酯酶是催化内外源物结构中酯键断裂的水解酶, 广泛分布于小至细菌、大至人体内。酯酶按其代谢化学键断裂方式不同可分为A、B和C族酯酶[1], 其中羧酸酯酶(carboxylesterases, CESs)、乙酰胆碱酯酶、丁酰胆碱酯酶同属于B族酯酶, 是重要的Ⅰ相代谢酶[2]。按序列同源性分类, CESs可分为CES1、CES2、CES3、CES4和CES5[3], 其中CES1和CES2在药物代谢中发挥主要作用, 研究得最为深入。CESs主要分布在哺乳动物组织的内质网中, 通常应用人肝S9和微粒体进行CESs的体外研究[4]。CESs作用底物广泛, 对维持机体正常生理功能具有重要作用。
CESs参与众多内、外源物及药物的代谢, 如血管紧张素转换酶抑制剂、抗癌药、免疫调节剂、有机磷农药等[5]。不同种属CESs表达水平不一, 导致药物在不同种属的代谢特性存在明显差异。此外, 由于羧酸酯酶是多基因家族, 其表达与活性会受单核苷酸多态性(single nucleotide polymorphism, SNP)影响, 使不同个体对同种药物反应也不尽相同[6]。
药物成酯因其可提高难溶性药物的溶解性、增加透膜能力、提高生物利用度的优点获得广泛关注, 越来越多的酯类前药被开发研究[7]。酯类药物进入机体后, 酯酶对其代谢动力学过程产生不可替代的作用。本文就CESs的分类、分布、功能和作用机制、种属差异性与基因多态性以及对药物代谢动力学的影响等最新研究进展进行总结, 以期为相关研究与药物设计提供借鉴。
1 CESs分类与分布药物代谢酶广泛存在于机体中, 参与內源物与外源物的生物转化, 在机体解毒与代谢中发挥重要作用。CESs作为体内重要的Ⅰ相药物代谢酶, 隶属于酯酶家族, 主要催化酯键、酰胺键、硫酯键等[7]水解, 其底物广泛, 且不同亚型间具有底物交叉性。1998年, Satoh等[3]根据对比编码基因的氨基酸序列, 在充分考虑底物特异性、组织分布、免疫特性以及基因调控差异后, 将哺乳动物羧酸酯酶分为5个家族, 即CES1、CES2、CES3、CES4和CES5。其中, CES1和CES2是CESs中主要同工酶, 也是人体内主要表达的亚型, 研究最为深入。所以本文以CES1与CES2为主, 讨论CESs对于药物代谢动力学的影响。
CESs在体内分布广泛, 主要在具有屏障功能的组织中表达, 如肺、肠、肝脏、肾脏、皮肤等。不同同工酶在体内分布也不相同, CES1在肝脏中高表达, 也存在于单核细胞、巨噬细胞、肺上皮细胞、心脏、睾丸等组织中, 在胃肠道表达较低[8]。与CES1相反, CES2主要在小肠、结肠、肾、肝脏、心脏和脑组织中表达[9]。CESs分布的不同初步决定了其代谢特异性。
2 结构与作用机制 2.1 CESs的结构特点CESs的结构特点与其活性和功能紧密相关。CESs主要位于内质网, 与内质网内很多蛋白质相似, CESs依靠C-末端的His-X-Glu-Leu (HXEL)序列通过KDEL受体与内质网膜结合从而储存在内质网内[10]。此外, 由17至20个氨基酸残基(包括疏水性氨基酸)组成的信号肽也有助于其在内质网的储存。CESs由3个不同的区域构成, 包括含催化中心(催化三连体)的中心区、调控区和α/β区。在3个区域的交界面存在一个10~15Å深的疏水口袋, 由催化三连体残基构成, 也包括C末端的螺旋结构, 是建立质子转移链的重要活化位点[11], 4个半胱氨酸组成特定二硫键以维持活性中心的空间构型。调控区主要由螺旋结构组成, 较高的热取代参数提示这个区域具有较好的动态流动性, 推测与调控底物的结合和产物的释放相关[12]。α/β区则与催化区和调控区毗连。此外, 在N79位有一个N-糖苷键多糖, 在芳香族和脂肪族酯催化过程中起重要作用[13]。1993年, 研究发现人CES基因跨度约为30 kb, 包含14个外显子, CES1和CES2基因有48%序列同源[7]。经互变分析确认, 人CES1催化三聚体主要由Ser221、Glu354和His467组成[14]。这些特殊位点的互变极大程度减少了CESs对许多底物的催化活性, 进一步确定催化三连体的组成及在形成假定电荷中断系统中发挥重要作用。
2.2 CESs的作用机制近年来研究发现, His和Glu形成的低阻性氢键可促进丝氨酸β-OH基团的亲核攻击[15]。Ser将质子经His传递给底物羰基基团, 从而形成四面体中间物。低阻性氢键可有效催化三连体稳定在四面体中间物中, 同时, Gly123与Gly124和N-H肽键形成的弱氢键同低阻性氢键共同使四面体中间物处于过渡状态[16], 随后His450中移除一个质子使四面体中间物释放醇产物形成酰基-酶复合物。此时, 水分子作为亲核试剂, 代替原始底物醇基团发生脱酰反应。此外, 除了水之外的化合物也可以攻击酰基-酶中间体, 引起酯交换反应而非水解反应[17]。据文献报道, 人CES催化区残基与其他种属高度同源, 提示不同种属间催化机制可能相近。具体反应过程如图 1所示。
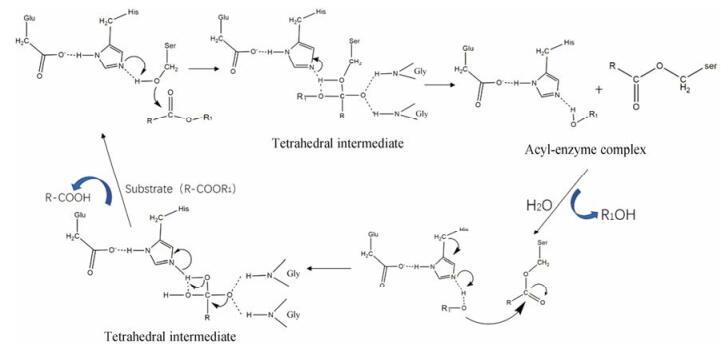
|
Figure 1 The hydrolysis mechanism of carboxylesterases |
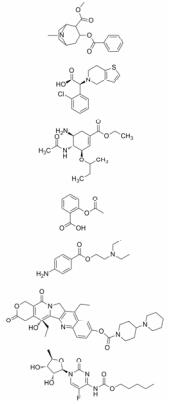
药物的结构决定参与代谢的水解酶类型和药代动力学差异。CESs的底物广泛, 不同结构药物与酶作用种类与难易程度也不相同[18]。CES1 X射线晶体结构显示, CES1结构中包含一个大型灵活口袋和一个小型刚性口袋, 柔性大口袋有水解多种不同结构化合物的能力, 而刚性口袋对于底物更有选择性[19]。CES1偏向于水解含有较小乙醇基和较大乙酰基的底物, 如可卡因、氯吡格雷和奥司他韦等[19-21]。而CES2因为活性区的构象干扰, 酰基-CES2结合物的性质限制了CES2的底物特异性[22, 23]。因此CES2偏向于水解含有较大乙醇基和较小乙酰基底物, 如阿司匹林、普鲁卡因、伊立替康和卡培他滨[24]等。此外, 根据CESs底物特异性, 还可设计特异结合CES1或CES2的探针底物用来检测不同生物基质中的酶活性[25-31]。因此, 设计药物时应考虑结构对代谢的影响, 以得到药代性质良好的候选药物。表 1列举了CES1与CES2特征底物结构。
