药物代谢是指药物分子被机体吸收后, 在机体作用下发生化学结构转化的过程。药物代谢对药物的药效、毒性及临床合并用药时的药物相互作用等具有重要影响。作为体内最重要的Ⅰ相药物代谢酶, 细胞色素P450酶参与约75%的药物代谢反应。临床上基于代谢的药物相互作用大多都是通过对P450酶亚型的抑制或诱导实现的, 因此这类酶及相关氧化代谢反应一直是新药研发中关注的焦点, 相应的研究技术和评价策略也已趋于成熟。
近年来, 随着组合化学、高通量筛选等技术的发展, 结构新颖的药物层出不穷, 非P450酶催化的氧化代谢途径逐渐进入新药研发科学家的视线。然而, 由于对非P450酶催化的特殊代谢途径认识不足, 导致临床前实验动物种属选择失误, 使临床试验阶段药物被终止开发的案例越来越多。例如, 美国因赛特制药公司开发的第一代高效、高选择性c-MET抑制剂SGX523, 在Ⅰ期临床剂量上升至 > 80 mg时, 患者出现急性肾功能衰竭, 表现为血清肌酸酐升高。代谢研究发现SGX523结构中的喹啉环能够被醛氧化酶 (AO) 代谢生成2-喹啉酮, 这一代谢物水溶性差, 在肾小管内形成结晶。安全性评价中使用的动物醛氧化酶的催化活性低, 因而未发现SGX523的肾毒性[1]。此外, 日本山之内制药公司开发的新型腺苷A1/2双重抑制剂FK3453, 到了临床阶段才发现人体的系统暴露量极低, 达不到药效浓度, 而被终止开发, 究其原因是人体内AO介导了2-氨基嘧啶环的氧化代谢[2]。这些案例引发了人们对非P450酶及其介导的特殊代谢途径的关注。
常见的催化药物氧化代谢的非P450酶包括黄素单氧化酶 (FMO)、单胺氧化酶 (MAO)、醛氧化酶 (AO)、黄嘌呤氧化酶 (XO)、乙醇脱氢酶 (ADH) 和乙醛脱氢酶 (ALDH)[3-8]。本文将从催化反应类型、常见底物、基因多态性及药物相互作用等方面对以上非P450酶进行介绍。
1 黄素单氧化酶 (FMO) 1.1 底物类型FMO为微粒体中除P450酶外的另一种氧化酶。与P450酶类似, FMO催化的氧化反应需要辅助因子NADPH的参与。FMO和P450均能催化杂环类化合物发生N-氧化和S-氧化。不同的是, FMO的底物通常为含有N或S原子的弱亲核试剂, 而P450酶的底物并不局限于亲核试剂[9-11]。与P450酶相比, FMO对温度更敏感。在不加NADPH时, 45 ℃条件下预孵5 min会使FMO失活, 而P450酶活性不受影响[12]。
一般认为, pKa值为8~11的碱性胺的N-氧化反应主要由FMO催化, 而弱碱由P450和FMO共同催化[13]。此外, FMO一般不催化伯胺的氧化, 而主要催化仲胺的N-氧化过程, 形成羟胺代谢产物, 如抗真菌药酮康唑的脱乙酰基代谢物[14] (deacetyl-ketoconazole, 图 1A)。叔胺类化合物也可经FMO催化发生N-氧化, 如治疗乳腺癌的药物他莫昔芬[15] (tamoxifen, 图 1B)。除了N-氧化外, FMO也催化S-氧化反应, 如抗组胺药西咪替丁[16, 17] (cimetidine, 图 1C)。近年来, 也有文献[18]报道FMO能催化C-氧化反应, 如MRX-I在FMO5催化下发生Baeyer-Villiger 氧化反应。

|
Figure 1 Flavin-containing monooxygenase (FMO)-mediated oxidative metabolism of deacetyl-ketoconazole (A), tamoxifen (B) and cimetidine (C) |
目前, FMO共有5种亚型被鉴定, 分别为FMO1、FMO2、FMO3、FMO4和FMO5。在成人肝中, FMO3和FMO5是最主要的两种亚型[17]。FMO1在成人肝中表达量低, 但在胎儿肝脏中是最主要的亚型[19]。
文献[20]报道FMO3具有基因多态性。在正常人体内, 三甲胺经FMO3代谢产生三甲胺氮氧化物, 尿中回收的氮氧化物与原形含量的比值为97:3, 而在FMO3基因表达异常的人体内, 三甲胺代谢严重不足, 氮氧化物与原形含量的比值仅为1:9, 从而引发“鱼腥综合征”[21, 22]。
1.3 药物相互作用与P450酶不同, 有关FMO抑制剂和诱导剂的报道较少。FMO的经典抑制剂有吲哚甲醇[23, 24]和甲硫基咪唑[25]等。Zaragoza等[26]报道苯巴比妥可以提高FMO介导的硫代乙酰胺生物活化的程度, 从而导致硫代乙酰胺在大鼠体内肝毒性增加。此外, 性激素等内源性物质可以诱导FMO3活性[27], 孕妇在服用经FMO3代谢的药物时应考虑到这一点。
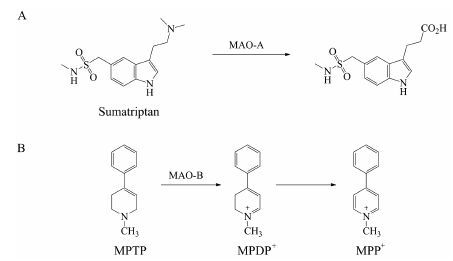
2 单胺氧化酶 (MAO) 2.1 底物类型MAO是存在于线粒体外膜上的一类氧化酶, 可以催化胺类化合物 (伯、仲、叔胺) 发生氧化脱氨或脱氢反应[28, 29]。MAO催化的氧化反应不需要辅助因子的参与。人的脑、肝和胎盘组织中均存在MAO[30]。MAO共有两个亚型, 分别为MAO-A和MAO-B[29]。M AO-A的常见底物有肾上腺素和5-羟色胺[29]等内源性物质。此外, 与5-羟色胺具有类似结构的药物如舒马曲坦 (sumatriptan, 图 2A) 也可被MAO-A广泛代谢[4, 31]。MAO-B的常见底物有β-苯乙胺、苄胺和司来吉兰等[32]。与P450酶类似, MAO能够介导化合物发生氧化代谢产生毒性代谢物。例如, MAO-B氧化1-甲基-4-苯基-1, 2, 3, 6-四氢吡啶 (MPTP, 图 2B) 形成1-甲基-4-苯基-2, 3-二氢吡啶 (MPDP+) 中间体, 最终生成1-甲基-4-苯基吡啶 (MPP+)[33]。MPP+是一种神经毒素, 可导致脑黑质线粒体中毒, 产生与帕金森症类似的症状[34]。
|
Figure 2 Monoamine oxidase (MAO)-mediated oxidative metabolism of sumatriptan (A) and MPTP (B) |
由于MAO能催化神经递质的代谢, 寻找MAO的可逆或不可逆抑制剂, 增加神经递质在体内的水平, 对于抑郁症及帕金森症的治疗具有重要意义[30, 35, 36]。文献[6, 37]报道的MAO抑制剂有氯吉兰、苯乙肼、艾梦克酮和拉扎贝胺等。此外, Fowler等[38]报道烟草中的尼古丁能够降低吸烟者体内的MAO水平, 增加体内去甲肾上腺素水平, 进而引发一系列中枢及外周神经效应。
目前, 关于MAO被外源性物质诱导的报道较少。Sarabia等[39]指出雌二醇可以诱导仓鼠肾脏中的MAO-B水平, 增加儿茶酚胺类化合物的脱胺代谢过程, 该过程产生过氧化氢和羟基自由基等有毒物质, 可能导致肾肿瘤的发生。
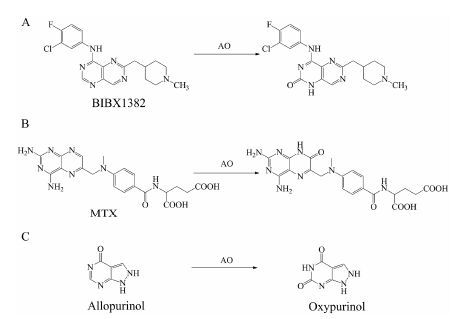
3 醛氧化酶 (AO) 3.1 底物类型AO是一类广泛存在于胞浆中的高度保守的钼-黄素蛋白, 除了催化醛生成羧酸外, 还能催化含氮杂环类化合物的氧化过程, 以及一些化合物, 如芳硝基化合物[40, 41]、环氧化物[42]、氮氧化物[43, 44]和亚砜[45]等的还原过程。本文重点阐述AO催化的含氮杂环类化合物的氧化反应。这类反应通常表现为芳香环中杂原子邻位的碳被氧化。表皮生长因子受体抑制剂BIBX1382 (图 3A)、抗肿瘤药甲氨蝶呤 (MTX, 图 3B), 以及治疗痛风的药物别嘌呤醇 (allopurinol, 图 3C) 等[5, 46-48]均为AO的常见底物。
|
Figure 3 AO-mediated oxidative metabolism of BIBX1382 (A), MTX (B) and allopurinol (C) |
AO存在明显的种属及个体差异[1, 49, 50]。目前, 共有4种亚型被鉴定, 分别为AOX1、AOX3、AOX4和AOX3l1[51, 52]。Garattini等[52]总结了人、黑猩猩和常用实验动物的肝及其他组织表达的AO亚型 (表 1) 。
| Table 1 The AO isoforms expressed in the liver and other tissues of humans, chimpanzees and popular animal experimental models[52] |
由于AO具有明显的种属差异, 当候选药物主要经AO代谢时, 临床前实验动物种属的选择至关重要。由于实验动物选择不当而被终止开发的药物除了前言中提到的c-MET抑制剂SGX523和新型腺苷A1/2双重抑制剂FK3453外, 还有c-MET抑制剂JNJ-38877605[53]和表皮生长因子受体抑制剂BIBX1382[47]等。一般认为, 人和猴肝中AO活性最高, 其次为大鼠和小鼠, 而犬中几乎没有AO活性[1, 2, 47, 54]。然而, 不同种属体内AO活性的相对高低并不是绝对的, 而是取决于催化的底物。例如, Schofield等[55]发现N-[(2'-二甲氨基)乙基] 吖啶-4-甲酰胺 (DACA) 作为底物时, 豚鼠肝胞浆中的AO催化活性最高, 其次为大鼠, 人肝胞浆中的AO催化活性最低。此外, Choughule等[56]报道甲氨蝶呤作为底物时, 兔肝胞浆中的AO表现出最高的催化活性, 接下来依次为猴、豚鼠和大鼠, 人肝胞浆中AO催化活性最低。因此, 当候选药物确定经AO代谢时, 可以比较该药物在不同种属肝胞浆中的代谢情况, 选择与人体代谢最接近的种属作为临床前安全性评价及药动学性质评价的动物模型。
