2. 洛阳市活载体生物材料与动物疫病防控重点实验室, 洛阳 471023
2. Key Laboratory of Live Vector Biomaterials and Animal Disease Prevention and Control, Luoyang 471023, China
RNA结合蛋白(RNA binding protein, RBP)是转录后基因调控的关键调节因子,据估计,哺乳动物的基因组编码了超过1 500个RBP,这些RBP通过与靶RNA结合后,在转录后基因调控过程中发挥重要的作用,包括选择性剪接、多聚腺苷酸化、转运、亚细胞定位、丰度和翻译等[1-2]。RBP与靶mRNA之间的互相结合作用,对维持内环境稳态十分重要[3]。胚胎致死异常视觉蛋白1(embryonic lethal abnormal vision like 1, ELAVL1)为ELAV家族的一员,也称为人类抗原R(human antigen R, HuR),是一种典型的RBP[4]。ELAVL1在多种细胞中广泛表达,主要存在于正常哺乳细胞的细胞核中,受到外源刺激时或在肿瘤细胞中则主要表达于细胞质[5]。与哺乳动物不同的是,伊蚊的细胞质中含有大量的ELAVL1同源物蛋白,这反映了ELAVL1在昆虫和脊椎动物中的作用方式存在先天差异[6]。ELAVL1是基因表达的关键效应因子,通过与3′非翻译区(3′untranslated element, 3′UTR)的AU富含元件(AU-rich element, ARE)结合,发挥稳定mRNA的作用[7]。另有研究表明,ELAVL1还能促进和抑制靶mRNA的翻译[8]。ELAVL1的核质转运和高表达与多种肿瘤的发生发展及病毒感染相关[9-10]。病毒种类繁多,多数对宿主有致病作用,常引起疫病流行,造成重大经济损失。病毒复制是一个十分复杂的过程,必须借助宿主因子在宿主细胞内完成病毒的生命周期,因此,寻找参与病毒复制的宿主因子及其调控机制,对于深入了解病毒与机体互作的分子机制,研究新的抗病毒策略具有重要意义[11]。
近年来,ELAVL1蛋白的功能研究是研究热点之一,除了研究其在肿瘤细胞中发挥的重要作用,国内外关于ELAVL1对多种病毒复制影响的研究也日益增多。本文总结了近年来ELAVL1的相关研究,主要综述ELAVL1的生物学特点、功能、调控机制及其在病毒复制过程中的调控作用,同时对未来ELAVL1的研究方向进行了展望,为进一步研究ELAVL1与病毒复制之间互作的机制提供参考。
1 ELAVL1的生物学特点ELAVL1蛋白由326个氨基酸组成,其编码基因位于人的第19号染色体,位于小鼠的第8号染色体,位于山羊的第7号染色体,位于鸡的第28号染色体[12]。ELAVL1首先在果蝇中被发现,于1996年被鉴定和克隆,在大部分恶性肿瘤中高表达[4, 13-14]。ELAVL1在各种组织中普遍表达,而哺乳动物中的其他ELAV家族成员,包括ELAVL2(HuB)、ELAVL3(HuC)和ELAV4(HuD),几乎只存在于神经元组织中[4, 9]。ELAVL1包含3个保守的RNA识别基序(RNA recognition motif,RRM)和1个铰链区,与其它ELAV蛋白具有高度同源性。其中,前两个位于N端的结构域(RRM1和RRM2)通过10个残基接头连接,与3′UTR的ARE结合;而位于C端的RRM3通过与靶mRNA的3′poly(A)尾结合,介导典型RNA相互作用,并在RRM3的α-螺旋面存在一个二聚界面(图 1)[15-16]。ARE的序列长度约为17~20个核苷,根据核心AUUUA五聚体的数量和分布,可将ARE分为3类:其一是在富AU区含有AUUUA基序的几个分散拷贝的基因;其二是具有至少2个重叠的UUAUUA(U/A)(U/A)基序的基因;其三是特征较少的不包含典型的AUUUA基序[17]。ARE介导的mRNA快速降解是哺乳动物细胞转录后基因调控的重要机制[18]。此外,研究发现ELAVL1还能与靶基因的5′UTR相互作用,从而调控翻译[8]。
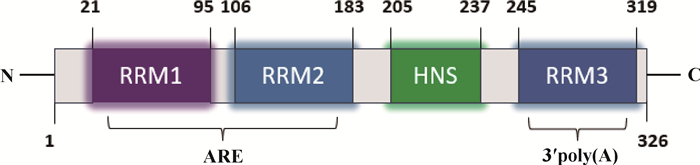
|
图 1 ELAVL1结构简示图 Fig. 1 Schematic diagram of ELAVL1 structure |
正常健康条件下,ELAVL1蛋白主要位于细胞核中,在刺激因子作用下,ELAVL1与靶mRNA结合,形成ELAVL1-mRNA复合物,该复合物通过核孔被转运到细胞质,以发挥其稳定靶mRNA和调节蛋白质翻译的功能[4]。这种核质转运是通过位于铰链区的HuR核质穿梭序列(HuR nuclear-cytoplasmic shuttling sequence, HNS)来实现,该序列长52个氨基酸,是ELAVL1发生翻译后修饰的主要基序[19]。也有研究表明,在增殖活跃的未分化细胞中,ELAVL1主要定位于细胞核,但在诱导和分化过程中,其在细胞质中的丰度显著增加,在细胞分化过程完成后又恢复到核定位[20]。此外,有研究发现ELAVL1在S期向M期转变时主要以细胞质的形式存在,而在T细胞中,ELAVL1在早期G1期主要存在于细胞质中[21-22]。
2 ELAVL1的功能和调控机制ELAVL1作为一种反式调节因子,与不稳定的顺式作用元件3′UTR和5′UTR中的ARE结合,在转运蛋白的帮助下通过HNS或染色体区域维持蛋白1(chromosome maintenance protein 1, CRM1)从细胞核转移到细胞质中,从而增加mRNA的稳定性或蛋白的翻译来实现转录后基因调控作用[4]。
2.1 ELAVL1与核质转运ELAVL1蛋白具有核质穿梭的功能,其核质易位和细胞质表达对许多疾病中的活性和功能是必要的[2, 23]。据报道,许多应激刺激因子如紫外线辐射、脂多糖、化学化合物、微环境改变、细胞因子、热休克、病毒感染和激素治疗等可诱导ELAVL1穿梭[24]。通常,铰链区处的磷酸化可以调节ELAVL1亚细胞定位,转运CRM1(也称为输出蛋白1)、转运蛋白1和2以及输入蛋白α1等组分,也参与ELAVL1的核质穿梭[5]。目前已知至少有3种信号通路与ELAVL1蛋白的核质转运相关。第1种是p38丝裂原活化蛋白激酶(p38 mitogen-activated protein kinase, p38-MAPK)通路,目前还不确定这条通路的具体机制,推测是由于p38-MAPK使ELAVL1甲基化,从而影响其核质转运;第2种是蛋白激酶C(protein kinase C,PKC)通路,血管紧张素II(Angiotension II,AngII)和ATP类似物通过结合特异性受体,之后与PKC结合,使PKC输入到细胞核与ELAVL1结合并发生磷酸化,从而导致ELAVL1向细胞质转运;第3种是AMP激活激酶(adenosine 5′-monophosphate -activated protein kinase, AMPK)通路,AMPK通过引起输入蛋白α1的磷酸化和乙酰化来阻断ELAVL1的核质穿梭[23-24]。特定的细胞刺激引起信号通路级联,促使ELAVL1由细胞核向细胞质输出,从而保护mRNA免受降解[25]。ELAVL1的核质转运增加了靶mRNA的稳定性,促进了mRNA的转录和蛋白翻译(图 2)[9]。

|
图 2 ELAVL1发生的核质转运示意图 Fig. 2 Schematic representation of the nucleoplasmic transport occurring with ELAVL1 |
ELAVL1通过与靶mRNA 3′UTR的ARE结合,发挥稳定靶mRNA的作用[8]。ELAVL1稳定靶mRNA确切机制尚未完全阐明,但人们广泛地认为是ELAVL1与靶mRNA的结合阻断了mRNA募集到衰变位点[26]。mRNA的稳定性取决于它与顺式元件和反式调节因子的相互作用并高度依赖于poly(A)尾。顺式元件存在于mRNA转录本中,不仅包括非翻译区,也包括编码区,ARE是许多mRNA中研究最多的顺式元件[27]。ELAVL1则是作为反式调节因子。Poly(A)尾是一长串保护转录物3′端的腺苷残基,去除poly(A)尾部,称为去腺苷酸化,是mRNA降解的第一步[28]。去除mRNA 5′端“帽”结构也是mRNA降解的重要部分[29]。因此,稳定mRNA可以保护其免受降解。ELAVL1可以稳定大量的靶mRNA,如环氧合酶-2(cyclooxygenase-2,COX-2)[30-31]、p53[32]、基质金属蛋白酶(metalloproteinase,MMP)-9[33]和TNF-α[34]等超过80种mRNA[8, 16],从而增加相应的蛋白产物和转录产物,参与病理过程[19]。
