人DNA拓扑异构酶,包括DNA拓扑异构酶1 (topoisomerase 1,Top1) 和拓扑异构酶2 (topoisomerase 2,Top2),广泛存在于人体细胞中,参与许多重要的细胞过程,例如DNA的复制、转录和染色质重塑等[1]。部分化疗药物和放射过程可造成拓扑异构酶介导的DNA损伤,从而杀死肿瘤细胞[4],成为肿瘤治疗的重要手段。近年来发现,酪氨酰-DNA磷酸二酯酶1 (tyrosyl-DNA phosphodiesterase 1,TDP1) 和酪氨酰- DNA磷酸二酯酶2 (tyrosyl-DNA phosphodiesterase 2,TDP2) 可以分别特异性识别和修复由Top1或Top2介导的DNA损伤,是潜在的肿瘤治疗靶点,其抑制剂可增强化疗药物或放疗措施的功效。本文从酪氨 酰-DNA磷酸二酯酶1/2的发现、结构、催化机制和生物学功能出发,讨论了其作为肿瘤治疗靶点的理论依据,并综述了其小分子抑制剂的最新研究进展。
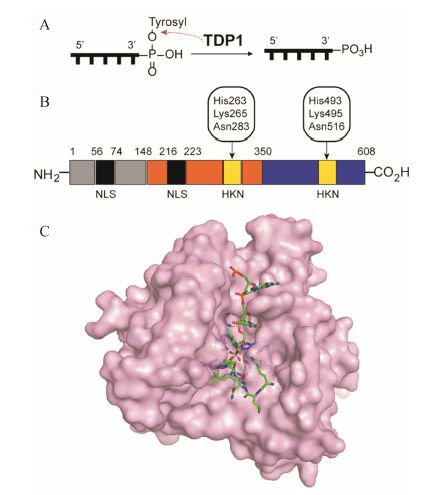
1 酪氨酰-DNA磷酸二酯酶11996年,Nash等[7]在Saccharomyces cerevisiae酵母菌中发现了一种DNA修复酶,它可以特异性地水解DNA的3'-磷酸与酪氨酸酚羟基之间的酪氨酰磷酸二酯键 (图 1A)。由于Top1是迄今发现的唯一可以形成DNA 3'-酪氨酰磷酸二酯键的酶,因此这个新发现的酶具有修复Top1介导的DNA损伤 (Top1-mediated DNA damage,Top1-DD) 的活性,被命名为酪氨酰- DNA磷酸二酯酶1[8, 9] (图 1B,C)。
|
图 1 (A) 酪氨酰-DNA磷酸二酯酶1 (TDP1) 的催化水解反应。 (B) 人TDP1的结构[9]。 (C) 人TDP1的单晶结构 (PDB code: 1NOP),催化中心 (HKN) 以蓝色棒状显示,DNA-钒酸盐-多肽复合物以红绿蓝色棒状显示 |
人TDP1属于磷脂酶D家族,是一条68 kDa的多肽[10],共有17个外显子,其中2个非编码外显子和15个编码外显子。人TDP1由2条α-β-α结构域组成[11],分别为由氨基酸残基1~350组成的氮端域,以及由氨基酸残基351~608组成的碳端域 (图 1B)。
人TDP1的氮端域含有2条核定位基序 (nuclear localization sequence,NLS)。氮端域由两部分组成[12]: 氨基酸残基为1~148的基序和149~350的基序。基序1~148不影响酶的活性,而是通过各种修饰过程,起着调控蛋白-蛋白相互作用的重要功能[13]。这些修饰过程包括: 在氮端域的聚ADP糖基化 (poly ADP glycosylation)、Ser81的磷酸化以及Lys111的小分子类泛素化[14, 15]。基序149~350中含有一段参与构成TDP1活性中心的HKN (组氨酸-赖氨酸-天冬酰胺) 催化基序[16]。在HKN基序中,组氨酸及赖氨酸对于酶的活性非常重要[10, 13, 17, 18]。TDP1共含有2条HKN催化基序,分别位于氮端域和碳端域。
人TDP1的晶体结构 (图 1C,PDB code: 1NOP) 显示[19],TDP1的每个HKN基序的氨基酸残基都面 向活性中心口袋。在口袋中,氮端域的His263与碳端域的Ser514及Glu538靠近,而碳端域的His493则与氮端域的Thr281及Gln294靠近,共同构成一个键合环道[11, 19]。在该晶体结构中,TDP1与底物钒酸共价结合,形成TDP1-DNA-钒酸盐-多肽四元复合物。单链DNA结合在一条狭长的、不对称分布着正电荷的沟槽内。沟槽表面的正电荷有助于TDP1与DNA的磷酸骨架通过静电作用相互结合。此外,狭缝内Ser400、Ser403、Lys469、Ser518、Lys519及Ala520与共价复合物中DNA的3'-酪氨酰磷酸二酯键上游3个核苷酸的磷酸基团共同构成一个极性网络,通过极性相互作用使DNA被稳定在结合沟槽内; 而Phe259、Pro461和Trp590则通过疏水作用稳定DNA[20]。另一方面,多肽则结合在与之方向相反的一条较宽的、碗口状的沟槽内。
1.2 TDP1的催化机制及其生物学功能研究认为,TDP1的催化机制可能包含两步SN2亲核进攻过程,是由HKN基序中具有催化活性的组氨酸介导完成 的[9, 21, 22]。首先,His263的咪唑氮亲核进攻底物的3'-酪氨酰磷酸二酯键。His493作为广义酸,向苯酚负离子提供一个质子,促使酪氨酸残基离去 (图 2A)。同时,His263与DNA 3'端形成一个磷酰胺键,形成一个短暂的TDP1-DNA共价中间体 (图 2B); 第二步,His493作为广义碱,激活一个水分子亲核进攻TDP1-DNA共价中间体的3'-磷酰胺键 (图 2C),使之水解,并释放出一条含有3'-磷酸基的DNA链 (图 2D)。

|
图 2 人TDP1可能的催化水解酪氨酰磷酸二酯键的机制,包括两步的亲核进攻过程[9] |
已知Top1是唯一可形成DNA 3'-酪氨酰磷酸二酯键的酶。在Top1的催化过程中,Top1活性中心的酪氨酸残基进攻DNA单链的3'末端,断开DNA单链,酪氨酸残基与DNA 3'端的磷酸以可逆的共价键——酪氨酰磷酸二酯键结合,形成Top1-DNA断裂共价复合物 (Top1 cleavage complex,Top1cc,图 3A)[23]。在正常生理过程中,Top1cc仅瞬间存在。在多种外源性因素的影响下,如使用Top1毒剂[3]、烷化剂[24]、DNA嵌入剂[27, 28]或核苷类链终止剂[29],以及辐射[30]等过程,Top1cc被捕捉,造成Top1-DD (图 3B),最终 导致细胞死亡。Top1cc的蛋白必须被 降解为寡肽后 (图 3C),TDP1才能识别并水解该复合物的酪氨酰磷酸二酯键。研究认为,TDP1是与PARP1 (poly ADP- ribose polymerase 1) 共同发挥水解作用的,并得到
DNA 3' 端为磷酸基的中间体 (图 3D)[15]。由于DNA缺口必须为3'-羟基和5'-磷酸时才能在各种酶的作用下重新连接、修复得到完整的DNA。因此,该断裂DNA中间体 (图 3D) 首先在聚核苷酸激酶3'-磷酸酶 (polynucleotide kinase 3'-phosphatase,PNKP) 的作用下,DNA缺口两端分别被修饰为3'-羟基和5'-磷酸[33]。PNKP与XRCC1 (X-ray repair cross-complementing protein 1) 相互作用,并与其他修复蛋白,包括DNA聚合酶β (polymerase β,Polβ)、DNA连接酶3 (ligase 3,Lig3) 以及PARP1等,一起形成一个多蛋白的DNA修复复合物[36],共同作用下修复DNA (图 3E)。

|
图 3 Top/TDP介导的DNA损伤和修复过程[37] |
这一修复过程预示,在TDP1过度表达或者活性增加的肿瘤细胞中,Top1-DD得以修复,使得肿瘤细胞对与之相关的药物治疗和放疗措施耐受。在正常 细胞中,有两条冗长的通路可以修复Top1-DD: TDP1依赖的修复通路[37] (图 3A~E) 和细胞周期检测点 依赖的修复通路。该细胞周期检测点修复通路涉及BRCA1、53BP1和MDC1等检测点蛋白[38]。但在许多肿瘤细胞中,检测点蛋白常常缺失[39],这时,TDP1依赖的通路成为主要的修复Top1-DD的通路。
以上的理论分析表明,TDP1是一个潜在的肿瘤治疗靶点: 第一、高表达TDP1的肿瘤细胞出现耐药。TDP1可修复化疗试剂 (包括Top1毒剂、烷基化试 剂和核苷试剂等) 引起的Top1-DD,降低肿瘤细胞中
DNA损伤水平[12,26,40],使得肿瘤细胞对这些化疗药物耐受[12,41,42,42,43]。研究发现,TDP1在多种肿瘤细胞或组织中高表达,如肺癌[42, 45]、横纹肌肉瘤[44]、甲状腺癌、乳腺癌、肝癌、子宫内膜癌和卵巢癌等[46]。在高表达TDP1的非小细胞肺癌组织中,TDP1活性与肿瘤的体积呈正相关[45]。TDP1成为肿瘤耐药的主要原因。第二、TDP1抑制剂可逆转肿瘤耐药,与某些化疗药物 (包括Top1毒剂、烷化剂、DNA嵌入剂和核苷类药物等) 以及放疗措施具有协同作用。研究发现,Tdp1基因突变[47, 48]或耗尽TDP1的细胞对Top1毒剂、烷基化试剂和核苷类链终止剂高度敏感[26, 40, 44, 49, 50];