3.3 药物相互作用目前文献报道的具有AO抑制活性的化合物有: 甲萘醌[1, 50]、雷洛昔芬[57, 58]、氯丙嗪[58, 59]、西咪替丁[59]、喹硫平[58]、美沙酮[60]、雌二醇[41, 58, 61]和肼屈嗪[59, 62]等。Nirogi等[61]考察了雷洛昔芬、氯丙嗪、肼屈嗪、雌二醇和喹硫平对AO及其他酶的抑制能力, 结果发现仅雌二醇是AO的特异性抑制剂。
AO是一种可以被诱导的酶, 内源性物质褪黑素以及外源性化合物哌嗪和2, 3, 7, 8-四氯二苯并二噁英 (TCDD) 等均能诱导AO[63-66]。
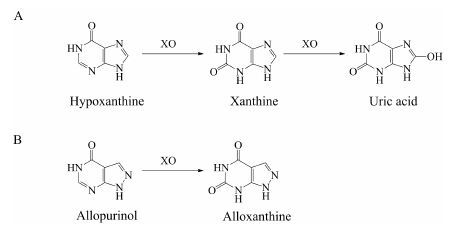
4 黄嘌呤氧化酶(XO) 4.1 底物类型XO也是一类存在于胞浆中的钼-黄素蛋白, 与AO具有高度同源性。不同的是, AO主要分布在脑和肝中, 而XO主要存在于小肠中[6]。除了催化一些化合物的还原过程外[40], XO的主要功能为催化嘌呤类化合物的代谢。例如, XO催化次黄嘌呤 (hypoxanthine) 氧化形成黄嘌呤 (xanthine), 进一步催化黄嘌呤形成尿酸 (uric acid)[6] (图 4A)。此外, XO也可以催化别嘌呤醇 (allopurinol) 氧化形成别黄嘌呤 (alloxanthine)[6] (图 4B)。
|
Figure 4 Xanthine oxidase (XO)-mediated oxidative metabolism of hypoxanthine (A) and allopurinol (B) |
别嘌呤醇是XO的经典抑制剂, 临床上可以用于治疗体内尿酸过多引起的高尿酸血症及痛风[67, 68]。此外, 文献报道的XO抑制剂还有甲氨蝶呤和奥昔嘌醇[1, 69]等。Barr等[70]报道在由人的肝组织制备肝胞浆的过程中会使用含有别嘌呤醇的肝灌流液, 别嘌呤醇在AO作用下产生大量奥昔嘌醇 (图 3C), 奥昔嘌醇显著抑制XO活性, 因此作者认为在利用人肝胞浆进行XO底物筛查实验前应先利用阳性底物考察人肝胞浆是否具有XO活性, 避免出现假阴性结果。
XO是一种可以被诱导的酶。文献报道干扰素、细菌脂多糖和TCDD均能够诱导部分啮齿类动物体内的XO活性[65, 71-73]。此外, 也有文献[74]报道低氧条件 (0.5%~3% O2) 可以增加XO活性, 但此时XO的mRNA和蛋白表达水平没有发生改变。
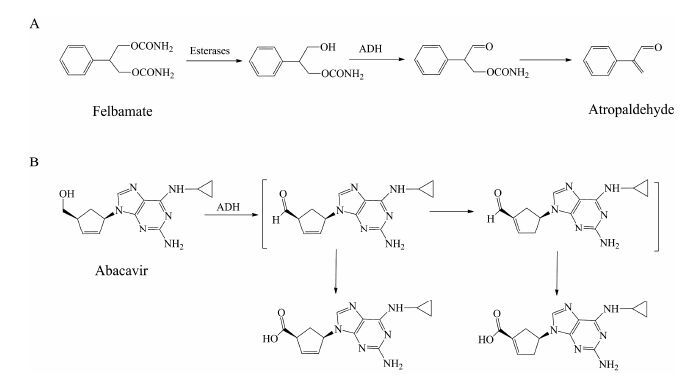
5 乙醇脱氢酶 (ADH) 5.1 底物类型ADH是一类存在于胞浆中, 主要催化醇类化合物氧化形成相应的醛或酮的酶。ADH催化的反应是可逆的, 需要辅助因子NAD+或NADH的参与。最常见的ADH底物为乙醇。此外, ADH还参与非甾体抗炎药塞来昔布[75]、治疗脑缺血药丁苯酞[76]、抗癫痫药非尔氨脂 (felbamate)[77]、新型酪氨酸激酶抑制剂tivantinib[7], 以及逆转录酶抑制剂阿巴卡韦 (abacavir)[78]等药物的代谢过程。ADH催化的代谢反应可能与药物在临床上出现的不良反应有关。例如, 非尔氨脂经酯酶 (esterases) 水解形成羟基代谢物, 进一步经ADH氧化形成醛, 最后自发形成阿托醛 (图 5A)[77]。该代谢活化过程被认为与非尔氨脂在临床上出现的再生障碍性贫血及肝毒性等不良反应有关[79]。类似的例子还有阿巴卡韦, 其在ADH催化下氧化产生醛中间体, 进一步发生异构化形成α, β-不饱和醛 (图 5B)。α, β-不饱和醛若与体内蛋白结合, 可能产生过敏反应[80]。值得注意的是, 与P450酶不同, ADH在催化一些药物 (如阿巴卡韦) 的代谢反应时, pH大于7.4条件下的催化活性要高于pH 7.4条件下的催化活性[78], 因此, pH 7.4的孵育条件可能导致ADH在一些药物代谢中的作用被低估。
|
Figure 5 Alcohol dehydrogenase (ADH)-mediated oxidative metabolism of felbamate (A) and abacavir (B) |
根据分子组成的不同,在人体内共鉴定出5种ADH亚型, 分别为ADH1、ADH2、ADH3、ADH4和ADH5[81]。除了ADH4主要分布于胃黏膜细胞及其他上皮细胞外, 其余亚型在肝脏中均有分布[82]。ADH1、ADH2和ADH4均参与乙醇的代谢过程[81]。ADH3几乎不催化乙醇的代谢, 它主要通过与GSH形成复合物发挥甲醛脱氢酶的活性[81]。此外, 文献报道ADH1和ADH2是催化塞来昔布代谢的两种亚型[75], 而ADH4是催化tivantinib代谢的主要亚型[7]。目前, 关于ADH5催化的底物尚不清楚。ADH2和ADH3具有基因多态性[83]。Yamauchi等[84]报道ADH2的基因多态性可能与酒精性肝硬化在不同人群中的发生率有关。
5.3 药物相互作用4-甲基吡唑 (4-MP) 是ADH 的经典抑制剂, 已广泛应用于ADH底物的筛查实验[78, 85, 86]。ADH也是一种可以被诱导的酶, 雌二醇以及雄性激素可以诱导大鼠或小鼠体内的ADH水平[87-89]。
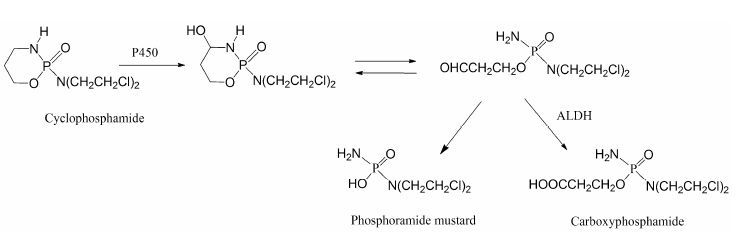
6 乙醛脱氢酶 (ALDH) 6.1 底物类型ALDH主要催化醛类化合物氧化生成羧酸, 该反应通常是不可逆的。ALDH主要存在于线粒体中, 其次为胞浆。线粒体中ALDH催化的反应倾向于以NAD+ 作为辅助因子 (NADP+ 也可以作为辅助因子参与反应); 而胞浆中ALDH催化的反应则依赖辅助因子NADP+[90, 91]的参与。药物在代谢过程中产生的醛中间体一般具有较高的反应活性, 可与蛋白发生共价结合导致毒性, 因此, ALDH催化醛的氧化反应通常被认为是药物的解毒过程。例如, 抗肿瘤药环磷酰胺 (cyclophosphamide) 在P450酶催化下氧化形成4-羟基环磷酰胺, 该代谢物能够自发开环形成醛磷酰胺, 后者在缺氧的肿瘤细胞中能分解产生活性代谢物磷酰胺氮芥 (phosphoramide mustard), 发挥杀死肿瘤细胞的作用, 而在正常细胞中则经ALDH催化形成无毒性的代谢物羧基磷酰胺 (carboxyphosphamide)[6, 13] (图 6) 。
|
Figure 6 Aldehyde dehydrogenase (ALDH)-mediated oxidative metabolism of cyclophosphamide |
目前,在人类基因组中共鉴定出17种ALDH基因[8]。线粒体乙醛脱氢酶ALDH2被认为是催化乙醛氧化形成乙酸的主要亚型[92]。ALDH2基因在亚洲人群中缺失的现象较为常见, 这部分人在饮酒后体内产生大量乙醛, 无法及时代谢, 使得毛细血管扩张, 引起脸红反应[92, 93]。此外, 乙醛若与心脏内的蛋白结合可能导致酒精性心肌病的发生[94]。近年来, 越来越多的文献报道ALDH2基因多态性与急性心肌梗死面积和冠状动脉疾病发病率紧密相关[95, 96]。
6.3 药物相互作用双硫仑是ALDH的经典抑制剂,已广泛应用于ALDH底物的筛查实验[97, 98]。ALDH也是一种可以被诱导的酶。Vasiliou等[99]报道苯巴比妥和甲基胆蒽能诱导大鼠、小鼠及豚鼠等动物肝胞浆中的ALDH活性。此外, 苯并芘和TCDD也能诱导大鼠肝中的ALDH活性[100, 101]。
7 其他类氧化酶除了上述非P450酶外, 其他氧化酶也能介导药物的氧化代谢, 如长链L-α-羟基酸氧化酶、髓过氧化物酶、NADPH氧化酶、谷胱甘肽过氧化物酶、嗜酸性粒细胞过氧化物酶[102]等。其中, 长链L-α-羟基酸氧化酶可以介导L-α-羟基酸类物质的代谢[103]; 髓过氧化物酶能够催化一些药物的代谢活化过程, 如氯氮平[104]、绿原酸[105]等。
8 结语近年来, 非P450酶在催化药物氧化代谢方面的作用日益受到重视。与P450酶类似, 非P450酶既可直接催化药物发生氧化代谢, 也可进一步氧化经其他酶催化产生的代谢物。表 2总结了常见的非P450酶的催化反应类型、常见底物、诱导剂、抑制剂和基因多态性。
| Table 2 Non-P450-mediated drug oxidative metabolism |
在药物发现和先导化合物优化过程中, 通常使用P450酶相关的体外模型进行代谢研究, 非P450酶在药物代谢中的贡献往往被低估。与P450酶不同, 大部分非P450酶, 如MAO、AO、XO、ADH和ALDH, 主要存在于胞浆或线粒体中。此外, P450酶催化反应的辅助因子为NADPH, 而ADH和ALDH催化反应的辅助因子为NAD+或NADP+。结合作者实验室的研究经验以及文献[46], 提出了一个决策树, 用来判断候选药物代谢的酶表型, 具体见图 7。

|
Figure 7 Decision tree to guide decision-making on the enzymes involved in drug oxidation |