2.3 ELAVL1转录水平的调控转录调控发生在两个相互关联的水平上:转录因子和染色质表观调控因子[29]。转录因子结合到特定的靶基因上后,促进或是抑制其表达,是调控转录的核心。而染色质表观因子通过影响DNA甲基化、组蛋白修饰等多种事件,增加或干扰转录因子对启动子的结合,影响基因的转录[35]。研究表明,ELAVL1的启动子通常包含多个转录起始位点,而且存在可替代的启动子,这些启动子在细胞受到应激时可被激活或抑制[36]。Kang等[37]发现,在胃癌中,通过PI3K/AKT/NF-κB途径激活ELAVL1,有助于肿瘤进展,并证明ELAVL1是NF-κB的直接转录靶标。Jeyaraj等[36]发现,ELAVL1 mRNA表达为两种交替转录物,在其5′UTR区域中变化,较长的约150个碱基,较短的约20个碱基,并发现NF-κB位点下游约100 bp处的Smad1/5/8结合位点是ELAVL1短mRNA的正调节因子。此外,Govindaraju和Lee[38]发现,Krüppel样转录因子8(Krüppel-like factor 8,KLF8)有特异性结合ELAVL1启动子的能力,引起ELAVL1长mRNA的增加。
2.4 ELAVL1转录后水平的调控转录后调控主要体现在对mRNA前体核内不均一核糖核蛋白(heterogeneous nuclear ribonucleoprotein, hnRNP)的剪接和加工、mRNA由细胞核转至细胞质的过程及定位、mRNA的稳定性及其降解过程等多个环节进行的调控,在调节细胞基因表达和形成转录组及蛋白质组中起着重要作用,也是哺乳动物细胞在应激反应时控制基因表达的主要机制[39-41]。研究发现,miRNA和ELAVL1在转录后基因调控中能够协作,如vegfa 3′UTR中miR-200b的结合位点与ELAVL1的结合位点重叠,也有具有相同的功能位点;而miR-548c、miR-122、miR-331、miR-16和miR-519,与ELAVL1则有拮抗作用,降低其与靶mRNA的结合[9, 42]。另外,ELAVL1的丰度也可以由miRNA调节,已发现miR-125、miR-519、miR-9、291B-3P、miR-570-3、miR-16、miR-22靶向ELAVL1 mRNA的编码区和3′UTR,抑制其表达。此外,ELAVL1 3′-UTR与miRNA-155-5P还有一个结合位点,该位点稳定转录本,有利于细胞迁移[16]。除了miRNA,circRNA能充当蛋白海绵,吸附RNA结合蛋白ELAVL1,隔离靶蛋白阻止其发挥生物学功能[40]。ELAVL1通过与FOXO(Forkhead Box O,FOXO)-1 3′UTR内的ARE结合特异性地调控FoxO1 mRNA的表达并通过转录后调节FoxO1介导5-氟脲嘧啶(5-fluorouracil,5-FU)诱导的细胞凋亡[43]。Phillips等[44]鉴定了PABPN1 3′UTR中的多个顺式调节元件,并在体外和体内鉴定了ELAVL1介导的PABPN1调节,证明ELAVL1是PABPN1转录和蛋白水平的转录后调节因子。有些RBP如丁酸盐反应因子1(butyrate response factor 1,BRF1)、富含AU的RNA结合蛋白1(AUrich element RNA-binding factor 1,AUF1)、RNA结合蛋白TTP(tristetraprolin,TTP)对于ELAVL1和靶基因的结合也有拮抗作用,它们通过竞争性结合细胞因子mRNA,降低其稳定性,从而参与炎性细胞因子的转录后调控[45]。
2.5 ELAVL1的翻译后修饰蛋白质是细胞生命活动的实际功能执行者,前体蛋白是没有生物学活性的,通常需要一系列的翻译后加工,才能成为具有功能的成熟蛋白[46]。翻译后修饰(post-translational modifications, PTMs)是细胞精细调控诸多生物学过程的关键方式之一,对细胞的生长、分化、凋亡、代谢、免疫等生命过程均具有一定的作用[47]。PTMs通常指蛋白质合成后发生的酶促反应,包括小分子基团或生物分子与一个氨基酸的共价加成、化学修饰和肽键的裂解[48]。ELAVL1的功能可以通过PTMs来调节,这些修饰包括磷酸化、甲基化、泛素化、类泛素化修饰和蛋白水解等,改变ELAVL1的亚细胞定位和结合靶RNA的能力[1]。其中研究最多的是磷酸化修饰。周期素依赖性激酶(cyclin-dependent kinases,cdks)-1可在铰链区丝氨酸(serine, S)202处磷酸化ELAVL1,增强ELAVL1的核定位[49]。CDK5与神经胶质瘤细胞中心体的ELAVL1相互作用,使ELAVL1在S202处磷酸化,降低了ELAVL1与靶mRNA的结合能力[50]。在细胞应激情况下,检查点激酶2(checkpoint kinase 2,CHK2)在S88、S100和苏氨酸(threonine,T)118处磷酸化ELAVL1,并降低其与靶mRNA的结合能力[51]。p38-MAPK在T118磷酸化ELAVL1,增强了ELAVL1的细胞质定位,提高与靶mRNA的结合[52]。PKCα在S158和S221处磷酸化ELAVL1,导致ELAVL1由细胞核转运到细胞质,但PKCδ在S318磷酸化ELAVL1却不影响其核质转运,而是增强ELAVL1与COX-2 mRNA的结合[53-54]。
3 ELAVL1调控病毒复制近年来研究表明,ELAVL1可参与多种病毒的复制,其对于病毒的调控机制的研究主要在于其控制靶mRNA的稳定性、亚细胞定位及RNA干扰方面。然而更具体的调控机制在很大程度上还未被探索。
3.1 ELAVL1调控腺病毒的复制腺病毒(Adenovirus,Ad)是一种无包膜的双链DNA病毒,腺病毒载体具有很强的免疫原性,能够激发哺乳动物宿主细胞的先天免疫和特异性免疫,是将外源基因送入宿主细胞的最有效载体之一[53-56]。有研究表明,腺病毒感染诱导ELAVL1向宿主细胞的细胞质重新定位,并提高了ARE IVa2 mRNA的稳定性,通过敲除ELAVL,抑制了IVa2 mRNA的稳定可以减少腺病毒的产生,说明ELAVL1可以促进腺病毒的复制[57]。另有研究发现E4ORF6缺失的腺病毒DL355具有溶瘤活性,而ELAVL1核质转运促进了腺病毒复制,并且ELAVL1的核质转运可由许多刺激因素介导。因此,Ahmed等[58]用乙醇处理DL355并作用于癌细胞,发现与未处理相比,促进了ELAVL1的细胞质含量,提高了靶mRNA的稳定性,也增加了溶瘤活性。顺铂(Cis-diamminedichloroplatinum,CDDP)是治疗人类恶性肿瘤常用的抗癌药物之一。Habiba等[59]将DL355和CDDP联合用于肿瘤细胞并进行了体内试验,发现二者联用能诱导ELAVL1向细胞质转运,使ARE mRNA稳定,增强溶瘤腺病毒DL355的复制,从而发挥协同抗肿瘤作用。以上研究表明,通过一定方式促进ELAVL1向细胞质转运可以促进腺病毒的复制(表 1)。
|
|
表 1 ELAVL1对病毒复制的影响 Table 1 Effect of ELAVL1 on viral replication |
肝炎病毒有甲、乙、丙、丁、戊、庚六型[72]。其中,乙型肝炎病毒(hepatitis B virus, HBV)是双链DNA病毒,属于嗜肝DNA病毒科;丙型肝炎病毒(hepatitis C virus,HCV)是正义单链RNA病毒,属于黄病毒科;丁型肝炎病毒(hepatitis D virus,HDV)是负义单链RNA病毒,为缺陷型病毒[73]。Luo[74]首次发现ELAVL1与HCV 3′UTR中的富U区域相互作用,Spångberg等[28]发现3′UTR主要与病毒的有义链结合,5′UTR主要与病毒的反义链结合。随后Korf等[61]发现,敲除ELAVL1可抑制HCV复制,蛋白酶体亚基α7(proteasome α-subunit 7,PSMA7)可调节HCV内部核糖体进入位点(internal ribosomal entry site,IRES)的活性,IRES能招募核糖体对mRNA进行翻译,而敲除ELAVL1与敲除PSMA 7联合使用时可增强抑制HCV复制的效果。Rivas-Aravena等[64]发现,转染ELAVL1 RNA或质粒增加了HCV IRES的翻译,通过内源性途径敲除ELAVL1导致HCV IRES蛋白降低,从而调节病毒复制周期。La自身抗原(La autoantigen,La)已被证明通过与HCV结合来调节HCV的翻译。Shwetha等[62]发现,ELAVL1在HCV感染宿主后从细胞核重定位到细胞质,与NS5B相互作用,并将ELAVL1募集到病毒RNA合成的区域,促进病毒复制。此外,ELAVL1与LA相互作用,促进LA与3′UTR结合,增强LA介导的HCV基因组循环并与聚嘧啶区结合蛋白(polypyrimidine tract binding protein,PTB)(PTB是病毒复制的负调节因子)竞争结合3′UTR,从而促进病毒复制。之后更深入的研究表明,ELAVL1通过稳定和增加miR-122水平促进HCV增殖,抗癌药物Rigosertib具有miR-122调节能力,通过靶向polo样激酶1(polo-like kinase 1,PLK1)并随后调节ELAVL1/miR-122信号传导发挥抗HCV活性[60]。ELAVL1还能影响HBV mRNA稳定性,敲除ELAVL1可抑制HBV复制,而且还能抑制肝癌细胞生长[63]。另外,HBV编码的X蛋白(X protein,HBX)在HBV诱导的肝细胞癌中起着重要作用。HBX通过上调ELAVL1增强HER2 mRNA的稳定性来增强HER2蛋白的表达,从而增强肝癌细胞的迁移能力[75]。HDV是迄今为止发现的最小的人类病原体,它是一种卫星病毒,没有独立的生命周期,只能依靠HBV的功能进行复制和表达[73]。ELAVL1与HDV的小大δ抗原(small and large delta antigens,S-HDAg和L-HDAg)于体内外相互作用,在病毒复制和生命周期中发挥关键作用[76]。以上研究表明,ELAVL1可参与调控肝炎病毒的复制,通过敲除/低ELAVL1的表达,能达到抑制病毒复制的效果。