| Table 1 The representative substrates of CES1 and CES2 |
猕猴、鼠类、猪、狗、兔及人的血浆酯酶活性及表达水平存在显著差异。对氧磷酶(paraoxonase, PON)和丁酰胆碱酯酶(butyrylcholi nesterase, BChE)普遍存在于所有物种, 在灵长类动物与狗中高表达, 而CESs在兔子和鼠类中表达丰富[32]。酯酶表达水平的差异导致不同种属对药物的水解活性不同[33]。普萘洛尔(propranolol, PL)及其衍生物在小鼠和大鼠血浆中被高活性CESs迅速水解, 但兔血浆中的CESs只水解PL衍生物[34]。与啮齿类动物相比, 人体内CESs活性较低[35]。此外, CESs在不同种属的表达与功能也存在差异, 如CES3、CES4和CES5主要在猫或小鼠体内存在, 负责附睾中脂质转移、肠首过代谢等[36], 而在人体内表达较少[8]。因此, 临床前研究时应充分考虑上述差异对药物代谢的影响, 可重点参考非啮齿类动物数据, 以与人接近的灵长类动物最佳, 从而选择合适的动物模型。
3.3 基因多态性和性别差异CESs是多基因家族酶系。近年来大量研究表明, 人体内CES1和CES2的表达水平存在个体差异, 影响药物代谢动力学过程, 引起药物疗效改变甚至产生严重不良反应。
奥司他韦是无活性抗流感病毒前药, 通过CES1代谢生成活性代谢产物。由于CES1是唯一参与其代谢的催化酶, 所以CES1功能的改变是临床奥司他韦治疗差异的主要原因[37]。研究表明, 基因多态性在CES1功能调控中发挥重要作用。无论是CES1蛋白质表达还是对奥司他韦的催化活性都与rs2244613、rs8192935、-816A>C或CES1P1/CES1P1VAR等基因型密切相关[38]。例如, CES1突变体G143E (rs71647871)是一个源于外显子4由G替代A的非同义突变, 导致了143编码位置甘氨酸变成了谷氨酸, 此变异最初见于盐酸哌甲酯控释片的代谢研究[39]。与143G/G基因型相比, 基因型为143G/E时约40%奥司他韦在肝内水解, 基因突变改变了药物在体内的药效与代谢特性。体外CES1-转染细胞研究结果显示, G143E的催化活性可改变多种CES1底物的代谢, 包括奥司他韦、哌甲酯、氯吡格雷、沙库比曲和血管转化酶抑制剂。临床研究也证实, 哌甲酯治疗注意力不足多动障碍症(ADHD)时对G143E携带者所需的剂量明显降低[40], 与非携带者相比氯吡格雷血浆中活性代谢产物水平和药效显著提高。
与CES1相似, CES2基因多态性对药物代谢也会产生显著影响。Song等[41]发现, 利福平血浆药物浓度的明显差异与CES2三个紧密相关的变异基因型(C-2263a > G、c.269-965a > G和c.1612 + 136g > A)和c.1872*302_304delGAA有关。随着3个紧密相关变异基因型的风险等位基因数目增多, 利福平平均血药浓度显著升高, 而随着c.1872*302_304delgaa风险等位基因数目增加其血药浓度下降[42]。启动子分析结果表明, 启动子区变体c.-2263A > G影响了酶的转录。A到G的改变可导致荧光素酶持续下降, 引起药物代谢水平和血药浓度的改变。总之, CES1的基因多态性可影响达比加群、哌醋甲酯、奥司他韦、咪达普利和氯吡格雷等多种药物代谢; CES2基因变异则会改变利福平、阿司匹林和伊立替康等药物处置[43]。此外, CESs新型基因突变体可预测严重的早发型卡培他滨毒性[44], 临床治疗意义重大。虽然只是初步研究结果, 但也提示CESs多态性在未来临床应用中的意义。
以往临床研究中, 多个CES1底物的药代动力学特征都显示出性别差异, 包括洛伐他汀、辛伐他汀、哌甲酯和奥司他韦[45]。文献[46]曾报道, 奥司他韦服药女性肝中CES1蛋白表达比男性高17.3%, 同时女性肝内活化速率也较男性高27.8%, 导致药代特性发生改变。研究[47]也曾报道单剂量给药(0.3 mg·kg-1)消旋哌甲酯后, d-哌甲酯的AUC值男性明显高于女性。虽然大量临床研究表明了CESs的性别差异, 但是分子机制与应用还有待深入研究。此外, 疾病也会影响CESs的表达, 如CES1与CES2在LPS诱导的免疫性肝损伤中表达降低[48]。
4 结语随着酯类前药的大量出现, 药物在机体的处置过程及代谢特征成为值得关注的热点。对于亲脂性强、水溶性差的化合物, 非CYP介导酶(UGTs、酯酶和醛氧化酶等)可能成为主要代谢酶[49]。其中CESs更是酯类药物水解的关键酶, 其在体内分布广泛, 底物广泛而多样, 作用机制复杂, 具有基因多态性。药物经过CESs代谢后可导致活化或失活, 也常被认为是一种解毒机制。除此之外, CESs还可与有机磷酸酯类化合物(organophosphorus compounds, OPs)以化学计量方式相互作用导致OPs失活, 可用于减缓OPs的毒性, 保护环境安全[18]。虽然目前CESs的研究与日俱增, 但是有关种属差异、性别差异的机制与应用还有待深入研究。因此, 充分了解CESs的作用机制与生物学特性在药物代谢动力学和药物-药物相互作用中具有重要意义, 并可在新药设计与研发中提供指导。
| [1] | Masaki K, Hashimoto M, Imai T. Intestinal first-pass metabolism via carboxylesterase in rat jejunum and ileum[J]. Drug Metab Dispos, 2007, 35: 1089–1095. DOI:10.1124/dmd.106.013862 |