首先, 比较候选药物在人肝细胞和微粒体中代谢物的生成量, 若两种体系中代谢物的生成量较为一致, 提示为P450酶或FMO介导的代谢。接下来, 可以将微粒体在45 ℃条件下预孵5 min使FMO失活, 以及重组酶孵化等方法区分P450酶和FMO的作用。若代谢物在人肝细胞中的生成量明显高于微粒体, 提示有非P450酶的参与, 可以根据反应类型判断可能的催化酶, 再选择合适的亚细胞器进行孵化。例如, MAO主要催化胺类化合物发生氧化脱氨或脱氢反应, AO主要催化含氮杂环类化合物杂原子邻位碳的氧化。当推测代谢途径由ADH或ALDH介导时, 可以考察药物在人肝S9 (加入辅助因子NAD+和NADP+) 中的代谢情况, 若产生该代谢物, 则进一步向孵化体系中加入ADH或ALDH的特异性抑制剂以区分ADH和ALDH的作用。当推测由AO或XO介导时, 可以考察候选药物在不加辅助因子的人肝胞浆中的代谢情况, 若产生该代谢物, 则进一步向孵化体系中加入AO或XO的特异性抑制剂, 以区分AO和XO的作用。当推测由MAO介导时, 可以考察候选药物在不加辅助因子的人肝线粒体中的代谢情况。若候选药物同时经P450酶和非P450酶代谢时, 可以先向肝细胞中加入特异性抑制剂考察这两种类型酶的相对贡献, 再选择合适的亚细胞器进行酶表型实验。
在过去几十年里, 人们对P450酶介导的药物氧化代谢进行了大量且深入的研究, 对不同亚型P450酶的特征底物、选择性诱导剂和抑制剂, 以及种属差异已经有了充分的了解。然而, 人们对非P450酶的研究尚处于起步阶段。对不同亚型的非P450酶是否具有特征底物以及种属差异尚不清楚。此外, 由于一些非P450酶 (如MAO和XO) 参与内源性物质的代谢过程, 对这类酶选择性抑制剂的研究主要集中在治疗内源性物质水平异常导致的疾病中的作用[35, 36, 106, 107], 而不是基于临床可能发生的药物相互作用。
由于对P450酶介导的药物氧化代谢充分认识, 目前, 药物化学家在进行药物结构改造的一个策略是减少P450酶介导的代谢活性, 以提高候选药物的代谢稳定性或减少临床发生药物相互作用风险, 但这也往往提高了其他非P450酶参与体内代谢的可能性。因此深入研究非P450酶介导的氧化代谢机制、种属差异和药物相互作用, 对指导候选化合物的结构优化、合理选择安全性评价动物种属、判断药物是否有继续开发的价值、预测临床可能的药物相互作用等多方面均具有重要价值。
| [1] | Diamond S, Boer J, Maduskuie TP Jr., et al. Species-specific metabolism of SGX523 by aldehyde oxidase and the toxicological implications[J]. Drug Metab Dispos, 2010, 38:1277-1285. |
| [2] | Akabane T, Tanaka K, Irie M, et al. Case report of extensive metabolism by aldehyde oxidase in humans:pharmacokinetics and metabolite profile of FK3453 in rats, dogs, and humans[J]. Xenobiotica, 2011, 41: 372–384. DOI:10.3109/00498254.2010.549970 |
| [3] | Vannelli TA, Dykman A, Ortiz de Montellano PR. The antituberculosis drug ethionamide is activated by a flavoprotein monooxygenase[J]. J Biol Chem, 2002, 277: 12824–12829. DOI:10.1074/jbc.M110751200 |
| [4] | Dixon CM, Park GR, Tarbit MH. Characterization of the enzyme responsible for the metabolism of sumatriptan in human liver[J]. Biochem Pharmacol, 1994, 47: 1253–1257. DOI:10.1016/0006-2952(94)90397-2 |
| [5] | alvie D, Zientek M. Metabolism of xenobiotics by aldehyde oxidase[J]. Curr Protoc Toxicol, 2015. DOI:10.1002/0471140856.tx0441s63 |
| [6] | Strolin Benedetti M, Whomsley R, Baltes E. Involvement of enzymes other than CYPs in the oxidative metabolism of xenobiotics[J]. Expert Opin Drug Metab Toxicol, 2006, 2: 895–921. DOI:10.1517/17425255.2.6.895 |
| [7] | Nishiya Y, Nakai D, Urasaki Y, et al. Stereoselective hydroxylation by CYP2C19 and oxidation by ADH4 in the in vitro metabolism of tivantinib[J]. Xenobiotica, 2016, 46: 967–976. DOI:10.3109/00498254.2016.1144896 |
| [8] | Vasiliou V, Pappa A, Estey T. Role of human aldehyde dehydrogenases in endobiotic and xenobiotic metabolism[J]. Drug Metab Rev, 2004, 36: 279–299. DOI:10.1081/DMR-120034001 |
| [9] | Ziegler DM. Recent studies on the structure and function of multisubstrate flavin-containing monooxygenases[J]. Annu Rev Pharmacol Toxicol, 1993, 33: 179–199. DOI:10.1146/annurev.pa.33.040193.001143 |
| [10] | Ziegler DM. Flavin-containing monooxygenases:catalytic mechanism and substrate specificities[J]. Drug Metab Rev, 1988, 19: 1–32. DOI:10.3109/03602538809049617 |
| [11] | Cashman JR. Some distinctions between flavin-containing and cytochrome P450 monooxygenases[J]. Biochem Biophys Res Commun, 2005, 338: 599–604. DOI:10.1016/j.bbrc.2005.08.009 |
| [12] | Yanni SB, Annaert PP, Augustijns P, et al. Role of flavincontaining monooxygenase in oxidative metabolism of voriconazole by human liver microsomes[J]. Drug Metab Dispos, 2008, 36: 1119–1125. DOI:10.1124/dmd.107.019646 |
| [13] | Beedham C. The role of non-P450 enzymes in drug oxidation[J]. Pharm World Sci, 1997, 19: 255–263. DOI:10.1023/A:1008668913093 |
| [14] | Rodriguez RJ, Acosta D Jr. Metabolism of ketoconazole and deacetylated ketoconazole by rat hepatic microsomes and flavin-containing monooxygenases[J]. Drug Metab Dispos, 1997, 25: 772–777. |
| [15] | Mani C, Hodgson E, Kupfer D. Metabolism of the antimammary cancer antiestrogenic agent tamoxifen. II. Flavin-containing monooxygenase-mediated N-oxidation[J]. Drug Metab Dispos, 1993, 21:657-661. |