3.3 ELAVL1调控辛德比斯病毒的复制辛德比斯病毒(Sindbis virus,SinV)是甲病毒属的模式病毒,该病毒的复制只发生在细胞质中。研究发现,SinV以及甲病毒属的西方马脑炎病毒、罗斯河病毒和基孔肯亚病毒在感染细胞时ELAVL1都发生了重定位,并且其机制与细胞应激时不同(应激时主要为Caspase介导的蛋白裂解)[67]。敲除ELAVL导致SinV RNA的衰变率显著增加,并抑制了病毒复制[66]。研究还发现,设计SinV突变体,停止了绝大多数病毒基因的表达和复制,随后感染细胞,发现大多数ELAVL位于细胞核中,说明ELAVL1的重定位需要SinV的表达和复制。此外,在SinV感染时,ELAVL1发生了去磷酸化,也影响了细胞核内前体mRNA的选择性多聚腺苷酸化和剪接,这一过程与ELAVL被留在细胞质有关[40]。以上研究表明,ELAVL1通过核质转运可调控SinV的复制,且ELAVL1的翻译后修饰调控其核质转运。之前的研究发现,细胞应激可以通过改变ELAVL1残基(如Ser-88、Ser-100、Thr 118、Ser-158、Ser-202、Ser-221、Ser-242、Ser-318)的磷酸化而影响其核质穿梭的关键因素[1]。然而在SinV感染后,ELAVL1蛋白发生的修饰不同于在细胞应激反应下发生的变化,而是发生了去磷酸化。因此,还需明确SinV引起的ELAVL1核质转运的翻译后修饰方式及其调控机制。
3.4 ELAVL1调控其它病毒的复制研究发现ELAVL1直接与人类免疫缺陷病毒1型(human immunodeficiency virus type 1,HIV-1)逆转录酶(reverse transcriptase,RT)的核糖核酸酶H(Ribonuclease H,RNase H)区域相互作用影响HIV-1的复制[65]。随后有研究人员重复试验,发现ELAVL不是直接通过与RT复合物的相互作用来干扰HIV-1的复制,而是可能通过不明宿主因子或RNA介导的间接作用来干扰HIV-1的复制[77]。ELAVL1是茎-环II(stem loop II,SL-II)控制病毒翻译的关键IRES相关反式作用因子(IRES-associated trans-acting factors,ITAFs),有研究表明,ELAVL1与肠道病毒71型(Enterovirus 71, EV71)5′UTR的SL-II结合,促进IRES依赖的复制和翻译。病毒衍生的小RNA(virus-derived small RNA 1,VSRNA1)可增强ELAVL1与SL-II的结合,从而更加促进EV71的复制和翻译。敲除ELAVL1能降低EV71滴度约1 000倍,当与敲除Argonaute 2联合使用时,病毒滴度降低了近100万倍[69, 78]。而在柯萨奇B3病毒(coxsackie virus B3, CVB3)中,发现ELAVL1不直接影响翻译并且不直接与IRES相互作用,而是通过置换病毒复制的正调节因子细胞多聚(rC)结合蛋白2(poly(rC)binding protein 2,PCBP-2)来抑制病毒复制[70]。此外,ELAVL1在寨卡病毒(Zika virus,ZIKV)上的作用不似其它病毒,敲除ELAVL1增加了ZIKV的蛋白和RNA水平以及病毒滴度,促进了病毒复制,同时ELAVL1在ZIKV感染时也发生了核质转运[10]。这表明同一宿主蛋白在不同病毒感染中的功能具有多样性,也表明ELAVL1在不同病毒感染中对病毒复制的影响是有差异的。
4 展望ELAVL1以往的研究多集中于其在肿瘤方面的作用,它在几乎所有类型癌症中均呈异常高表达,可以影响肿瘤细胞的增殖、侵袭与迁移,是治疗各种癌症的潜在治疗靶标,并参与调节编码关键过程(如器官发育和组织稳态)的蛋白质和mRNA的水平,这体现了其与整个生物体的相关性[23, 79-81]。近年来,ELAVL1在病毒方面的研究逐渐增加,发现其在黄病毒科、腺病毒科、嗜肝DNA病毒科、疱疹病毒科、逆转录病毒科、正黏病毒科、小RNA病毒科、乳头瘤病毒科、披膜病毒科等均发挥一定的作用,并且主要表现在病毒复制方面。虽然ELAVL1的表达对于病毒复制有着十分重要的作用,但其具体机制的相关研究还不够深入。确定ELAVL1在病毒感染中发挥的作用及其具体机制,将为病毒-宿主相互作用提供新的见解[84]。ELAVL1的核质转运及其定位对病毒感染有一定影响,深入研究ELAVL1核质转运的调节机制,进行定向突变,改变其定位,可为病毒相关研究提供更多的可能。目前已有ELAVL1的小分子抑制剂MS-444、KH3和CMLD-2等,并且已在多种体外和体内模型中使用,相信未来能为病毒的防治提供更加合理、高效的方案[15, 79, 83]。
目前,ELAVL1在感染畜禽的动物病毒上的功能和作用机制方面还鲜有研究。Du等[84]发现,感染猪繁殖与呼吸综合征病毒(porcine reproductive and respiratory syndrome virus,PRRSV)会激活MEK1-ERK1/2-C/EBPβ信号通路,从而上调小胶质细胞中COX-2和前列腺素E2(prostaglandin E2,PGE2)的产生。Sajiki等[85]发现,在牛感染牛白血病病毒(bovine leukemia virus,BLV)期间,COX-2的表达升高,血浆中PGE2的浓度也出现升高。这表明COX-2在某些畜禽病毒感染中也发挥一定的作用。家禽养殖中,最常见的致肿瘤性病毒有3种,即马立克病病毒(Marek’s disease virus,MDV)、禽白血病病毒(Avian Leukosis virus,ALV)和禽网状内皮组织增殖病毒(reticuloendotheliosis virus,REV),导致家禽的生产性能下降,甚至死亡,给养殖户造成较大的经济损失[86-87]。Kamble等[88]发现,MDV强毒株激活COX-2/PGE2途径,其以EP2和EP4(PGE2的受体)依赖性方式调节T细胞增殖,口服COX-2和PGE2抑制剂可恢复MDV感染鸡的T细胞增殖,表明COX-2和PGE2的表达在MDV感染致肿瘤发生中具有重要的作用。目前,已有多项研究表明ELAVL1和COX-2的表达有一定的相关性,二者在肿瘤中都呈高表达[89-91]。此外,ELAVL1还能稳定COX-2 mRNA的稳定性[92]。因此推测,ELAVL1在畜禽病毒感染中具有重要的调控作用。ELAVL1能否影响家禽致肿瘤性病毒的复制及其致肿瘤的发生是值得探讨的。
| [1] |
GRAMMATIKAKIS I, ABDELMOHSEN K, GOROSPE M. Posttranslational control of HuR function[J]. WIREs RNA, 2017, 8(1): e1372. DOI:10.1002/wrna.1372 |
| [2] |
WANG H, DING N N, GUO J, et al. Dysregulation of TTP and HuR plays an important role in cancers[J]. Tumor Biol, 2016, 37(11): 14451-14461. DOI:10.1007/s13277-016-5397-z |
| [3] |
SCHULTZ C W, PREET R, DHIR T, et al. Understanding and targeting the disease-related RNA binding protein human antigen R (HuR)[J]. WIRev RNA, 2020, 11(3): e1581. DOI:10.1002/wrna.1581 |
| [4] |
GOUTAS D, PERGARIS A, GIAGINIS C, et al. HuR as therapeutic target in cancer: what the future holds[J]. Curr Med Chem, 2022, 29(1): 56-65. DOI:10.2174/0929867328666210628143430 |
| [5] |
WANG J, GUO Y, CHU H L, et al. Multiple functions of the RNA-binding protein HuR in cancer progression, treatment responses and prognosis[J]. Int J Mol Sci, 2013, 14(5): 10015-10041. DOI:10.3390/ijms140510015 |