TDP1抑制剂可提高乳腺癌肿瘤细胞对喜树碱 (CPT) 的敏感性[46]。第三、PARP1抑制剂的成功上市[51],证明“合成致死”(synthetic lethality) 理论的可行 性[52, 53]。鉴于TDP1功能与PARP1功能的相似性,其抑制剂可能具有“合成致死”的功能。值得注意 的是,由于正常细胞存在两条平行的修复通路,抑制TDP1对正常细胞的影响可能不大,预示着TDP1抑制剂的毒副作用可能较低[46]。
1.3 TDP1小分子抑制剂尽管类生物大分子钒酸盐和钨酸盐可以抑制TDP1活性[22],但是该类抑制剂不具有成药性。自从人TDP1-DNA-多肽-钒酸盐复合物晶体被发现后[19],TDP1小分子抑制剂成为研究热点。理论上,TDP1小分子抑制剂可以作用在催化过程的每个阶段。例如,抑制剂可以阻止His263或His493的亲核进攻,分别抑制酪氨酰磷酸二酯键 (图 2A) 或磷酰胺键 (图 2C) 的水解; 能够稳定TDP1-DNA中间体 (图 2B),使TDP1被耗尽。其中,稳定TDP1- DNA中间体的方式类似于稳定Top1cc[4, 54],似乎更具可能性[55]。
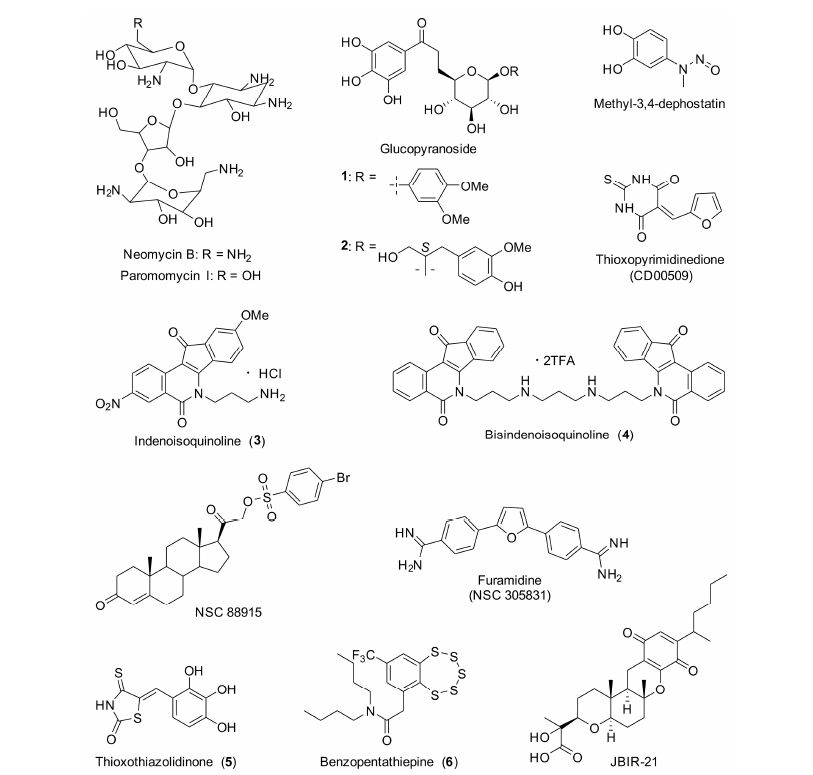
美国国立卫生研究院的Pommier课题组[56]在TDP1小分子抑制剂的研究中做了大量的工作。首先发现了氨基糖苷类小分子抑制剂——新霉素B和巴龙霉素I (neomycin B,paromomycin I,图 4)。遗憾的是,该类抑制剂的活性较低。新霉素B和巴龙霉素I的IC50值分别为8 和21 mmol×L-1。动力学研究发现,新霉素B是一个可逆的TDP1抑制剂,它可能是直接作用于TDP1或TDP1-DNA复合物而发挥抑制活性的。
|
图 4 TDP1小分子抑制剂的结构 |
最近,Tian等[57]从澳大利亚热带雨林植物Macropteranthes leichhardtii的皮中分离得到了2个吡喃葡萄糖苷类化合物 (图 4,化合物1和2),具有良好的TDP1抑制活性,其IC50值均为1 μmol·L-1。
Pommier课题组建立了2种TDP1高通量筛选方法,并先后发现了呋喃二脒类 (NSC 305831) 和methyl-3,4-dephostatin小分子抑制剂 (图 4)。
NSC 305831的抑制活性在微摩尔浓度水平,其IC50值约为90 μmol·L-1[58]。呋喃二脒类化合物是DNA沟槽结合试剂。在生理条件下,NSC 305831质子化后可与DNA的磷酸骨架发生静电相互作用。研究发现,NSC 305831与单链DNA的解离平衡常数 (Kd) 约为70 μmol·L-1,其TDP1抑制活性可能与其DNA的结合能力 相关。
Methyl-3,4-dephostatin的TDP1抑制活性在亚 微摩尔浓度 (submicromolar) 水平,其IC50值为0.36 μmol·L-1。Pommier等[59]认为,methyl-3,4-dephostatin 作用于TDP1催化水解酪氨酰磷酸二酯键阶段,抑制TDP1的水解功能。Methyl-3,4-dephostatin的化学结构稳定,成为工具药或先导药物的可能性较大。
甾体类化合物 (NSC 88915,图 4) 是该课题组发现的另一类TDP1抑制剂,其IC50值为7.7 μmol·L-1[33]。但是,它对细胞粗提液 的TDP1 (whole cell extract,WCE TDP1) 抑制活性显著下降,表明它在细胞环境下有脱靶效应 (off-target effects)[60]。进一步的机制研究表明,NSC 88915是竞争性抑制剂。作为底物类似物,NSC 88915首先作用于酶,阻止TDP1与底物的结合。
另外,他们与美国普渡大学的Cushman课题组合作,首次发现了一类茚并异喹啉类TDP1/Top1双效抑制剂[60]。该类化合物直接作用于酶,属于竞争性抑制剂,而且对于TDP1和WCE TDP1的抑制活性相近。其代表化合物3 (图 4) 对TDP1的IC50值在1~12 μmol·L-1之间[61],对Top1的抑制活性与CPT相 似,具有良好的细胞毒活性。他们测试了化合物3抑制NCI60的活性 (美国肿瘤研究所建立的一种抗肿瘤药物筛选方法,测试药物对60株临床肿瘤细胞的细胞毒活性[64]),其平均GI50为0.027 μmol·L-1[65]。TDP1抑制活性最高的是一个双茚并异喹啉化合物4(图 4),IC50值为1.52 μmol·L-1[60]; 同样具有良好Top1抑制活性和细胞毒活性,Top1抑制活性与CPT相近,对NCI60的平均GI50为0.394 μmol·L-1[66]。
Sirivolu等[67]发现,thioxothiazolidinone类抑制 剂具有较高的TDP1抑制活性 (图 4)。其代表化合 物5的IC50值为0.87 μmol·L-1,但对WCE TDP1 的抑制活性降低了约40倍 (IC50 35 μmol·L-1),表明该化合物具有严重的脱靶效应。另外,化合物5也 是TDP2抑制剂,其IC50值为17 μmol·L-1。表面等 离子共振实验 (surface plasmon resonance) 表明,化合物5与核酸底物没有作用,而是作用于TDP1[67]。Thioxothiazolidinone类抑制剂不仅有脱靶效应,其化学结构也不稳定[68],大大降低了其成药性。
Dean等[46]发现了thioxopyrimidinedione类TDP1抑制剂CD00509 (图 4),其IC50值为0.71 μmol·L-1。CD00509可以增加CPT引起的DNA损伤,提高乳腺癌肿瘤细胞MCF-7对CPT的敏感性,二者联用使得MCF-7的增殖降低了25%。另外,单独使用CD00509 (10 μmol·L-1) 不影响人乳腺上皮细胞的生长,表明它的细胞毒性很低。
Takagi等[69]从变形真菌Anamorphicfungus次级代谢产物中分离得到了JBIR-21 (图 4),其抑制TDP1的IC50为18 μmol·L-1。JBIR-21具有较好的细胞毒活性,对4株肿瘤细胞的IC50值在3.5~13 μmol·L -1之间; 并可有效抑制小鼠结肠癌 (HT-29) 的生长,用药21天后,肿瘤体积降低了49%,而且没有明显的毒副作用。
最近,Zakharenko等[70]报道了一类苯并五硫平 类化合物,具有良好的TDP1抑制活性。化合物6 (图 4) 为迄今报道的抑制活性最佳的化合物,其IC50为0.22 μmol·L-1。该类化合物可引起MCF-7和肝癌细胞HepG2的凋亡。
在以上报道的TDP1小分子抑制剂中,大多未说明这些抑制剂的选择性 (如是否抑制TDP2)。本课题组发现了一类异喹啉类抑制剂,其抑制活性在低微摩尔浓度水平。尤其值得注意的是,该类抑制剂在111 μmol·L-1时对TDP2没有抑制活性,属于TDP1选择性抑制剂。
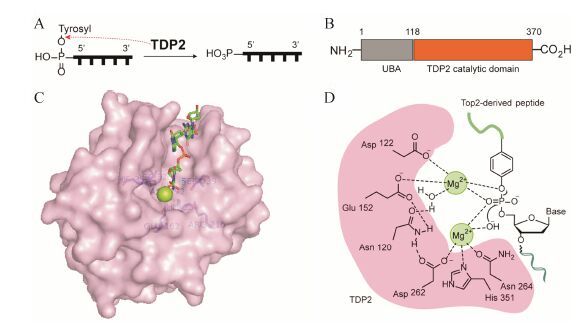
2 酪氨酰-DNA磷酸二酯酶2相比于TDP1,酪氨酰-DNA磷酸二酯酶2的发现较晚。2000年,Pype等[71]在研究CD40信号转导通路时发现了一种蛋白,作为细胞内肿瘤坏死因子受体结合蛋白,被命名为TTRAP (TRAF and TNF receptor- associated protein),但此时并未发现它的5'-酪氨酰磷酸二酯酶活性。2003年,Pei等[72]在酵母中也发现了此种蛋白,主要功能为转录因子ETS1 (E-twenty six 1 protein) 和AP1 (APETALA 1) 的负转录调控因子,并命名为EAP II (ETS1-associated protein 2)。2009年,Cortes Ledesma等[73]在人的细胞中再次发现了这种蛋白,并发现其可特异性地水解DNA的5'-磷酸与酪氨酸酚羟基形成的酪氨酰磷酸二酯键。如图 5A所示,TDP2水解DNA 5'-端的酪氨酰磷酸二酯键,清除酪氨酸残基,使5'-磷酸基裸露,以便与其他DNA片段连接,恢复DNA的完整性。为了与TDP1名称相对应,将其命名为TDP2。
|