| [16] | Cashman JR, Park SB, Yang ZC, et al. Chemical, enzymatic, and human enantioselective S-oxygenation of cimetidine[J]. Drug Metab Dispos, 1993, 21: 587–597. |
| [17] | Cashman JR, Park SB, Berkman CE, et al. Role of hepatic flavin-containing monooxygenase 3 in drug and chemical metabolism in adult humans[J]. Chem Biol Interact, 1995, 96: 33–46. DOI:10.1016/0009-2797(94)03581-R |
| [18] | Meng J, Zhong D, Li L, et al. Metabolism of MRX-I, a novel antibacterial oxazolidinone, in humans:the oxidative ring opening of 2,3-dihydropyridin-4-one catalyzed by non-P450 enzymes[J]. Drug Metab Dispos, 2015, 43: 646–659. DOI:10.1124/dmd.114.061747 |
| [19] | Koukouritaki SB, Simpson P, Yeung CK, et al. Human hepatic flavin-containing monooxygenases 1(FMO1) and 3(FMO3) developmental expression[J]. Pediatr Res, 2002, 51: 236–243. DOI:10.1203/00006450-200202000-00018 |
| [20] | Cashman JR, Akerman BR, Forrest SM, et al. Populationspecific polymorphisms of the human FMO3 gene:signifycance for detoxication[J]. Drug Metab Dispos, 2000, 28: 169–173. |
| [21] | Ayesh R, Mitchell SC, Zhang A, et al. The fish odour syndrome:biochemical, familial, and clinical aspects[J]. BMJ, 1993, 307: 655–657. DOI:10.1136/bmj.307.6905.655 |
| [22] | Al-Waiz M, Ayesh R, Mitchell SC, et al. A genetic polymorphism of the N-oxidation of trimethylamine in humans[J]. Clin Pharmacol Ther, 1987, 42: 588–594. DOI:10.1038/clpt.1987.201 |
| [23] | Larsen-Su S, Williams DE. Dietary indole-3-carbinol inhibits FMO activity and the expression of flavin-containing monooxygenase form 1 in rat liver and intestine[J]. Drug Metab Dispos, 1996, 24: 927–931. |
| [24] | Katchamart S, Stresser DM, Dehal SS, et al. Concurrent flavin-containing monooxygenase down-regulation and cytochrome P-450 induction by dietary indoles in rat:implications for drug-drug interaction[J]. Drug Metab Dispos, 2000, 28: 930–936. |
| [25] | Uehara S, Uno Y, Inoue T, et al. Activation and deactivation of 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine by cytochrome P450 enzymes and flavin-containing monooxygenases in common marmosets (Callithrix jacchus)[J]. Drug Metab Dispos, 2015, 43: 735–742. DOI:10.1124/dmd.115.063594 |
| [26] | Zaragoza A, Andres D, Sarrion D, et al. Potentiation of thioacetamide hepatotoxicity by phenobarbital pretreatment in rats. Inducibility of FAD monooxygenase system and age effect[J]. Chem Biol Interact, 2000, 124:87-101. |
| [27] | Hukkanen J, Dempsey D, Jacob P 3rd, et al. Effect of pregnancy on a measure of FMO3 activity[J]. Br J Clin Pharmacol, 2005, 60: 224–226. DOI:10.1111/bcp.2005.60.issue-2 |
| [28] | Kanazawa I. Short review on monoamine oxidase and its inhibitors[J]. Eur Neurol, 1994, 34(Suppl 3): 36–39. |
| [29] | Youdim MB, Finberg JP. New directions in monoamine oxidase A and B selective inhibitors and substrates[J]. Biochem Pharmacol, 1991, 41: 155–162. DOI:10.1016/0006-2952(91)90471-G |
| [30] | Thorpe LW, Westlund KN, Kochersperger LM, et al. Immunocytochemical localization of monoamine oxidases A and B in human peripheral tissues and brain[J]. J Histochem Cytochem, 1987, 35: 23–32. DOI:10.1177/35.1.3025289 |
| [31] | Scott AK. Sumatriptan clinical pharmacokinetics[J]. Clin Pharmacokinet, 1994, 27: 337–344. DOI:10.2165/00003088-199427050-00002 |
| [32] | Cesura AM, Imhof R, Galva MD, et al. Interactions of the novel inhibitors of MAO-B Ro 19-6327 and Ro 16-6491 with the active site of the enzyme[J]. Pharmacol Res Commun, 1988, 20(Suppl 4): 51–61. |
| [33] | Castagnoli N Jr., Chiba K, Trevor AJ. Potential bioactivation pathways for the neurotoxin 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP)[J]. Life Sci, 1985, 36:225-230. |
| [34] | Chiba K, Trevor AJ, Castagnoli N Jr. Active uptake of MPP+, a metabolite of MPTP, by brain synaptosomes[J]. Biochem Biophys Res Commun, 1985, 128: 1228–1232. DOI:10.1016/0006-291X(85)91071-X |
| [35] | Van der Walt MM, Terre'Blanche G, Petzer JP, et al. Benzyloxynitrostyrene analogues-a novel class of selective and highly potent inhibitors of monoamine oxidase B[J]. Eur J Med Chem, 2016, 125: 1193–1199. |
