反刍动物瘤胃微生物菌群是一个庞大的群体,包括细菌、原虫、真菌、产甲烷菌和噬菌体,在瘤胃中的浓度分别高达1011、106、106、109和1010个·mL-1瘤胃液[1]。细菌通常以群居的方式存在,无论是在非生物表面还是在瘤胃内,细菌都会在复杂的微生物群落中聚集。在这些动态聚集中,细菌密集堆积、相互依赖以提供碳代谢物保证自身的正常繁殖和生长。为了在复杂多变的环境中生存,细菌会通过群体感应(quorum sensing,QS)感知周围微生物的存在,并与它们密切交流,从而达到协调各种生理活动的目的[2]。
1 LuxS/AI-2介导的群体感应系统QS由三个不同类型的系统组成,这三个系统使用不同类型的自体诱导物(autoinducer molecule,AI),即QS信号分子。第一个系统通过酰基高丝氨酸内酯(acylated homoserine lactones,AHLs)类信号分子感知细胞的密度,一般认为这是一种革兰阴性菌进行物种内部通讯的QS系统[3];第二个系统利用自诱导肽(autoinducing peptides,AIPs)类分子作为AI,其信号识别系统是AIP经过加工修饰后,与跨膜受体结合触发双组分磷酸化信号转导过程,从而响应调控蛋白来诱导相关基因的表达[4];第三个系统由自体诱导物-2(autoinducer-2, AI-2)介导,其产生受到luxS基因编码的影响,在革兰阴性菌和革兰阳性菌中均发现了luxS基因的同源物,因此LuxS/AI-2 QS常被认为是微生物进行种间交流的通讯方式[5]。近些年研究发现,QS普遍存在于瘤胃微生物中,而瘤胃微生物中的丁酸弧菌、普雷沃氏菌、瘤胃球菌和假丁酸弧菌这4个菌属含有的luxS基因数量最多,暗示这些细菌可能具有AI-2信号转导能力;多组学研究发现,瘤胃内普雷沃氏菌的LuxS合成酶表达水平较高[6],表明瘤胃内该属主要是以LuxS/AI-2 QS作为通讯方式。
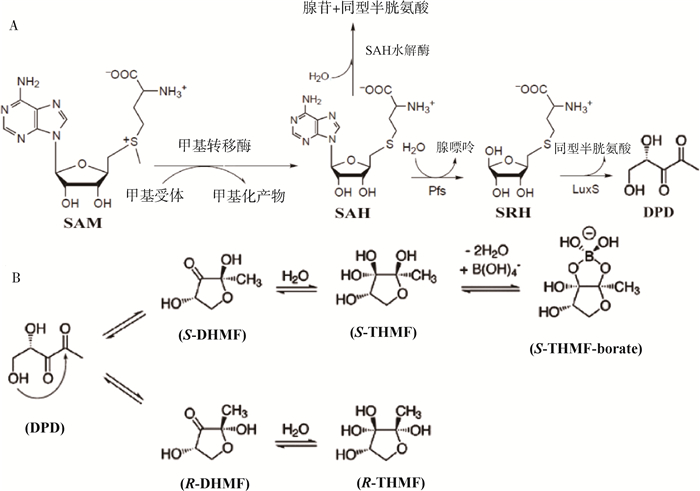
1.1 AI-2的合成AI-2的生物合成途径如图 1A所示。AI-2是由4, 5-二羟基-2, 3-戊二酮(DPD)自发环化形成的一种物质,是LuxS催化裂解S-核糖基高半胱氨酸的产物[7]。DPD的合成在所有细菌中非常相似,起始代谢物是S-腺苷蛋氨酸(SAM),它作为甲基转移酶的甲基供体,通过甲基化转化为S-腺苷高半胱氨酸(SAH);SAH被Pfs酶分解为S-核苷酸同型半胱氨酸(SRH)和腺嘌呤,SRH最终作为LuxS酶的底物,用于合成DPD和副产品同型半胱氨酸[7]。DPD自发的环化作用导致了两个嵌合型呋喃酮的形成,分别是(2S, 4S)-2, 4-二羟基-2-甲基-二氢呋喃-3-酮(S-DHMF)和(2R, 4S)-2, 4-二羟基-2-甲基-二氢呋喃-3-酮(R-DHMF)(图 1B);S-DHMF和R-DHMF经水合作用得到相应的(2S, 4S)-和(2R, 4S)-2-甲基-2, 3, 3, 4-四羟基-四氢呋喃(分别为S-THMF和R-THMF);随着B(OH)4-的加入,S-THMF会自发形成S-THMF硼酸盐[7]。DPD的所有构象都可以相互转化,并且处于平衡状态,这也导致了AI-2的普遍性[7]。AI-2的生物活性形式包括R-THMF(存在于大肠杆菌或伤寒沙门菌等肠杆菌中)和S-THMF硼酸盐(存在于哈维弧菌等海洋细菌中)[8]。
|