图 5 (A) 酪氨酰-DNA磷酸二酯酶2 (TDP2) 的催化水解反应。(B) 小鼠TDP2的结构[76]。(C) 小鼠TDP2的单晶结构 (PDB code: 4GZ1),催化中心以蓝色棒状显示,Mg2+离子以绿色球状显示,DNA底物以红绿蓝色棒状显示。(D) 人TDP2催化水解酪氨酰磷酸二酯键可能的机制[75] |
TDP2属于二价金属离子 (Mg2+/ Mn2+) 依赖的磷酸二酯酶[73]。2012年,Aihara和Williams课题组[37, 76]同时报道了鼠、线虫和斑马鱼的TDP2晶体结构。如图 5B所示,鼠TDP2是一种双结构域DNA修复蛋白,其氮端为泛素相关 (ubiquitin- associated,UBA) 结构域,可能与TDP2的细胞信号转导和应激反应功能有关; 其碳端为核酸外切酶-核酸内切酶-磷酸酶 (exonuclease-endonuclease-phosphatase) 催化结构域,是TDP2发挥其磷酸二酯酶活性的关键区域[76]。如图 5C所示,TDP2 (PDB code: 4GZ1) 有一个深而窄的DNA底物结合沟槽。保守的疏水性残基Trp307、Leu315、Ile317和Phe325形成一个高度精密的结合平面,与DNA的碱基发生堆积
作用,并与脱氧核糖相互作用,“捕捉”单链DNA的5'端,使其5'-酪氨酸的芳香环结合在疏水口袋中,限制了非专一性的核酸内切或外切过程。氨基酸残基Glu162、Arg216、His236、Ser239、Asp272、Asp326、His359和一个Mg2+离子共同组成了TDP2的活性中心[76]。
目前,人TDP2的单晶结构尚未报道。序列分析发现,人TDP2和APE1 (apurinic endonuclease 1) 具有30% 的序列相似性以及14% 的序列一致性。Gao等[75]以人APE1为基础,通过在线蛋白预测程序I-Tasser模拟了人TDP2结构。预测的结构显示,作为TDP2的催化中心,其碳端的核心区域由2个α螺旋夹着3个β折叠组成,这很可能就是DNA的结合沟槽。在这个区域的中心,4个关键残基: Asn120、Glu152、Asp262和His351组成了TDP2的催化口袋。
2.2 TDP2的催化机制及其生物学功能尽管小鼠和斑马鱼的TDP2单晶结构均显示,活性中心包含一个Mg2+ 离子[37, 76],但Gao等[75]通过生物化学方法证实,人的TDP2需要两个Mg2+参与。他们认为,人TDP2催化水解DNA 5'-酪氨酰磷酸二酯键是一步完成的。如图 5D所示,其中一个Mg2+离子与Asp262、His351、Asn264以及一个OH-离子络合,OH-离子亲核进攻P原子; 另一个Mg2+离子与Asp122和Glu152络合,辅助酪氨酰磷酸酯键断裂; Asn120通过桥键间接连接两个Mg2+金属离子。在协同作用下水解酪氨酰磷酸二酯键,去除酪氨酸残基,获得末端为5'-磷酸的缺口DNA[75]。
含有5'-酪氨酰磷酸二酯键的缺口DNA主要是由Top2引起的。与Top1不同,Top2同时断开DNA的两条链,并分别生成一个5'-酪氨酰磷酸二酯键,形成Top2-DNA断裂共价复合物Top2cc (Top2 cleavage complex)。在外源性或内源性因素刺激下,Top2cc被“捕捉”(图 3G),造成Top2介导的DNA损伤 (Top2- mediated DNA damage,Top2-DD),最终导致细胞死亡。造成外源性损伤的因素包括使用Top2毒剂[5]、DNA嵌入剂[77]和AraC等药物治疗[78],以及辐射[79]等 (图 3F)。类似于TDP1,TDP2必须在Top2cc的蛋白被水解代谢后才能识别Top2来源的多肽-DNA复合物 (图 3H)[80],并水解该底物的5'-酪氨酰磷酸二酯键,获得含有3'-OH和5'-磷酸末端的缺口DNA (图 3I)。然后在XRCC4/Lig4、Ku70/Ku80/DNA PK等修复酶的参与下,通过非同源末端连接 (nonhomologous end joining,NHEJ) 通路,进行双链断裂DNA的无错修复 (error-free repair)[81],获得没有碱基缺失的、完整的DNA (图 3J)。
从上述通路可知,TDP2主要在修复Top2-DD的起始阶段发挥作用,而泛素化则能避免将正常复制转录的DNA误认为可裂解底物[82]。这种准确、精细的修复功能使得细胞基因组在面对内源性或外源性的威胁时,依然能保持DNA的完整性和稳定性[83]。但是对于肿瘤细胞来说,这样的修复功能使得它们对上述外源性治疗方法不再敏感[37]。研究证实,在肺癌细胞和组织中高表达TDP2可促进肿瘤细胞增殖以及肿瘤组织生长[84],并使肺癌细胞对Top2毒剂——依托泊苷耐药[85]。
在正常细胞中,同样存在两条通路修复Top2-DD: TDP2依赖的修复通路 (如图 3F~J) 和细胞周期检测点依赖的修复通路。如前所述,在许多肿瘤细胞 中,检测点蛋白常常缺失[39],TDP2依赖的通路成为主要的修复通路。理论上分析,在相关检测点蛋白缺失的肿瘤细胞中,抑制TDP2活性将对这些肿瘤细胞增殖产生很大的影响,其抑制剂可以增强Top2毒剂、DNA嵌入剂或AraC类药物的疗效,甚至提高放疗效果。研究证实,删除Tdp2基因的小鼠对Top2-DD敏感[81]; 如果抑制TDP2的活性或敲除TDP2,可增加细胞对依托泊苷和阿霉素的敏感性[85, 86]。
通过以上的理论分析和研究发现可知,TDP2是一个潜在的肿瘤治疗靶点。其抑制剂与Top2毒剂、DNA嵌入剂或核苷类链终止剂联用,具有协同治疗肿瘤的功效。
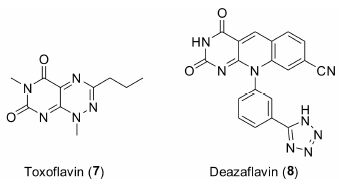
2.3 TDP2小分子抑制剂由于TDP2的发现时间尚短,其抑制剂的报道较少。2013年,Raoof等[87]发现了两类TDP2选择性抑制剂 —— toxoflavin类化合物和deazaflavin类化合物。如图 6所示,代表化合物7和8的TDP2抑制活性IC50值分别为1.51和0.59 μmol·L-1。然而,toxoflavin类化合物具有很强的氧化还原活性,而deazaflavin类化合物则细胞膜透过性不佳。这些性质限制了它们成为药物的可能性。
|
图 6 TDP2小分子抑制剂的结构 |
本课题组发现了一类呋喃并喹啉二酮类化合物,具有良好的TDP2选择性抑制活性。目前,该研究正在进行中。总之,TDP2抑制剂的研究尚处于起始阶段,有待更多的研究发现。
3 问题与展望作为近年来才发现的DNA修复酶,TDP1和TDP2具有特殊的生物学功能,是潜在的肿瘤治疗靶点。但是,目前对其研究和认识尚处于初级阶段,需要进行深入研究: 如它们是如何专一性识别底物的; 水解酪氨酰磷酸二酯键的详细过程; 当前发现的抑制剂种类较少,尤其是TDP2抑制剂,仅报道了两类。对于药物化学家来说,发现新的抑制剂,尤其是高活性的、选择性抑制剂将为解决以上问题提供有力帮助,甚至发现以其为靶点的创新药物,证实其作为肿瘤治疗靶点的可行性。
TDP1和TDP2除了作为肿瘤治疗靶点外,还具有更加广泛的功能。如研究发现: ① 低剂量的伊立替康和TDP1抑制剂呋喃二脒NSC 305831联用,可显著改善狼疮肾炎小鼠的生存率,预示着Top1/TDP1可能是系统性红斑狼疮的联合治疗靶点[88]; ② TDP1和TDP2可调控人类乳头瘤病毒 (human papillomavirus) 游离基因的稳定性[89]; ③ TDP2还是小核糖 核酸病毒 (picornaviruses) 的断裂酶 (unlinkase)[90]。一旦感染小核糖核酸病毒,宿主的TDP2会将其RNA 5' 端的一段小的病毒蛋白移除,病毒才能开始翻译 自己的蛋白[90, 91]。另外,宿主TDP2在乙肝病毒的感染中也扮演着重要角色,可能与乙肝病毒cccDNA (covalently closed circular DNA) 的形成有关[92]。因 此,TDP1和TDP2不仅是潜在的肿瘤治疗靶点,还可能在感染性疾病的治疗中扮演重要角色。
| [1] | Wang JC. DNA topoisomerases[J]. Annu Rev Biochem, 1996, 65:635-692. |