| [6] |
KEENE J D. RNA regulons: coordination of post-transcriptional events[J]. Nat Rev Genet, 2007, 8(7): 533-543. DOI:10.1038/nrg2111 |
| [7] |
SIDALI A, TEOTIA V, SOLAIMAN N S, et al. AU-rich element RNA binding proteins: at the crossroads of post-transcriptional regulation and genome integrity[J]. Int J Mol Sci, 2022, 23(1): 96. |
| [8] |
SRIKANTAN S, GOROSPE M. HuR function in disease[J]. Front Biosci (Landmark Ed), 2012, 17(1): 189-205. DOI:10.2741/3921 |
| [9] |
曹楠婧, 王九菊. RNA结合蛋白HuR的研究进展[J]. 生命的化学, 2021, 41(6): 1222-1229. CAO N J, WANG J J. Research progress of RNA binding protein HuR[J]. Chemistry of Life, 2021, 41(6): 1222-1229. (in Chinese) |
| [10] |
BONENFANT G, WILLIAMS N, NETZBAND R, et al. Zika virus subverts stress granules to promote and restrict viral gene expression[J]. J Virol, 2019, 93(12): e00520-19. |
| [11] |
冯晓辉, 汤承, 朱鑫, 等. 核不均一核糖核蛋白与病毒复制[J]. 畜牧兽医学报, 2015, 46(6): 882-888. FENG X H, TANG C, ZHU X, et al. Heterogeneous nuclear ribonucleoprotein and virus replication[J]. Acta Veterinaria et Zootechnica Sinica, 2015, 46(6): 882-888. (in Chinese) |
| [12] |
孙严金, 薛亚男, 仲涛, 等. HuR的功能及其对肌肉生长发育的调控作用[J]. 畜牧兽医学报, 2022, 53(5): 1345-1353. SUN Y J, XUE Y N, ZHONG T, et al. The function of HuR and its regulation on muscle growth and development[J]. Acta Veterinaria et Zootechnica Sinica, 2022, 53(5): 1345-1353. (in Chinese) |
| [13] |
MA W J, CHENG S, CAMPBELL C, et al. Cloning and characterization of HuR, a ubiquitously expressed Elav-like protein[J]. J Biol Chem, 1996, 271(14): 8144-8151. DOI:10.1074/jbc.271.14.8144 |
| [14] |
SUBRAMANIAN P, GARGANI S, PALLADINI A, et al. The RNA binding protein human antigen R is a gatekeeper of liver homeostasis[J]. Hepatology, 2022, 75(4): 881-897. DOI:10.1002/hep.32153 |
| [15] |
PAPATHEOFANI V, LEVIDOU G, SARANTIS P, et al. HuR protein in hepatocellular carcinoma: implications in development, prognosis and treatment[J]. Biomedicines, 2021, 9(2): 119. DOI:10.3390/biomedicines9020119 |
| [16] |
LACHIONDO-ORTEGA S, DELGADO T C, BAÑOS-JAIME B, et al. Hu antigen R (HuR) protein structure, function and regulation in hepatobiliary tumors[J]. Cancers (Basel), 2022, 14(11): 2666. DOI:10.3390/cancers14112666 |
| [17] |
YOO J, KANG J, LEE H N, et al. Kaposin-B enhances the PROX1 mRNA stability during lymphatic reprogramming of vascular endothelial cells by Kaposi's sarcoma herpes virus[J]. PLoS Pathog, 2010, 6(8): e1001046. DOI:10.1371/journal.ppat.1001046 |
| [18] |
SONG X Q, SHI X, LI W J, et al. The RNA-binding protein HuR in digestive system tumors[J]. Biomed Res Int, 2020, 2020: 9656051. |
| [19] |
LEVIDOU G, KOTTA-LOIZOU I, TASOULAS J, et al. Clinical significance and biological role of HuR in head and neck carcinomas[J]. Dis Markers, 2018, 2018: 4020937. |
| [20] |
GOMEZ-SANTOS L, VAZQUEZ-CHANTADA M, MATO J M, et al. SAMe and HuR in liver physiology: usefulness of stem cells in hepatic differentiation research[J]. Methods Mol Biol, 2012, 826: 133-149. |
| [21] |
ATASOY U, WATSON J, PATEL D, et al. ELAV protein HuA (HuR) can redistribute between nucleus and cytoplasm and is upregulated during serum stimulation and T cell activation[J]. J Cell Sci, 1998, 111(21): 3145-3156. DOI:10.1242/jcs.111.21.3145 |
| [22] |
KIM H H, GOROSPE M. Phosphorylated HuR shuttles in cycles[J]. Cell Cycle, 2008, 7(20): 3124-3126. DOI:10.4161/cc.7.20.6884 |
| [23] |
陈禹鑫, 殷文婕, 蒋壮, 等. RNA结合蛋白ELAVL1与肿瘤进展的关系研究[J]. 中国现代医学杂志, 2021, 31(13): 65-70. CHEN Y X, YIN W J, JIANG Z, et al. Review on relationship between RNA-binding protein ELAVL1 and tumor progression[J]. China Journal of Modern Medicine, 2021, 31(13): 65-70. (in Chinese) |
| [24] |
DOLLER A, PFEILSCHIFTER J, EBERHARDT W. Signalling pathways regulating nucleo-cytoplasmic shuttling of the mRNA-binding protein HuR[J]. Cell Signal, 2008, 20(12): 2165-2173. DOI:10.1016/j.cellsig.2008.05.007 |
| [25] |