| [36] | Abbas N, Zaib S, Bakht SM, et al. Symmetrical aryl linked bis-iminothiazolidinones as new chemical entities for the inhibition of monoamine oxidases:synthesis, in vitro biological evaluation and molecular modelling analysis[J]. Bioorg Chem, 2016, 11: 004. DOI:10.1016/j.bioorg.2016.11.004 |
| [37] | Strolin Benedetti M. FAD-dependent enzymes involved in the metabolic oxidation of xenobiotics[J]. Ann Pharm Fr, 2011, 69: 45–52. DOI:10.1016/j.pharma.2010.10.004 |
| [38] | Fowler JS, Logan J, Wang GJ, et al. Comparison of monoamine oxidase a in peripheral organs in nonsmokers and smokers[J]. J Nucl Med, 2005, 46: 1414–1420. |
| [39] | Sarabia SF, Liehr JG. Induction of monoamine oxidase B by 17 beta-estradiol in the hamster kidney preceding carcinogenesis[J]. Arch Biochem Biophys, 1998, 355: 249–253. DOI:10.1006/abbi.1998.0727 |
| [40] | Ueda O, Sugihara K, Ohta S, et al. Involvement of molybdenum hydroxylases in reductive metabolism of nitro polycyclic aromatic hydrocarbons in mammalian skin[J]. Drug Metab Dispos, 2005, 33: 1312–1318. DOI:10.1124/dmd.105.005306 |
| [41] | Zhou L, Pang X, Xie C, et al. Chemical and enzymatic transformations of nimesulide to GSH conjugates through reductive and oxidative mechanisms[J]. Chem Res Toxicol, 2015, 28: 2267–2277. DOI:10.1021/acs.chemrestox.5b00290 |
| [42] | Hirao Y, Kitamura S, Tatsumi K. Epoxide reductase activity of mammalian liver cytosols and aldehyde oxidase[J]. Carcinogenesis, 1994, 15: 739–743. DOI:10.1093/carcin/15.4.739 |
| [43] | Dick RA, Kanne DB, Casida JE. Identification of aldehyde oxidase as the neonicotinoid nitroreductase[J]. Chem Res Toxicol, 2005, 18: 317–323. DOI:10.1021/tx049737i |
| [44] | Sugihara K, Kitamura S, Tatsumi K. Involvement of mammalian liver cytosols and aldehyde oxidase in reductive metabolism of zonisamide[J]. Drug Metab Dispos, 1996, 24: 199–202. |
| [45] | Tatsumi K, Kitamura S, Yamada H. Involvement of liver aldehyde oxidase in sulfoxide reduction[J]. Chem Pharm Bull (Tokyo), 1982, 30: 4585–4588. DOI:10.1248/cpb.30.4585 |
| [46] | Pryde DC, Dalvie D, Hu Q, et al. Aldehyde oxidase:an enzyme of emerging importance in drug discovery[J]. J Med Chem, 2010, 53: 8441–8460. DOI:10.1021/jm100888d |
| [47] | Dittrich C, Greim G, Borner M, et al. Phase I and pharmacokinetic study of BIBX 1382 BS, an epidermal growth factor receptor (EGFR) inhibitor, given in a continuous daily oral administration[J]. Eur J Cancer, 2002, 38: 1072–1080. DOI:10.1016/S0959-8049(02)00020-5 |
| [48] | Sanoh S, Tayama Y, Sugihara K, et al. Significance of aldehyde oxidase during drug development:effects on drug metabolism, pharmacokinetics, toxicity, and efficacy[J]. Drug Metab Pharmacokinet, 2015, 30: 52–63. DOI:10.1016/j.dmpk.2014.10.009 |
| [49] | Hutzler JM, Yang YS, Brown C, et al. Aldehyde oxidase activity in donor-matched fresh and cryopreserved human hepatocytes and assessment of variability in 75 donors[J]. Drug Metab Dispos, 2014, 42: 1090–1097. DOI:10.1124/dmd.114.057984 |
| [50] | Sahi J, Khan KK, Black CB. Aldehyde oxidase activity and inhibition in hepatocytes and cytosolic fractions from mouse, rat, monkey and human[J]. Drug Metab Lett, 2008, 2: 176–183. DOI:10.2174/187231208785425818 |
| [51] | Garattini E, Fratelli M, Terao M. Mammalian aldehyde oxidases:genetics, evolution and biochemistry[J]. Cell Mol Life Sci, 2008, 65: 1019–1048. DOI:10.1007/s00018-007-7398-y |
| [52] | Garattini E, Terao M. The role of aldehyde oxidase in drug metabolism[J]. Expert Opin Drug Metab Toxicol, 2012, 8: 487–503. DOI:10.1517/17425255.2012.663352 |
| [53] | Lolkema MP, Bohets HH, Arkenau HT, et al. The c-Met tyrosine kinase inhibitor JNJ-38877605 causes renal toxicity through species-specific insoluble metabolite formation[J]. Clin Cancer Res, 2015, 21: 2297–2304. DOI:10.1158/1078-0432.CCR-14-3258 |