| [2] | Champoux JJ. DNA topoisomerases:structure, function, and mechanism[J]. Annu Rev Biochem, 2001, 70:369-413. |
| [3] | Pommier Y. Topoisomerase I inhibitors:camptothecins and beyond[J]. Nat Rev Cancer, 2006, 6:789-802. |
| [4] | Pommier Y. DNA topoisomerase I inhibitors:chemistry, biology, and interfacial inhibition[J]. Chem Rev, 2009, 109:2894-2902. |
| [5] | Nitiss JL. Targeting DNA topoisomerase Ⅱ in cancer chemo-therapy[J]. Nat Rev Cancer, 2009, 9:338-350. |
| [6] | Ashour ME, Atteya R, El-Khamisy SF. Topoisomerase-mediated chromosomal break repair:an emerging player in many games[J]. Nat Rev Cancer, 2015, 15:137-151. |
| [7] | Yang SW, Burgin AB, Huizenga BN, et al. A eukaryotic enzyme that can disjoin dead-end covalent complexes between DNA and type I topoisomerases[J]. Proc Natl Acad Sci U S A, 1996, 93:11534-11539. |
| [8] | Pouliot JJ, Yao KC, Robertson CA, et al. Yeast gene for a Tyr-DNA phosphodiesterase that repairs topoisomerase I complexes[J]. Science, 1999, 286:552-555. |
| [9] | Dexheimer TS, Antony S, Marchand C, et al. Tyrosyl-DNA phosphodiesterase as a target for anticancer therapy[J]. Anti-cancer Agent Med Chem, 2008, 8:381-389. |
| [10] | Interthal H, Pouliot JJ, Champoux JJ. The tyrosyl-DNA phosphodiesterase Tdp1 is a member of the phospholipase D superfamily[J]. Proc Natl Acad Sci U S A, 2001, 98:12009-12014. |
| [11] | Davies DR, Interthal H, Champoux JJ, et al. The crystal structure of human tyrosyl-DNA phosphodiesterase, Tdp1[J]. Structure, 2002, 10:237-248. |
| [12] | Barthelmes HU, Habermeyer M, Christensen MO, et al. TDP1 overexpression in human cells counteracts DNA damage mediated by topoisomerases I and Ⅱ[J]. J Biol Chem, 2004, 279:55618-55625. |
| [13] | He XP, van Waardenburg RCAM, Babaoglu K, et al. Muta-tion of a conserved active site residue converts tyrosyl-DNA phosphodiesterase I into a DNA topoisomerase I-dependent poison[J]. J Mol Biol, 2007, 372:1070-1081. |
| [14] | Das BB, Antony S, Gupta S, et al. Optimal function of the DNA repair enzyme TDP1 requires its phosphorylation by ATM and/or DNA-PK[J]. EMBO J, 2009, 28:3667-3680. |
| [15] | Das BB, Huang SYN, Murai J, et al. PARP1-TDP1 coupling for the repair of topoisomerase I-induced DNA damage[J]. Nucleic Acids Res, 2014, 42:4435-4449. |
| [16] | Waite M. The PLD superfamily:insights into catalysis[J]. Biochim Biophy Acta, 1999, 1439:187-197. |
| [17] | Raymond AC, Rideout MC, Staker B, et al. Analysis of human tyrosyl-DNA phosphodiesterase I catalytic residues[J]. J Mol Biol, 2004, 338:895-906. |
| [18] | Gajewski S, Comeaux EQ, Jafari N, et al. Analysis of the active-site mechanism of tyrosyl-DNA phosphodiesterase I:a member of the phospholipase D superfamily[J]. J Mol Biol, 2012, 415:741-758. |
| [19] | Davies DR, Interthal H, Champoux JJ, et al. Crystal structure of a transition state mimic for Tdp1 assembled from vanadate, DNA, and a topoisomerase I-derived peptide[J]. Chem Biol, 2003, 10:139-147. |
| [20] | Pommier Y, Huang SYN, Gao R, et al. Tyrosyl-DNA-phosphodiesterases (TDP1 and TDP2)[J]. DNA Repair, 2014, 19:114-129. |
| [21] | Comeaux EQ, van Waardenburg RCAM. Tyrosyl-DNA phosphodiesterase I resolves both naturally and chemically induced DNA adducts and its potential as a therapeutic target[J]. Drug Metab Rev, 2014, 46:494-507. |
| [22] | Davies DR, Interthal H, Champoux JJ, et al. Insights into substrate binding and catalytic mechanism of human tyro-syl-DNA phosphodiesterase (Tdp1) from vanadate and tungstate-inhibited structures[J]. J Mol Biol, 2002, 324:917-932. |