| [2] | Zhu QG, Hu JH, Zeng HW. Stereoselectivity of skin carboxylesterase metabolism[J]. Acta Pharm Sin (药学学报), 2005, 40: 322–326. |
| [3] | Satoh T, Hosokawa M. The mammalian carboxylesterases:from molecules to functions[J]. Annu Rev Pharmacol Toxicol, 1998, 38: 257. DOI:10.1146/annurev.pharmtox.38.1.257 |
| [4] | Trapa PE, Beaumont K, Atkinson K, et al. In vitro-in vivo extrapolation of intestinal availability for carboxylesterase substrates using portal vein-cannulated monkey[J]. J Pharm Sci, 2017, 106: 898–905. DOI:10.1016/j.xphs.2016.12.001 |
| [5] | Makhaeva GF, Rudakova EV, Serebryakova OG, et al. Esterase profiles of organophosphorus compounds in vitro predict their behavior in vivo[J]. Chem Biol Interact, 2016, 259: 332–342. DOI:10.1016/j.cbi.2016.05.002 |
| [6] | Wang X, Wang G, Shi J, et al. CES1 genetic variation affects the activation of angiotensin-converting enzyme inhibitors[J]. Pharmacogenomics J, 2016, 16: 220–230. DOI:10.1038/tpj.2015.42 |
| [7] | Hui DY, Howles PN. Carboxyl ester lipase structure-function relationship and physiological role in lipoprotein metabolism and atherosclerosis[J]. J Lipid Res, 2002, 43: 2017–2030. DOI:10.1194/jlr.R200013-JLR200 |
| [8] | Satoh T, Hosokawa M. Structure, function and regulation of carboxylesterases[J]. Chem Biol Interact, 2006, 162: 195–211. DOI:10.1016/j.cbi.2006.07.001 |
| [9] | Hatfield MJ, Umans RA, Hyatt JL, et al. Carboxylesterases:general detoxifying enzymes[J]. Chem Biol Interact, 2016, 259: 327–331. DOI:10.1016/j.cbi.2016.02.011 |
| [10] | Lian J, Nelson R, Lehner R. Carboxylesterases in lipid metabolism:from mouse to human[J]. Protein Cell, 2017. DOI:10.1007/s13238-017-0437-z |
| [11] | Hosokawa M, Furihata T, Yaginuma Y, et al. Genomic structure and transcriptional regulation of the rat, mouse, and human carboxylesterase genes[J]. Drug Metab Rev, 2007, 39: 1–15. |
| [12] | Imai T. Human carboxylesterase isozymes:catalytic properties and rational drug design[J]. Drug Metab Pharmacokinet, 2006, 21: 173–185. DOI:10.2133/dmpk.21.173 |
| [13] | Arena de Souza V, Scott DJ, Nettleship JE, et al. Comparison of the structure and activity of glycosylated and aglycosylated human carboxylesterase 1[J]. PLoS One, 2015, 10: e0143919. DOI:10.1371/journal.pone.0143919 |
| [14] | Yu X, Sigler SC, Hossain D, et al. Global and local molecular dynamics of a bacterial carboxylesterase provide insight into its catalytic mechanism[J]. J Mol Model, 2012, 18: 2869–2883. DOI:10.1007/s00894-011-1308-9 |