| [54] | Austin NE, Baldwin SJ, Cutler L, et al. Pharmacokinetics of the novel, high-affinity and selective dopamine D3 receptor antagonist SB-277011 in rat, dog and monkey:in vitro/in vivo correlation and the role of aldehyde oxidase[J]. Xenobiotica, 2001, 31: 677–686. DOI:10.1080/00498250110056531 |
| [55] | Schofield PC, Robertson IGC, Paxton JW. Inter-species variation in the metabolism and inhibition of N-[(2'-dimethylamino) ethyl] acridine-4-carboxamide (DACA) by aldehyde oxidase[J]. Biochem Pharmacol, 2000, 59:161-165. |
| [56] | Choughule KV, Joswig-Jones CA, Jones JP. Interspecies differences in the metabolism of methotrexate:an insight into the active site differences between human and rabbit aldehyde oxidase[J]. Biochem Pharmacol, 2015, 96: 288–295. DOI:10.1016/j.bcp.2015.05.010 |
| [57] | Obach RS. Potent inhibition of human liver aldehyde oxidase by raloxifene[J]. Drug Metab Dispos, 2004, 32: 89–97. DOI:10.1124/dmd.32.1.89 |
| [58] | Obach RS, Huynh P, Allen MC, et al. Human liver aldehyde oxidase:inhibition by 239 drugs[J]. J Clin Pharmacol, 2004, 44: 7–19. DOI:10.1177/0091270003260336 |
| [59] | Lake BG, Ball SE, Kao J, et al. Metabolism of zaleplon by human liver:evidence for involvement of aldehyde oxidase[J]. Xenobiotica, 2002, 32: 835–847. DOI:10.1080/00498250210158915 |
| [60] | Robertson IG, Gamage RS. Methadone:a potent inhibitor of rat liver aldehyde oxidase[J]. Biochem Pharmacol, 1994, 47: 584–587. DOI:10.1016/0006-2952(94)90192-9 |
| [61] | Nirogi R, Kandikere V, Palacharla RC, et al. Identification of a suitable and selective inhibitor towards aldehyde oxidase catalyzed reactions[J]. Xenobiotica, 2014, 44: 197–204. DOI:10.3109/00498254.2013.819594 |
| [62] | Strelevitz TJ, Orozco CC, Obach RS. Hydralazine as a selective probe inactivator of aldehyde oxidase in human hepatocytes:estimation of the contribution of aldehyde oxidase to metabolic clearance[J]. Drug Metab Dispos, 2012, 40: 1441–1448. DOI:10.1124/dmd.112.045195 |
| [63] | Johnson C, Stubley-Beedham C, Stell JGP. Elevation of molybdenum hydroxylase levels in rabbit liver after ingestion of phthalazine or its hydroxylated metabolite[J]. Biochem Pharmacol, 1984, 33: 3699–3705. DOI:10.1016/0006-2952(84)90159-X |
| [64] | Beedham C, Padwick DJ, Al-Tayib Y, et al. Diurnal variation and melatonin induction of hepatic molybdenum hydroxylase activity in the guinea-pig[J]. Biochem Pharmacol, 1989, 38: 1459–1464. DOI:10.1016/0006-2952(89)90185-8 |
| [65] | Sugihara K, Kitamura S, Yamada T, et al. Aryl hydrocarbon receptor (AhR)-mediated induction of xanthine oxidase/xanthine dehydrogenase activity by 2,3,7,8-tetrachlorodibenzop-dioxin[J]. Biochem Biophys Res Commun, 2001, 281: 1093–1099. DOI:10.1006/bbrc.2001.4464 |
| [66] | Rivera SP, Choi HH, Chapman B, et al. Identification of aldehyde oxidase 1 and aldehyde oxidase homologue 1 as dioxin-inducible genes[J]. Toxicology, 2005, 207: 401–409. DOI:10.1016/j.tox.2004.10.009 |
| [67] | Peterson GM, Boyle RR, Francis HW, et al. Dosage prescribing and plasma oxipurinol levels in patients receiving allopurinol therapy[J]. Eur J Clin Pharmacol, 1990, 39: 419–421. DOI:10.1007/BF00315424 |
| [68] | Borges F, Fernandes E, Roleira F. Progress towards the discovery of xanthine oxidase inhibitors[J]. Curr Med Chem, 2002, 9: 195–217. DOI:10.2174/0929867023371229 |
| [69] | Pacher P, Nivorozhkin A, Szabo C. Therapeutic effects of xanthine oxidase inhibitors:renaissance half a century after the discovery of allopurinol[J]. Pharmacol Rev, 2006, 58: 87–114. DOI:10.1124/pr.58.1.6 |
| [70] | Barr JT, Choughule KV, Nepal S, et al. Why do most human liver cytosol preparations lack xanthine oxidase activity?[J]. Drug Metab Dispos, 2014, 42: 695–699. DOI:10.1124/dmd.113.056374 |
| [71] | Cribb AE, Renton KW. Dissociation of xanthine oxidase induction and cytochrome P450 depression during interferon induction in the rat[J]. Biochem Pharmacol, 1993, 46: 2114–2117. DOI:10.1016/0006-2952(93)90658-J |
| [72] | Moochhala SM, Renton KW. A role for xanthine oxidase in the loss of cytochrome P-450 evoked by interferon[J]. Can J Physiol Pharmacol, 1991, 69: 944–950. DOI:10.1139/y91-143 |