| [23] | Stewart L, Redinbo MR, Qui XY, et al. A model for the mechanism of human topoisomerase I[J]. Science, 1998, 279:1534-1541. |
| [24] | Pommier Y, Kohlhagen G, Pourquier P, et al. Benzo[a]pyrene diol epoxide adducts in DNA are potent suppressors of a normal topoisomerase I cleavage site and powerful inducers of other topoisomerase I cleavages[J]. Proc Natl Acad Sci U S A, 2000, 97:2040-2045. |
| [25] | Pourquie P, Waltman JL, Urasaki Y, et al. Topoisomerase I-mediated cytotoxicity of N-methyl-N'-nitro-N-nitrosoguanidine:trapping of topoisomerase I by the O6-methylguanine[J]. Cancer Res, 2001, 61:53-58. |
| [26] | Murai J, Huang SYN, Das BB, et al. Tyrosyl-DNA phos-phodiesterase 1(TDP1) repairs DNA damage induced by topoisomerases I and Ⅱ and base alkylation in vertebrate cells[J]. J Biol Chem, 2012, 287:12848-12857. |
| [27] | Pommier Y, Laco GS, Kohlhagen G, et al. Position-specific trapping of topoisomerase I-DNA cleavage complexes by intercalated benzo[a]-pyrene diol epoxide adducts at the 6-amino group of adenine[J]. Proc Natl Acad Sci U S A, 2000, 97:10739-10744. |
| [28] | Pommier Y, Kohlhagen G, Laco GS, et al. Different effects on human topoisomerase I by minor groove and intercalated deoxyguanosine adducts derived from two polycyclic aromatic hydrocarbon diol epoxides at or near a normal cleavage site[J]. J Biol Chem, 2002, 277:13666-13672. |
| [29] | Pourquier P, Takebayashi Y, Urasaki Y, et al. Induction of topoisomerase I cleavage complexes by 1-β-D-arabinofu-ranosylcytosine (ara-C) in vitro and in ara-C-treated cells[J]. Proc Natl Acad Sci U S A, 2000, 97:1885-1890. |
| [30] | Pourquier P, Ueng LM, Kohlhagen G, et al. Effects of uracil incorporation, DNA mismatches, and abasic sites on cleavage and religation activities of mammalian topoisomerase I[J]. J Biol Chem, 1997, 272:7792-7796. |
| [31] | Lanza A, Tornaletti S, Rodolfo C, et al. Human DNA topoi-somerase I-mediated cleavages stimulated by ultraviolet light-induced DNA damage[J]. J Biol Chem, 1996, 271:6978-6986. |
| [32] | Subramanian D, Rosenstein BS, Muller MT. Ultraviolet-induced DNA damage stimulates topoisomerase I-DNA complex formation in vivo:possible relationship with DNA repair[J]. Cancer Res, 1998, 58:976-984. |
| [33] | Dexheimer TS, Gediya LK, Stephen AG, et al. 4-Pregnen-21-ol-3,20-dione-21-(4-bromobenzenesulfonate) (NSC 88915) and related novel steroid derivatives as tyrosyl-DNA phosphodi-esterase (Tdp1) inhibitors[J]. J Med Chem, 2009, 52:7122-7131. |
| [34] | Beck DE, Agama K, Marchand C, et al. Synthesis and biological evaluation of new carbohydrate-substituted inde-noisoquinoline topoisomerase I inhibitors and improved syntheses of the experimental anticancer agents indotecan (LMP400) and indimitecan (LMP776)[J]. J Med Chem, 2014, 57:1495-1512. |
| [35] | Waters CA, Strande NT, Wyatt DW, et al. Nonhomologous end joining:a good solution for bad ends[J]. DNA Repair, 2014, 17:39-51. |
| [36] | Whitehouse CJ, Taylor RM, Thistlethwaite A, et al. XRCC1 stimulates human polynucleotide kinase activity at damaged DNA termini and accelerates DNA single-strand break repair[J]. Cell, 2001, 104:107-117. |
| [37] | Shi K, Kurahashi K, Gao R, et al. Structural basis for recog-nition of 5'-phosphotyrosine adducts by Tdp2[J]. Nat Struct Mol Biol, 2012, 19:1372-1377. |
| [38] | Zhou BBS, Elledge SJ. The DNA damage response:putting checkpoints in perspective[J]. Nature, 2000, 408:433-439. |
| [39] | Dasika GK, Lin SC, Zhao S, et al. DNA damage-induced cell cycle checkpoints and DNA strand break repair in development and tumorigenesis[J]. Oncogene, 1999, 18:7883-7899. |