SMITH M R, COSTA G. RNA-binding proteins and translation control in angiogenesis[J]. FEBS J, 2022, 289(24): 7788-7809. DOI:10.1111/febs.16286 |
| [26] |
SRIKANTAN S, TOMINAGA K, GOROSPE M. Functional interplay between RNA-binding protein HuR and microRNAs[J]. Curr Protein Pept Sci, 2012, 13(4): 372-379. DOI:10.2174/138920312801619394 |
| [27] |
FEIGERLOVÀ E, BATTAGLIA-HSU S F. Role of post-transcriptional regulation of mRNA stability in renal pathophysiology: focus on chronic kidney disease[J]. FASEB J, 2017, 31(2): 457-468. DOI:10.1096/fj.201601087RR |
| [28] |
SPÅNGBERG K, WIKLUND L, SCHWARTZ S. HuR, a protein implicated in oncogene and growth factor mRNA decay, binds to the 3' ends of hepatitis C virus RNA of both polarities[J]. Virology, 2000, 274(2): 378-390. DOI:10.1006/viro.2000.0461 |
| [29] |
何苗. 基于CRISPR-dCas9/Cas13a的转录及转录后调控体系构建与应用[D]. 合肥: 中国科学技术大学, 2022. HE M. The construction of CRISPR-dCas9/Cas13a-Based systems for transcriptional and post-Transcriptional regulation and its application[D]. Hefei: University of Science and Technology of China, 2022. (in Chinese). |
| [30] |
BARBISAN F, MAZZUCCHELLI R, SANTINELLI A, et al. Overexpression of ELAV-like protein HuR is associated with increased COX-2 expression in atrophy, high-grade prostatic intraepithelial neoplasia, and incidental prostate cancer in cystoprostatectomies[J]. Eur Urol, 2009, 56(1): 105-112. DOI:10.1016/j.eururo.2008.04.043 |
| [31] |
KAKUGUCHI W, KITAMURA T, KUROSHIMA T, et al. HuR knockdown changes the oncogenic potential of oral cancer cells[J]. Mol Cancer Res, 2010, 8(4): 520-528. DOI:10.1158/1541-7786.MCR-09-0367 |
| [32] |
ZOU T T, MAZAN-MAMCZARZ K, RAO J N, et al. Polyamine depletion increases cytoplasmic levels of RNA-binding protein HuR leading to stabilization of nucleophosmin and p53 mRNAs[J]. J Biol Chem, 2006, 281(28): 19387-19394. DOI:10.1074/jbc.M602344200 |
| [33] |
KRISHNAMURTHY P, RAJASINGH J, LAMBERS E, et al. IL-10 inhibits inflammation and attenuates left ventricular remodeling after myocardial infarction via activation of STAT3 and suppression of HuR[J]. Circ Res, 2009, 104(2): e9-e18. |
| [34] |
TSCHERNATSCH M M O, MLECNIK B, TRAJANOSKI Z, et al. LPL-mediated lipolysis of VLDL induces an upregulation of AU-rich mRNAs and an activation of HuR in endothelial cells[J]. Atherosclerosis, 2006, 189(2): 310-317. DOI:10.1016/j.atherosclerosis.2006.01.007 |
| [35] |
杨阳. Zfp217参与转录调控和转录后m6A修饰调控脂肪细胞生成的机制[D]. 武汉: 华中农业大学, 2019. YANG Y. Mechanism of Zfp217 regulating adipogenesis through transcriptional regulation and m6A posttranscriptional modification[D]. Wuhan: Huazhong Agricultural University, 2019. (in Chinese) |
| [36] |
JEYARAJ S C, SINGH M, AYUPOVA D A, et al. Transcriptional control of human antigen R by bone morphogenetic protein[J]. J Biol Chem, 2010, 285(7): 4432-4440. DOI:10.1074/jbc.M109.062216 |
| [37] |
KANG M J, RYU B K, LEE M G, et al. NF-κB activates transcription of the RNA-binding factor HuR, via PI3K-AKT signaling, to promote gastric tumorigenesis[J]. Gastroenterology, 2008, 135(6): 2030-2042.e3. DOI:10.1053/j.gastro.2008.08.009 |
| [38] |
GOVINDARAJU S, LEE B S. Krüppel -like factor 8 is a stress-responsive transcription factor that regulates expression of HuR[J]. Cell Physiol Biochem, 2014, 34(2): 519-532. DOI:10.1159/000363019 |
| [39] |
XIAO L, LI X X, CHUNG H K, et al. RNA-binding protein HuR regulates paneth cell function by altering membrane localization of TLR2 via post-transcriptional control of CNPY3[J]. Gastroenterology, 2019, 157(3): 731-743. DOI:10.1053/j.gastro.2019.05.010 |
| [40] |
BARNHART M D, MOON S L, EMCH A W, et al. Changes in cellular mRNA stability, splicing, and polyadenylation through HuR protein sequestration by a cytoplasmic RNA virus[J]. Cell Rep, 2013, 5(4): 909-917. DOI:10.1016/j.celrep.2013.10.012 |
| [41] |