| [15] | Chen Q, Luan ZJ, Cheng X, et al. Molecular dynamics investigation of the substrate binding mechanism in carboxylesterase[J]. Biochemistry, 2015, 54: 1841–1848. DOI:10.1021/bi5015612 |
| [16] | Redinbo MR, Bencharit S, Potter PM. Human carboxylesterase 1:from drug metabolism to drug discovery[J]. Biochem Soc Trans, 2003, 31: 620–624. DOI:10.1042/bst0310620 |
| [17] | Cashman J, Perroti B, Berkman C, et al. Pharmacokinetics and molecular detoxification[J]. Environ Health Perspect, 1996, 104: 23–40. DOI:10.1289/ehp.96104s123 |
| [18] | Hemmert AC, Otto TC, Chica RA, et al. Nerve agent hydrolysis activity designed into a human drug metabolism enzyme[J]. PLoS One, 2011, 6: e17441. DOI:10.1371/journal.pone.0017441 |
| [19] | Fleming CD, Edwards CC, Kirby SD, et al. Crystal structures of human carboxylesterase 1 in covalent complexes with the chemical warfare agents soman and tabun[J]. Biochemistry, 2007, 46: 5063–5071. DOI:10.1021/bi700246n |
| [20] | Thomsen R, Rasmussen HB, Linnet K, et al. In vitro drug metabolism by human carboxylesterase 1:focus on angiotensinconverting enzyme inhibitors[J]. Drug Metab Dispos, 2014, 42: 126–133. |
| [21] | Vistoli G, Pedretti A, Mazzolari A, et al. Influence of ionization state on the activation of temocapril by hCES1:a molecular-dynamics study[J]. Chem Biodivers, 2009, 6: 2092–2100. DOI:10.1002/cbdv.v6:11 |
| [22] | Ross MK, Crow JA. Human carboxylesterases and their role in xenobiotic and endobiotic metabolism[J]. J Biochem Mol Toxicol, 2007, 21: 187–196. DOI:10.1002/(ISSN)1099-0461 |
| [23] | Zhu QG, Hu JH, Liu JY, et al. Stereoselective characteristics and mechanisms of epidermal carboxylesterase metabolism observed in HaCaT keratinocytes[J]. Biol Pharm Bull, 2007, 30: 532–536. DOI:10.1248/bpb.30.532 |
| [24] | Fujita K, Kubota Y, Ishida H, et al. Irinotecan, a key chemotherapeutic drug for metastatic colorectal cancer[J]. World J Gastroenterol, 2015, 21: 12234–12248. DOI:10.3748/wjg.v21.i43.12234 |
| [25] | Du Z, Chen DW, Fu ZW, et al. The inhibition of carboxylesterases by praeruptorin C, D and E[J]. Acta Pharm Sin (药学学报), 2017, 52: 66–70. |
| [26] | Wang DD, Jin Q, Zou LW, et al. A bioluminescent sensor for highly selective and sensitive detection of human carboxylesterase 1 in complex biological samples[J]. Chem Commun, 2016, 52: 3183–3186. DOI:10.1039/C5CC09874B |