| [40] | Huang SYN, Murai J, Dalla Rosa I, et al. TDP1 repairs nuclear and mitochondrial DNA damage induced by chain-terminating anticancer and antiviral nucleoside analogs[J]. Nucleic Acids Res, 2013, 41:7793-7803. |
| [41] | Nivens MC, Felder T, Galloway AH, et al. Engineered resistance to camptothecin and antifolates by retroviral coex-pression of tyrosyl DNA phosphodiesterase-I and thymidylate synthase[J]. Cancer Chemother Pharmacol, 2004, 53:107-115. |
| [42] | Liu CY, Zhou SY, Begum S, et al. Increased expression and activity of repair genes TDP1 and XPF in non-small cell lung cancer[J]. Lung Cancer, 2007, 55:303-311. |
| [43] | Perego P, Cossa G, Tinelli S, et al. Role of tyrosyl-DNA phosphodiesterase 1 and inter-players in regulation of tumor cell sensitivity to topoisomerase I inhibition[J]. Biochem Pharmacol, 2012, 83:27-36. |
| [44] | Fam HK, Walton C, Mitra SA, et al. TDP1 and PARP1 deficiency are cytotoxic to rhabdomyosarcoma cells[J]. Mol Cancer Res, 2013, 11:1179-1192. |
| [45] | Jakobsen AK, Lauridsen KL, Samuel EB, et al. Correlation between topoisomerase I and tyrosyl-DNA phosphodi-esterase 1 activities in non-small cell lung cancer tissue[J]. Exp Mol Pathol, 2015, 99:56-64. |
| [46] | Dean RA, Fam HK, An JH, et al. Identification of a putative Tdp1 inhibitor (CD00509) by in vitro and cell-based assays[J]. J Biomol Screen, 2014, 19:1372-1382. |
| [47] | Interthal H, Chen HJ, Kehl-Fie TE, et al. SCAN1 mutant Tdp1 accumulates the enzyme-DNA intermediate and causes camptothecin hypersensitivity[J]. EMBO J, 2005, 24:2224-2233. |
| [48] | Miao ZH, Agama K, Sordet O, et al. Hereditary ataxia SCAN1 cells are defective for the repair of transcription-dependent topoisomerase I cleavage complexes[J]. DNA Repair, 2006, 5:1489-1494. |
| [49] | Alagoz M, Wells OS, El-Khamisy SF. TDP1 deficiency sensitizes human cells to base damage via distinct topoisomerase I and PARP mechanisms with potential applications for cancer therapy[J]. Nucleic Acids Res, 2014, 42:3089-3103. |
| [50] | Meisenberg C, Gilbert DC, Chalmers A, et al. Clinical and cellular roles for TDP1 and TOP1 in modulating colorectal cancer response to irinotecan[J]. Mol Cancer Ther, 2015, 14:575-585. |
| [51] | Bryant HE, Schultz N, Thomas HD, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase[J]. Nature, 2005, 434:913-917. |
| [52] | Farmer H, McCabe N, Lord CJ, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy[J]. Nature, 2005, 434:917-921. |
| [53] | Curtin NJ. DNA repair dysregulation from cancer driver to therapeutic target[J]. Nat Rev Cancer, 2012, 12:801-817. |
| [54] | Wang JC. Cellular roles of DNA topoisomerases:a mo-lecular perspective[J]. Nat Rev Mol Cell Biol, 2002, 3:430-440. |
| [55] | Comeaux EQ, Cuya SM, Kojima K, et al. Tyrosyl-DNA phosphodiesterase I catalytic mutants reveal an alternative nucleophile that can catalyze substrate cleavage[J]. J Biol Chem, 2015, 290:6203-6214. |
| [56] | Liao ZY, Thibaut L, Jobson A, et al. Inhibition of human tyrosyl-DNA phosphodiesterase by aminoglycoside antibiotics and ribosome inhibitors[J]. Mol Pharmacol, 2006, 70:366-372. |
| [57] | Tian LW, Feng YJ, Tran TD, et al. Tyrosyl-DNA phosphodi-esterase I inhibitors from the Australian plant Macropteranthes leichhardtii[J]. J Nat Prod, 2015, 78:1756-1760. |
| [58] | Antony S, Marchand C, Stephen AG, et al. Novel high-throughput electrochemiluminescent assay for identification of human tyrosyl-DNA phosphodiesterase (Tdp1) inhibi-tors and characterization of furamidine (NSC 305831) as an inhibitor of Tdp1[J]. Nucleic Acids Res, 2007, 35:4474-4484. |
