD, et al. Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors[J]. Plant J, 2008, 53: 312–323. |
| [106] | Mizuno T, Nomoto Y, Oka H, et al. Ambient temperature signal feeds into the circadian clock transcriptional circuitry through the EC night-time repressor in Arabidopsis thaliana[J]. Plant Cell Physiol, 2014, 55: 958–976. DOI:10.1093/pcp/pcu030 |
| [107] | Catala R, Medina J, Salinas J. Integration of low temperature and light signaling during cold acclimation response in Arabidopsis[J]. Proc Natl Acad Sci USA, 2011, 108: 16475–16480. DOI:10.1073/pnas.1107161108 |
| [108] | Alabadi D, Gallego-Bartolome J, Orlando L, et al. Gibberellins modulate light signaling pathways to prevent Arabidopsis seedling de-etiolation in darkness[J]. Plant J, 2008, 53: 324–335. |
| [109] | Li K, Yu R, Fan LM, et al. DELLA-mediated PIF degradation contributes to coordination of light and gibberellin signalling in Arabidopsis[J]. Nat Commun, 2016, 7: 11868. DOI:10.1038/ncomms11868 |
| [110] | Wang K, Dickinson RE. Contribution of solar radiation to decadal temperature variability over land[J]. Proc Natl Acad Sci USA, 2013, 110: 14877–14882. DOI:10.1073/pnas.1311433110 |
| [111] | Osterlund MT, Wei N, Deng XW. The roles of photoreceptor systems and the COP1-targeted destabilization of HY5 in light control of Arabidopsis seedling development[J]. Plant Physiol, 2000, 124: 1520–1524. DOI:10.1104/pp.124.4.1520 |
| [112] | Jang K, Lee HG, Jung SJ, et al. The E3 ubiquitin ligase COP1 regulates thermosensory flowering by triggering GI degradation in Arabidopsis[J]. Sci Rep, 2015, 5: 12071. DOI:10.1038/srep12071 |
| [113] | Legris M, Klose C, Burgie ES, et al. Phytochrome B integrates light and temperature signals in Arabidopsis[J]. Science, 2016, 354: 897–900. DOI:10.1126/science.aaf5656 |
| [114] | Findlay KM, Jenkins GI. Regulation of UVR8 photoreceptor dimer/monomer photo-equilibrium in Arabidopsis plants grown under photoperiodic conditions[J]. Plant Cell Environ, 2016, 39: 1706–1714. DOI:10.1111/pce.12724 |
| [115] | Miyazaki Y, Takase T, Kiyosue T. ZEITLUPE positively regulates hypocotyl elongation at warm temperature under light in Arabidopsis thaliana[J]. Plant Signaling Behav, 2015, 10: e998540. DOI:10.1080/15592324.2014.998540 |
| [116] | Zeng S, Wu M, Zou C, et al. Comparative analysis of anthocyanin biosynthesis during fruit development in two Lycium species[J]. Physiol Plant, 2014, 150: 505–516. DOI:10.1111/ppl.12131 |
| [117] | Huang W, Khaldun AB, Lv H, et al. Isolation and functional characterization of a R2R3-MYB regulator of the anthocyanin biosynthetic pathway from Epimedium sagittatum[J]. Plant Cell Rep, 2016, 35: 883–894. DOI:10.1007/s00299-015-1929-z |
| [118] | Enoki S, Hattori T, Ishiai S, et al. Vanillylacetone up-regulates anthocyanin accumulation and expression of anthocyanin biosynthetic genes by inducing endogenous abscisic acid in grapevine tissues[J]. J Plant Physiol, 2017, 219: 22–27. DOI:10.1016/j.jplph.2017.09.005 |
| [119] | Wan L, Li B, Pandey MK, et al. Transcriptome analysis of a new peanut seed coat mutant for the physiological regulatory mechanism involved in seed coat cracking and pigmentation[J]. Front Plant Sci, 2016, 7: 1491. |
| [120] | Hu J, Chen G, Zhang Y, et al. Anthocyanin composition and expression analysis of anthocyanin biosynthetic genes in kidney bean pod[J]. Plant Physiol Biochem, 2015, 97: 304–312. DOI:10.1016/j.plaphy.2015.10.019 |
| [121] | Dodd AN, Salathia N, Hall A, et al. Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage[J]. Science, 2005, 309: 630–633. DOI:10.1126/science.1115581 |
| [122] | Green RM, Tingay S, Wang ZY, et al. Circadian rhythms confer a higher level of fitness to Arabidopsis plants[J]. Plant Physiol, 2002, 129: 576–584. DOI:10.1104/pp.004374 |
| [123] | Yerushalmi S, Yakir E, Green RM. Circadian clocks and adaptation in Arabidopsis[J]. Mol Ecol, 2011, 20: 1155–1165. DOI:10.1111/j.1365-294X.2010.04962.x |
 2018, Vol. 53
2018, Vol. 53