| [27] | Wang DD, Jin Q, Hou J, et al. Highly sensitive and selective detection of human carboxylesterase 1 activity by liquid chromatography with fluorescence detection[J]. J Chromatogr B, 2016, 1008: 212–218. DOI:10.1016/j.jchromb.2015.11.046 |
| [28] | Lei W, Wang DD, Dou TY, et al. Assessment of the inhibitory effects of pyrethroids against human carboxylesterases[J]. Toxicol Appl Pharmacol, 2017, 321: 48–56. DOI:10.1016/j.taap.2017.02.018 |
| [29] | Feng L, Liu ZM, Xu L, et al. A highly selective longwavelength fluorescent probe for the detection of human carboxylesterase 2 and its biomedical applications[J]. Chem Commun, 2014, 50: 14519–14522. DOI:10.1039/C4CC06642A |
| [30] | Jin Q, Feng L, Wang DD, et al. A highly selective nearinfrared fluorescent probe for carboxylesterase 2 and its bioimaging applications in living cells and animals[J]. Biosens Bioelectron, 2016, 83: 193–199. DOI:10.1016/j.bios.2016.04.075 |
| [31] | Jin Q, Feng L, Wang DD, et al. A two-photon ratiometric fluorescent probe for imaging carboxylesterase 2 in living cells and tissues[J]. ACS Appl Mater Interfaces, 2015, 7: 28474–82481. DOI:10.1021/acsami.5b09573 |
| [32] | Bahar FG, Ohura K, Ogihara T, et al. Species difference of esterase expression and hydrolase activity in plasma[J]. J Pharm Sci, 2012, 101: 3979–3988. DOI:10.1002/jps.23258 |
| [33] | Taketani M, Shii M, Ohura K, et al. Carboxylesterase in the liver and small intestine of experimental animals and human[J]. Life Sci, 2007, 81: 924–932. DOI:10.1016/j.lfs.2007.07.026 |
| [34] | Godin SJ, Scollon EJ, Hughes MF, et al. Species differences in the in vitro metabolism of deltamethrin and esfenvalerate:differential oxidative and hydrolytic metabolism by humans and rats[J]. Drug Metab Dispos, 2006, 34: 1764–1771. DOI:10.1124/dmd.106.010058 |
| [35] | Merali Z, Ross S, Paré G. The pharmacogenetics of carboxylesterases:CES1 and CES2 genetic variants and their clinical effect[J]. Drug Metab Drug Interact, 2014, 29: 143–151. |
| [36] | Holmes RS, Wright MW, Laulederkind SJ, et al. Recommended nomenclature for five mammalian carboxylesterase gene families:human, mouse, and rat genes and proteins[J]. Mamm Genome, 2010, 21: 427–41. DOI:10.1007/s00335-010-9284-4 |
| [37] | Shi J, Wang X, Eyler RF, et al. Association of oseltamivir activation with gender and carboxylesterase 1 genetic polymorphisms[J]. Basic Clin Pharmacol Toxicol, 2016, 119: 555–561. DOI:10.1111/bcpt.2016.119.issue-6 |
| [38] | Oh J, Lee S, Lee H, et al. The novel carboxylesterase 1 variant c.662A > G may decrease the bioactivation of oseltamivir in humans[J]. PLoS One, 2017, 12: e0176320. DOI:10.1371/journal.pone.0176320 |