| [59] | Marchand C, Lea WA, Jadhav A, et al. Identification of phosphotyrosine mimetic inhibitors of human tyrosyl-DNA phosphodiesterase I by a novel AlphaScreen high-throughput assay[J]. Mol Cancer Ther, 2009, 8:240-248. |
| [60] | Nguyen TX, Morrell A, Conda-Sheridan M, et al. Synthesis and biological evaluation of the first dual tyrosyl-DNA phos-phodiesterase I (Tdp1)-topoisomerase I (Top1) inhibitors[J]. J Med Chem, 2012, 55:4457-4478. |
| [61] | Conda-Sheridan M, Reddy PVN, Morrell A, et al. Synthesis and biological evaluation of indenoisoquinolines that inhibit both tyrosyl-DNA phosphodiesterase I (Tdp1) and topoisom-erase I (Top1)[J]. J Med Chem, 2013, 56:182-200. |
| [62] | Lv PC, Agama K, Marchand C, et al. Design, synthesis, and biological evaluation of O-2-modified indenoisoquinolines as dual topoisomerase I-tyrosyl-DNA phosphodiesterase I inhibitors[J]. J Med Chem, 2014, 57:4324-4336. |
| [63] | Nguyen TX, Abdelmalak M, Marchand C, et al. Synthesis and biological evaluation of nitrated 7-, 8-, 9-, and 10-hydroxyindenoisoquinolines as potential dual topoisomerase I (Top1)-tyrosyl-DNA phosphodiesterase I (TDP1) inhibitors[J]. J Med Chem, 2015, 58:3188-3208. |
| [64] | Shoemaker RH. The NCI60 human tumour cell line anticancer drug screen[J]. Nat Rev Cancer, 2006, 6:813-823. |
| [65] | Morrell A, Placzek M, Parmley S, et al. Optimization of the indenone ring of indenoisoquinoline topoisomerase I inhibitors[J]. J Med Chem, 2007, 50:4388-4404. |
| [66] | Nagarajan M, Morrell A, Antony S, et al. Synthesis and biological evaluation of bisindenoisoquinolines as topoisomerase I inhibitors[J]. J Med Chem, 2006, 49:5129-5140. |
| [67] | Sirivolu VR, Vernekar SKV, Marchand C, et al. 5-Arylidenethioxothiazolidinones as inhibitors of tyrosyl-DNA phosphodiesterase I[J]. J Med Chem, 2012, 55:8671-8684. |
| [68] | Baell JB, Holloway GA. New substructure filters for re-moval of Pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays[J]. J Med Chem, 2010, 53:2719-2740. |
| [69] | Takagi M, Ueda JY, Hwang JH, et al. Tyrosyl-DNA phosphodiesterase 1 inhibitor from an anamorphic fungus[J]. J Nat Prod, 2012, 75:764-767. |
| [70] | Zakharenko A, Khomenko T, Zhukova S, et al. Synthesis and biological evaluation of novel tyrosyl-DNA phosphodi-esterase 1 inhibitors with a benzopentathiepine moiety[J]. Bioorg Med Chem, 2015, 23:2044-2052. |
| [71] | Pype S, Declercq W, Ibrahimi A, et al. TTRAP, a novel protein that associates with CD40, tumor necrosis factor (TNF) receptor-75 and TNF receptor-associated factors (TRAFs), and that inhibits nuclear factor-κB activation[J]. J Biol Chem, 2000, 275:18586-18593. |
| [72] | Pei HP, Yordy JS, Leng QX, et al. EAPII interacts with ETS1 and modulates its transcriptional function[J]. Oncogene, 2003, 22:2699-2709. |
| [73] | Cortes Ledesma F, El Khamisy SF, Zuma MC, et al. A human 5'-tyrosyl DNA phosphodiesterase that repairs topoi-somerase-mediated DNA damage[J]. Nature, 2009, 461:674-678. |
| [74] | Adhikari S, Karmahapatra SK, Karve TM, et al. Characteri-zation of magnesium requirement of human 5'-tyrosyl DNA phosphodiesterase mediated reaction[J]. BMC Res Notes, 2012, 5:134. |
| [75] | Gao R, Huang SYN, Marchand C, et al. Biochemical charac-terization of human tyrosyl-DNA phosphodiesterase 2(TDP2/TTRAP):a Mg2+/Mn2+-dependent phosphodiesterase specific for the repair of topoisomerase cleavage complexes[J]. J Biol Chem, 2012, 287:30842-30852. |