杨旭, 訾晶晶, 郭宏, 等. LncRNA参与转录后调控作用机制研究进展[J]. 天津农学院学报, 2022, 29(2): 85-87, 91. YANG X, ZI J J, GUO H, et al. Research progress on the mechanism of lncRNA involved in post-transcriptional regulation[J]. Journal of Tianjin Agricultural University, 2022, 29(2): 85-87, 91. (in Chinese) |
| [42] |
CHANG S H, HLA T. Post-transcriptional gene regulation by HuR and microRNAs in angiogenesis[J]. Curr Opin Hematol, 2014, 21(3): 235-240. DOI:10.1097/MOH.0000000000000040 |
| [43] |
LI Y B, YU J H, DU D H, et al. Involvement of post-transcriptional regulation of FOXO1 by HuR in 5-FU-induced apoptosis in breast cancer cells[J]. Oncol Lett, 2013, 6(1): 156-160. DOI:10.3892/ol.2013.1352 |
| [44] |
PHILLIPS B L, BANERJEE A, SANCHEZ B J, et al. Post-transcriptional regulation of Pabpn1 by the RNA binding protein HuR[J]. Nucleic Acids Res, 2018, 46(15): 7643-7661. DOI:10.1093/nar/gky535 |
| [45] |
GUO J, LEI M, CHENG F, et al. RNA-binding proteins tristetraprolin and human antigen R are novel modulators of podocyte injury in diabetic kidney disease[J]. Cell Death Dis, 2020, 11(6): 413. DOI:10.1038/s41419-020-2630-x |
| [46] |
刘兰香. 基于蛋白翻译后修饰和转录组改变的肠道微生物调节海马功能机制研究[D]. 重庆: 重庆医科大学, 2020. LIU L X. Study on the mechanism of intestinal microbes regulating hippocampal function based on protein post-translational modifications and transcriptome changes[D]. Chongqing: Chongqing Medical University, 2020. (in Chinese) |
| [47] |
LIU J, QIAN C, CAO X T. Post-translational modification control of innate immunity[J]. Immunity, 2016, 45(1): 15-30. DOI:10.1016/j.immuni.2016.06.020 |
| [48] |
刘静, 李亚超, 周梦岩, 等. 植物蛋白质翻译后修饰组学研究进展[J]. 生物技术通报, 2021, 37(1): 67-76. LIU J, LI Y C, ZHOU M Y, et al. Advances in the studies of plant protein post-translational modification[J]. Biotechnology Bulletin, 2021, 37(1): 67-76. (in Chinese) |
| [49] |
KIM H H, ABDELMOHSEN K, LAL A, et al. Nuclear HuR accumulation through phosphorylation by Cdk1[J]. Genes Dev, 2008, 22(13): 1804-1815. DOI:10.1101/gad.1645808 |
| [50] |
FILIPPOVA N, YANG X H, KING P, et al. Phosphoregulation of the RNA-binding protein Hu antigen R (HuR) by Cdk5 affects centrosome function[J]. J Biol Chem, 2012, 287(38): 32277-32287. DOI:10.1074/jbc.M112.353912 |
| [51] |
ABDELMOHSEN K, PULLMANN R Jr, LAL A, et al. Phosphorylation of HuR by Chk2 regulates SIRT1 expression[J]. Mol Cell, 2007, 25(4): 543-557. DOI:10.1016/j.molcel.2007.01.011 |
| [52] |
LAFARGA V, CUADRADO A, LOPEZ DE SILANES I, et al. p38 Mitogen-activated protein kinase- and HuR-dependent stabilization of p21Cip1 mRNA mediates the G1/S checkpoint[J]. Mol Cell Biol, 2009, 29(16): 4341-4351. DOI:10.1128/MCB.00210-09 |
| [53] |
DOLLER A, HUWILER A, MVLLER R, et al. Protein kinase Cα-dependent phosphorylation of the mRNA-stabilizing factor HuR: implications for posttranscriptional regulation of cyclooxygenase-2[J]. Mol Biol Cell, 2007, 18(6): 2137-2148. DOI:10.1091/mbc.e06-09-0850 |
| [54] |
DOLLER A, SCHLEPCKOW K, SCHWALBE H, et al. Tandem phosphorylation of serines 221 and 318 by protein kinase Cδ coordinates mRNA binding and nucleocytoplasmic shuttling of HuR[J]. Mol Cell Biol, 2010, 30(6): 1397-1410. DOI:10.1128/MCB.01373-09 |
| [55] |
LYNCH J P, FISHBEIN M, ECHAVARRIA M. Adenovirus[J]. Semin Respir Crit Care Med, 2011, 32(4): 494-511. DOI:10.1055/s-0031-1283287 |
| [56] |
郝彦霞. 腺病毒载体特异性单域抗体的筛选、制备及其鉴定[D]. 呼和浩特: 内蒙古农业大学, 2022. HAO Y X. Screening, preparation and identification of Adenoviral vector specific single domain antibodies[D]. Hohhot: Inner Mongolia Agricultural University, 2022. (in Chinese) |
| [57] |
JEHUNG J P, KITAMURA T, YANAGAWA-MATSUDA A, et al. Adenovirus infection induces HuR relocalization to facilitate virus replication[J]. Biochem Biophys Res Commun, 2018, 495(2): 1795-1800. DOI:10.1016/j.bbrc.2017.12.036 |
| [58] |
AHMED I, ALAM M T, YANAGAWA-MATSUDA A, et al. Enhanced oncolytic activity of E4orf6-deficient adenovirus by facilitating nuclear export of HuR[J]. Biochem Biophys Res Commun, 2020, 529(2): 494-499. DOI:10.1016/j.bbrc.2020.04.147 |
| [59] |