| [73] | Hassoun PM, Yu F-S, Cote CG, et al. Upregulation of xanthine oxidase by lipopolysaccharide, interleukin-1, and hypoxia:role in acute lung injury[J]. Am J Respir Crit Care Med, 1998, 158: 299–305. DOI:10.1164/ajrccm.158.1.9709116 |
| [74] | Linder N, Martelin E, Lapatto R, et al. Posttranslational inactivation of human xanthine oxidoreductase by oxygen under standard cell culture conditions[J]. Am J Physiol Cell Physiol, 2003, 285: C48–55. DOI:10.1152/ajpcell.00561.2002 |
| [75] | Sandberg M, Yasar U, Stromberg P, et al. Oxidation of celecoxib by polymorphic cytochrome P4502C9 and alcohol dehydrogenase[J]. Br J Clin Pharmacol, 2002, 54: 423–429. DOI:10.1046/j.1365-2125.2002.01660.x |
| [76] | Diao X, Deng P, Xie C, et al. Metabolism and pharmacokinetics of 3-n-butylphthalide (NBP) in humans:the role of cytochrome P450s and alcohol dehydrogenase in biotransformation[J]. Drug Metab Dispos, 2013, 41: 430–444. DOI:10.1124/dmd.112.049684 |
| [77] | Dieckhaus CM, Miller TA, Sofia RD, et al. A mechanistic approach to understanding species differences in felbamate bioactivation:relevance to drug-induced idiosyncratic reactions[J]. Drug Metab Dispos, 2000, 28: 814–822. |
| [78] | Walsh JS, Reese MJ, Thurmond LM. The metabolic activetion of abacavir by human liver cytosol and expressed human alcohol dehydrogenase isozymes[J]. Chem Biol Interact, 2002, 142: 135–154. DOI:10.1016/S0009-2797(02)00059-5 |
| [79] | Thompson CD, Kinter MT, Macdonald TL. Synthesis and in vitro reactivity of 3-carbamoyl-2-phenylpropionaldehyde and 2-phenylpropenal:putative reactive metabolites of felbamate[J]. Chem Res Toxicol, 1996, 9: 1225–1229. DOI:10.1021/tx9601566 |
| [80] | Grilo NM, Charneira C, Pereira SA, et al. Bioactivation to an aldehyde metabolite-possible role in the onset of toxicity induced by the anti-HIV drug abacavir[J]. Toxicol Lett, 2014, 224: 416–423. DOI:10.1016/j.toxlet.2013.10.036 |
| [81] | Hoog JO, Stromberg P, Hedberg JJ, et al. The mammalian alcohol dehydrogenases interact in several metabolic pathways[J]. Chem Biol Interact, 2003, 143-144: 175–181. DOI:10.1016/S0009-2797(02)00225-9 |
| [82] | Jornvall H, Hoog JO, Persson B, et al. Pharmacogenetics of the alcohol dehydrogenase system[J]. Pharmacology, 2000, 61: 184–191. DOI:10.1159/000028399 |
| [83] | Bosron WF, Lumeng L, Li TK. Genetic polymorphism of enzymes of alcohol metabolism and susceptibility to alcoholic liver disease[J]. Mol Aspects Med, 1988, 10: 147–158. DOI:10.1016/0098-2997(88)90019-2 |
| [84] | Yamauchi M, Maezawa Y, Mizuhara Y, et al. Polymorphisms in alcohol metabolizing enzyme genes and alcoholic cirrhosis in Japanese patients:a multivariate analysis[J]. Hepatology, 1995, 22: 1136–1142. |
| [85] | Ronis MJ, Korourian S, Blackburn ML, et al. The role of ethanol metabolism in development of alcoholic steatohepatitis in the rat[J]. Alcohol, 2010, 44: 157–169. DOI:10.1016/j.alcohol.2009.11.002 |
| [86] | Holm NB, Noble C, Linnet K. JWH-018ω-OH, a shared hydroxy metabolite of the two synthetic cannabinoids JWH-018 and AM-2201, undergoes oxidation by alcohol dehydrogenase and aldehyde dehydrogenase enzymes in vitro forming the carboxylic acid metabolite[J]. Toxicol Lett, 2016, 259: 35–43. DOI:10.1016/j.toxlet.2016.07.007 |
| [87] | Felder MR, Watson G, Huff MO, et al. Mechanism of induction of mouse kidney alcohol dehydrogenase by androgen. Androgen-induced stimulation of transcription of the Adh-1 gene[J]. J Biol Chem, 1988, 263:14531-14537. |
| [88] | Qulali M, Ross RA, Crabb DW. Estradiol induces class I alcohol dehydrogenase activity and mRNA in kidney of female rats[J]. Arch Biochem Biophys, 1991, 288: 406–413. DOI:10.1016/0003-9861(91)90213-3 |
| [89] | Ceci JD, Lawther R, Duester G, et al. Androgen induction of alcohol dehydrogenase in mouse kidney. Studies with a cDNA probe confirmed by nucleotide sequence analysis[J]. Gene, 1986, 41:217-224. |