| [76] | Schellenberg MJ, Appel CD, Adhikari S, et al. Mechanism of repair of 5'-topoisomerase Ⅱ-DNA adducts by mammalian tyrosyl-DNA phosphodiesterase 2[J]. Nat Struct Mol Biol, 2012, 19:1363-1371. |
| [77] | Khan QA, Kohlhagen G, Marshall R, et al. Position-specific trapping of topoisomerase Ⅱ by benzo[a]pyrene diol epoxide adducts:implications for interactions with intercalating anticancer agents[J]. Proc Natl Acad Sci U S A, 2003, 100:12498-12503. |
| [78] | Cline SD, Osheroff N. Cytosine arabinoside lesions are position-specific topoisomerase Ⅱ poisons and stimulate DNA cleavage mediated by the human type Ⅱ enzymes[J]. J Biol Chem, 1999, 274:29740-29743. |
| [79] | Corbett AH, Zechiedrich EL, Lloyd RS, et al. Inhibition of eukary-otic topoisomerase Ⅱ by ultraviolet-induced cyclobutane pyrimidine dimers[J]. J Biol Chem, 1991, 266:19666-19671. |
| [80] | Gao R, Schellenberg MJ, Huang SYN, et al. Proteolytic degradation of topoisomerase Ⅱ (Top2) enables the processing of Top2 DNA and Top2 RNA covalent complexes by tyrosyl-DNA-phosphodiesterase 2(TDP2)[J]. J Biol Chem, 2014, 289:17960-17969. |
| [81] | Gómez-Herreros F, Romero-Granados R, Zeng ZH, et al. TDP2-dependent non-homologous end-joining protects against topoisomerase Ⅱ-induced DNA breaks and genome instability in cells and in vivo[J]. PLoS Genet, 2013, 9:e1003226. |
| [82] | Nitiss JL, Nitiss KC. Tdp2:a means to fixing the ends[J]. PLoS Genet, 2013, 9:e1003370. |
| [83] | Gómez-Herreros F, Schuurs-Hoeijmakers JHM, McCormack M, et al. TDP2 protects transcription from abortive topoisomerase activity and is required for normal neural function[J]. Nat Genet, 2014, 46:516-521. |
| [84] | Li C, Fan S, Owonikoko TK, et al. Oncogenic role of EAPII in lung cancer development and its activation of the MAPK-ERK pathway[J]. Oncogene, 2011, 30:3802-3812. |
| [85] | Do PM, Varanasi L, Fan SQ, et al. Mutant p53 cooperates with ETS2 to promote etoposide resistance[J]. Genes Dev, 2012, 26:830-845. |
| [86] | Zeng ZH, Cortés-Ledesma F, El Khamisy SF, et al. TDP2/TTRAP is the major 5'-tyrosyl DNA phosphodiesterase activity in vertebrate cells and is critical for cellular resistance to topoisomerase Ⅱ-induced DNA damage[J]. J Biol Chem, 2011, 286:403-409. |
| [87] | Raoof A, Depledge P, Hamilton NM, et al. Toxoflavins and deazaflavins as the first reported selective small molecule inhibitors of tyrosyl-DNA phosphodiesterase Ⅱ[J]. J Med Chem, 2013, 56:6352-6370. |
| [88] | Keil A, Frese-Schaper M, Steiner SK, et al. The topoisom-erase I inhibitor irinotecan and the tyrosyl-DNA phosphodiesterase 1 inhibitor furamidine synergistically suppress murine lupus nephritis[J]. Arthritis Rheumatol, 2015, 67:1858-1867. |
| [89] | Edwards TG, Vidmar TJ, Koeller K, et al. DNA damage repair genes controlling human papillomavirus (HPV) episome levels under conditions of stability and extreme instability[J]. PLoS One, 2013, 8:e75406. |
| [90] | Virgen-Slane R, Rozovics JM, Fitzgerald KD, et al. An RNA virus hijacks an incognito function of a DNA repair enzyme[J]. Proc Natl Acad Sci U S A, 2012, 109:14634-14639. |
| [91] | Langereis MA, Feng Q, Nelissen FH, et al. Modification of picornavirus genomic RNA using ‘click’ chemistry shows that unlinking of the VPg peptide is dispensable for translation and replication of the incoming viral RNA[J]. Nucleic Acids Res, 2014, 42:2473-2482. |
| [92] | Königer C, Wingert I, Marsmann M, et al. Involvement of the host DNA-repair enzyme TDP2 in formation of the covalently closed circular DNA persistence reservoir of hepatitis B viruses[J]. Proc Natl Acad Sci U S A, 2014, 111:E4244-E4253. |
 2016, Vol. 51
2016, Vol. 51