HABIBA U, HOSSAIN E, YANAGAWA-MATSUDA A, et al. Cisplatin relocalizes RNA binding protein HuR and enhances the oncolytic activity of E4orf6 deleted adenovirus[J]. Cancers, 2020, 12(4): 809. DOI:10.3390/cancers12040809 |
| [60] |
SEO Y, KANG Y, HAM Y, et al. PLK1-ELAVL1/HuR-miR-122 signaling facilitates hepatitis C virus proliferation[J]. Proc Natl Acad Sci U S A, 2022, 119(51): e2214911119. DOI:10.1073/pnas.2214911119 |
| [61] |
KORF M, JARCZAK D, BEGER C, et al. Inhibition of hepatitis C virus translation and subgenomic replication by siRNAs directed against highly conserved HCV sequence and cellular HCV cofactors[J]. J Hepatol, 2005, 43(2): 225-234. DOI:10.1016/j.jhep.2005.02.046 |
| [62] |
SHWETHA S, KUMAR A, MULLICK R, et al. HuR displaces polypyrimidine tract binding protein to facilitate la binding to the 3' untranslated region and enhances hepatitis C virus replication[J]. J Virol, 2015, 89(22): 11356-11371. DOI:10.1128/JVI.01714-15 |
| [63] |
KANZAKI H, CHIBA T, KANEKO T, et al. The RNA-binding protein ELAVL1 regulates hepatitis B virus replication and growth of hepatocellular carcinoma cells[J]. Int J Mol Sci, 2022, 23(14): 7878. DOI:10.3390/ijms23147878 |
| [64] |
RIVAS-ARAVENA A, RAMDOHR P, VALLEJOS M, et al. The Elav-like protein HuR exerts translational control of viral internal ribosome entry sites[J]. Virology, 2009, 392(2): 178-185. DOI:10.1016/j.virol.2009.06.050 |
| [65] |
LEMAY J, MAIDOU-PEINDARA P, BADER T, et al. HuR interacts with human immunodeficiency virus type 1 reverse transcriptase, and modulates reverse transcription in infected cells[J]. Retrovirology, 2008, 5(1): 47. DOI:10.1186/1742-4690-5-47 |
| [66] |
SOKOLOSKI K J, DICKSON A M, CHASKEY E L, et al. Sindbis virus usurps the cellular HuR protein to stabilize its transcripts and promote productive infections in mammalian and mosquito cells[J]. Cell Host Microbe, 2010, 8(2): 196-207. DOI:10.1016/j.chom.2010.07.003 |
| [67] |
DICKSON A M, ANDERSON J R, BARNHART M D, et al. Dephosphorylation of HuR protein during alphavirus infection is associated with HuR relocalization to the cytoplasm[J]. J Biol Chem, 2012, 287(43): 36229-36238. DOI:10.1074/jbc.M112.371203 |
| [68] |
NILSSON K, ABDURAHMAN S, SCHWARTZ S. Influenza virus natural sequence heterogeneity in segment 8 affects interactions with cellular RNA-binding proteins and splicing efficiency[J]. Virology, 2020, 549: 39-50. DOI:10.1016/j.virol.2020.08.005 |
| [69] |
LI M L, BREWER G. Functional analyses of mammalian virus 5'UTR-derived, small RNAs that regulate virus translation[J]. Methods, 2020, 183: 13-20. DOI:10.1016/j.ymeth.2020.02.008 |
| [70] |
GAO H X, LIN Y X, HUANG C B, et al. A genome-wide CRISPR screen identifies HuR as a regulator of apoptosis induced by dsRNA and virus[J]. J Cell Sci, 2022, 135(6): jcs258855. DOI:10.1242/jcs.258855 |
| [71] |
GEORGE B, DAVE P, RANI P, et al. Cellular protein HuR regulates the switching of genomic RNA templates for differential functions during the coxsackievirus B3 life cycle[J]. J Virol, 2021, 95(21): e0091521. DOI:10.1128/JVI.00915-21 |
| [72] |
农汝, 吴俊仪, 黄宁, 等. 圈养灵长类动物肝炎病毒血清流行病学的调查[J]. 现代畜牧兽医, 2023(2): 65-68. NONG R, WU J Y, HUANG N, et al. Investigation of seroepidemiology of hepatitis virus in captive primates[J]. Modern Journal of Animal Husbandry and Veterinary Medicine, 2023(2): 65-68. (in Chinese) |
| [73] |
LIOU J W, MANI H, YEN J H. Viral hepatitis, cholesterol metabolism, and cholesterol-lowering natural compounds[J]. Int J Mol Sci, 2022, 23(7): 3897. DOI:10.3390/ijms23073897 |
| [74] |
LUO G X. Cellular proteins bind to the poly(U) tract of the 3' untranslated region of hepatitis C virus RNA genome[J]. Virology, 1999, 256(1): 105-118. DOI:10.1006/viro.1999.9639 |
| [75] |
HUNG C M, HUANG W C, PAN H L, et al. Hepatitis B virus X upregulates HuR protein level to stabilize HER2 expression in hepatocellular carcinoma cells[J]. Biomed Res Int, 2014, 2014: 827415. |
| [76] |