| [90] | Mukhopadhyay A, Wei B, Weiner H. Mitochondrial NAD dependent aldehyde dehydrogenase either from yeast or human replaces yeast cytoplasmic NADP dependent aldehyde dehydrogenase for the aerobic growth of yeast on ethanol[J]. Biochim Biophys Acta, 2013, 1830: 3391–3398. DOI:10.1016/j.bbagen.2013.02.010 |
| [91] | Remize F, Andrieu E, Dequin S. Engineering of the pyruvate dehydrogenase bypass in Saccharomyces cerevisiae:role of the cytosolic Mg2+ and mitochondrial K+ acetaldehyde dehydrogenases Ald6p and Ald4p in acetate formation during alcoholic fermentation[J]. Appl Environ Microbiol, 2000, 66: 3151–3159. DOI:10.1128/AEM.66.8.3151-3159.2000 |
| [92] | Crabb DW, Edenberg HJ, Bosron WF, et al. Genotypes for aldehyde dehydrogenase deficiency and alcohol sensitivity. The inactive ALDH22 allele is dominant[J]. J Clin Invest, 1989, 83:314-316. |
| [93] | Novoradovsky A, Tsai SJ, Goldfarb L, et al. Mitochondrial aldehyde dehydrogenase polymorphism in Asian and American Indian populations:detection of new ALDH2 alleles[J]. Alcohol Clin Exp Res, 1995, 19: 1105–1110. DOI:10.1111/acer.1995.19.issue-5 |
| [94] | Harcombe AA, Ramsay L, Kenna JG, et al. Circulating antibodies to cardiac protein-acetaldehyde adducts in alcoholic heart muscle disease[J]. Clin Sci (Lond), 1995, 88: 263–268. DOI:10.1042/cs0880263 |
| [95] | Li QY, Zhao NM, Ma JJ, et al. ALDH2*2 allele is a negative risk factor for cerebral infarction in Chinese women[J]. Biochem Genet, 2015, 53: 260–267. DOI:10.1007/s10528-015-9686-9 |
| [96] | Pang JJ, Barton LA, Chen YG, et al. Mitochondrial aldehyde dehydrogenase in myocardial ischemia-reperfusion injury:from bench to bedside[J]. Acta Physiol Sin, 2015, 67: 535–544. |
| [97] | Stagos D, Chen Y, Brocker C, et al. Aldehyde dehydrogenase 1B1:molecular cloning and characterization of a novel mitochondrial acetaldehyde-metabolizing enzyme[J]. Drug Metab Dispos, 2010, 38: 1679–1687. DOI:10.1124/dmd.110.034678 |
| [98] | Lipsky JJ, Shen ML, Naylor S. Overview-in vitro inhibition of aldehyde dehydrogenase by disulfiram and metabolites[J]. Chem Biol Interact, 2001, 130-132: 81–91. DOI:10.1016/S0009-2797(00)00224-6 |
| [99] | Vasiliou V, Torronen R, Malamas M, et al. Inducibility of liver cytosolic aldehyde dehydrogenase activity in various animal species[J]. Comp Biochem Physiol C, 1989, 94: 671–675. DOI:10.1016/0742-8413(89)90130-8 |
| [100] | Pappas P, Sotiropoulou M, Karamanakos P, et al. Acutephase response to benzo[a]pyrene and induction of rat ALDH3A1[J]. Chem Biol Interact, 2003, 143-144:55-62. |
| [101] | Dunn TJ, Lindahl R, Pitot HC. Differential gene expression in response to 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD). Noncoordinate regulation of a TCDD-induced aldehyde dehydrogenase and cytochrome P-450c in the rat[J]. J Biol Chem, 1988, 263:10878-10886. |
| [102] | Khan AA, Rahmani AH, Aldebasi YH, et al. Biochemical and pathological studies on peroxidases-an updated review[J]. Global J Health Sci, 2014, 6: 87–98. |
| [103] | Barawkar DA, Bandyopadhyay A, Deshpande A, et al. Discovery of pyrazole carboxylic acids as potent inhibitors of rat long chain L-2-hydroxy acid oxidase[J]. Bioorg Med Chem Lett, 2012, 22: 4341–4347. DOI:10.1016/j.bmcl.2012.05.020 |
| [104] | Gan J, Ma S, Zhang D. Non-cytochrome P450-mediated bioactivation and its toxicological relevance[J]. Drug Metab Rev, 2016, 48: 473–501. DOI:10.1080/03602532.2016.1225756 |
| [105] | Xie C, Zhong D, Chen X. Identification of the ortho-benzoquinone intermediate of 5-O-caffeoylquinic acid in vitro and in vivo:comparison of bioactivation under normal and pathological situations[J]. Drug Metab Dispos, 2012, 40: 1628–1640. DOI:10.1124/dmd.112.045641 |
| [106] | Ojha R, Singh J, Ojha A, et al. An updated patent review:xanthine oxidase inhibitors for the treatment of hyperuricemia and gout (2011-2015)[J]. Expert Opin Ther Pat, 2016: 1–35. |
| [107] | Wan Y, Zou B, Zeng H, et al. Inhibitory effect of verbascoside on xanthine oxidase activity[J]. Int J Biol Macromol, 2016, 93: 609–614. DOI:10.1016/j.ijbiomac.2016.09.022 |
 2017, Vol. 52
2017, Vol. 52