| [39] | Shi J, Wang X, Nguyen JH, et al. Dabigatran etexilate activetion is affected by the CES1 genetic polymorphism G143E (rs71647871) and gender[J]. Biochem Pharmacol, 2016, 119: 76–84. |
| [40] | Nemoda Z, Angyal N, Tarnok Z, et al. Carboxylesterase 1 gene polymorphism and methylphenidate response in ADHD[J]. Neuropharmacology, 2009, 57: 731–733. DOI:10.1016/j.neuropharm.2009.08.014 |
| [41] | Song SH, Chang HE, Jun SH, et al. Relationship between CES2 genetic variations and rifampicin metabolism[J]. J Antimicrob Chemother, 2013, 68: 1281–1284. DOI:10.1093/jac/dkt036 |
| [42] | Mendell J, Chen S, He L, et al. The effect of rifampin on the pharmacokinetics of edoxaban in healthy adults[J]. Clin Drug Investig, 2015, 35: 447–453. DOI:10.1007/s40261-015-0298-2 |
| [43] | Mukai M, Uchimura T, Zhang X, et al. Effects of rifampin on the pharmacokinetics of a single dose of istradefylline in healthy subjects[J]. J Clin Pharmacol, 2017. DOI:10.1002/jcph.1003 |
| [44] | Hamzic S, Kummer D, Milesi S, et al. Novel genetic variants in carboxylesterase 1 predict severe early-onset capecitabinerelated toxicity[J]. Clin Pharmacol Ther, 2017, 102: 796–804. DOI:10.1002/cpt.641 |
| [45] | Zhu HJ, Appel DI, Jiang Y, et al. Age- and sex-related expression and activity of carboxylesterase 1 and 2 in mouse and human liver[J]. Drug Metab Dispos, 2009, 37: 1819–1825. DOI:10.1124/dmd.109.028209 |
| [46] | Huang HJ, Tsai ML, Chen YW, et al. Quantitative shot-gun proteomics and MS-based activity assay for revealing gender differences in enzyme contents for rat liver microsome[J]. J Proteomics, 2011, 74: 2734–2744. DOI:10.1016/j.jprot.2011.01.015 |
| [47] | Stage C, Jürgens G, Guski LS, et al. The impact of CES1 genotypes on the pharmacokinetics of methylphenidate in healthy Danish subjects[J]. Br J Clin Pharmacol, 2017, 83: 1506–1514. DOI:10.1111/bcp.v83.7 |
| [48] | Shi J, Wang X, Nguyen J, et al. Sacubitril is selectively activated by carboxylesterase 1(CES1) in the liver and the activation is affected by CES1 genetic variation[J]. Drug Metab Dispos, 2016, 44: 554–559. DOI:10.1124/dmd.115.068536 |
| [49] | Argikar UA, Potter PM, Hutzler JM, et al. Challenges and opportunities with non-CYP enzymes aldehyde oxidase, carboxylesterase, and UDP-glucuronosyltransferase:focus on reaction phenotyping and prediction of human clearance[J]. AAPS J, 2016, 18: 1391–1405. DOI:10.1208/s12248-016-9962-6 |
 2018, Vol. 53
2018, Vol. 53