CASACA A, FARDILHA M, DA CRUZ E SILVA E, et al. In vivo interaction of the hepatitis delta virus small antigen with the ELAV-like protein HuR[J]. Open Virol J, 2011, 5: 12-21. DOI:10.2174/1874357901105010012 |
| [77] |
AHN J, BYEON I J L, DHARMASENA S, et al. The RNA binding protein HuR does not interact directly with HIV-1 reverse transcriptase and does not affect reverse transcription in vitro[J]. Retrovirology, 2010, 7: 40. DOI:10.1186/1742-4690-7-40 |
| [78] |
LIN J Y, BREWER G, LI M L. HuR and Ago2 bind the internal ribosome entry site of enterovirus 71 and promote virus translation and replication[J]. PLoS One, 2015, 10(10): e0140291. DOI:10.1371/journal.pone.0140291 |
| [79] |
ASSONI G, LA PIETRA V, DIGILIO R, et al. HuR-targeted agents: an insight into medicinal chemistry, biophysical, computational studies and pharmacological effects on cancer models[J]. Adv Drug Deliv Rev, 2022, 181: 114088. DOI:10.1016/j.addr.2021.114088 |
| [80] |
EBERHARDT W, BADAWI A, BIYANEE A, et al. Cytoskeleton-dependent transport as a potential target for interfering with post-transcriptional HuR mRNA regulons[J]. Front Pharmacol, 2016, 7: 251. |
| [81] |
RAGURAMAN R, SHANMUGARAMA S, MEHTA M, et al. Drug delivery approaches for HuR-targeted therapy for lung cancer[J]. Adv Drug Deliv Rev, 2022, 180: 114068. DOI:10.1016/j.addr.2021.114068 |
| [82] |
KOTTA-LOIZOU I, VASILOPOULOS S N, COUTTS R H A, et al. Current evidence and future perspectives on HuR and breast cancer development, prognosis, and treatment[J]. Neoplasia, 2016, 18(11): 674-688. DOI:10.1016/j.neo.2016.09.002 |
| [83] |
LIU Y B, LI X Z, ZHANG H, et al. HuR up-regulates cell surface PD-L1 via stabilizing CMTM6 transcript in cancer[J]. Oncogene, 2021, 40(12): 2230-2242. DOI:10.1038/s41388-021-01689-6 |
| [84] |
DU L, WANG H L, LIU F, et al. NSP2 is important for highly pathogenic porcine reproductive and respiratory syndrome virus to trigger high fever-related COX-2-PGE2 pathway in pigs[J]. Front Immunol, 2021, 12: 657071. DOI:10.3389/fimmu.2021.657071 |
| [85] |
SAJIKI Y, KONNAI S, OKAGAWA T, et al. Prostaglandin E2-induced immune exhaustion and enhancement of antiviral effects by anti-PD-L1 antibody combined with COX-2 inhibitor in bovine leukemia virus infection[J]. J Immunol, 2019, 203(5): 1313-1324. DOI:10.4049/jimmunol.1900342 |
| [86] |
崔燕, 吕茜, 史艺. 禽病毒性肿瘤病的诊断与防控[J]. 湖北畜牧兽医, 2013, 34(11): 19-20. CUI Y, LV X, SHI Y. Diagnosis and control of avian viral neoplastic diseases[J]. Hubei Journal of Animal and Veterinary Sciences, 2013, 34(11): 19-20. (in Chinese) |
| [87] |
SHI M Y, LI M, WANG W W, et al. The emergence of a vv+MDV can break through the protections provided by the current vaccines[J]. Viruses, 2020, 12(9): 1048. DOI:10.3390/v12091048 |
| [88] |
KAMBLE N, GURUNG A, KAUFER B B, P, et al. Marek's disease virus modulates T cell proliferation via activation of cyclooxygenase 2-dependent prostaglandin E2[J]. Front Immunol, 2021, 12: 801781. DOI:10.3389/fimmu.2021.801781 |
| [89] |
王梦雅. HuR、COX-2和VEGF-C在非小细胞肺癌中的表达及预后作用[D]. 济宁: 济宁医学院, 2022. WANG M Y. The expression and prognosis of HuR, COX-2 and VEGF-C in non-small cell lung cancer[D]. Jining: Jining Medical University, 2022. (in Chinese) |
| [90] |
MITSUNARI K, MIYATA Y, ASAI A, et al. Human antigen R is positively associated with malignant aggressiveness via upregulation of cell proliferation, migration, and vascular endothelial growth factors and cyclooxygenase-2 in prostate cancer[J]. Transl Res, 2016, 175: 116-128. DOI:10.1016/j.trsl.2016.04.002 |
| [91] |
LEIJON H, SALMENKIVI K, HEISKANEN I, et al. HuR in pheochromocytomas and paragangliomas-overexpression in verified malignant tumors[J]. Apmis, 2016, 124(9): 757-763. DOI:10.1111/apm.12571 |
| [92] |
ZHANG J, BOWDEN G T. UVB irradiation regulates Cox-2 mRNA stability through AMPK and HuR in human keratinocytes[J]. Mol Carcinog, 2008, 47(12): 974-983. DOI:10.1002/mc.20450 |
(编辑 范子娟)