A. DPD的合成途径; B. 哈维弧菌(上分支)和鼠伤寒沙门菌(下分支)的AI-2信号分子形成途径 A. The synthesis pathway of DPD; B. The formation pathways of AI-2 signaling molecules in Vibrio harveyi (upper branch) and Salmonella typhimurium (lower branch) 图 1 AI-2的生物合成途径[7, 9](核心过程已翻译成中文) Fig. 1 Biosynthesis pathway of AI-2[7, 9](Core process has been translated into Chinese) |
用于种间交流的AI-2信号分子在细菌中被细菌周质结合蛋白(bacterial periplasmic binding proteins, bPBPs)结合,导致构象发生变化,其中AI-2受体蛋白LuxP和LsrB分别与S-THMF硼酸盐和R-THMF结合[10]。
哈维弧菌的受体蛋白LuxP是第一个确定晶体结构的蛋白[11-12]。在对鼠伤寒沙门菌LsrB的结构进行研究后,发现这两种受体只能结合它们自己物种特有的AI-2,且LsrB中的呋喃糖环与LuxP中的硼酸盐环和大肠杆菌核糖结合蛋白(ribose binding protein, RBP)中的核糖环占据相同的位置[7, 12]。只有进行大的结构重新排列,LsrB结合位点才有足够的空间来容纳S-THMF硼酸盐[8]。此外,AI-2的结合会改变蛋白的构象,通过旋转结构域从具有开放结构的ApoLuxP和ApoLsrB转变为封闭结构的HoloLuxP和HoloLsrB[11]。
在哈维弧菌的LuxP中,有9个氨基酸参与AI-2的结合;在鼠伤寒沙门菌中,有6个氨基酸参与AI-2的结合,且每种自体诱导物会结合特定的受体蛋白[7, 12]。体内研究表明,S-THMF硼酸盐不能与受体LsrB结合[7]。体外研究发现,只有S-THMF硼酸盐可以与LuxP结合[13]。此外,Laganenka等[14]也鉴定了LsrB为鼠伤寒沙门菌中的AI-2信号分子受体。
1.3 AI-2的信号转导在海洋细菌哈维弧菌中,AI-1和AI-2进入同一下游信号通路,对光产生、III型分泌、金属蛋白酶产生、菌落形态和铁载体的产生具有协同作用[15-18]。研究发现,与其他自体诱导物不同,AI-2促进了种间信号的转导。一半以上的革兰阴性菌和革兰阳性菌中含有与luxS同源的基因,luxS是编码哈维弧菌AI-2合成酶的基因[8]。除luxS基因外,这些细菌还含有在AI-2合成过程中直接在luxS基因上游起作用的Pfs酶[19]。对哈维弧菌的检测发现,多数菌种可以释放有活性的AI-2,并且可对AI-2信号分子产生应答[20-22]。AI-2调节的细胞功能和基因非常广泛,例如在人类病原体霍乱弧菌中AI-2调节70多个基因的表达,包括编码霍乱毒素和毒素共调节菌毛以及生物膜形成所需的菌毛[16-17, 23-24]。
1.3.1 S-THMF的AI-2的信号转导 AI-2与受体LuxP结合后会激发哈维弧菌的LuxPQ感应系统(图 2)。Neiditch等[11]发现,无论是否存在AI-2,LuxP和LuxQ都会结合形成复合物,这种复合物不仅具有两种相反酶活性——激酶和磷酸酶,两者之间形成的结构域还可以用来结合不同的配体且两者结合的紧密程度会影响细菌对AI-2的敏感性。AI-2信号分子通过含有保守组氨酸(H)和天冬氨酸(D)残基的双组分蛋白质来介导磷酸转移,从而将信息传递给LuxO[25-26]。这种混合的双组分传感器激酶,由周质传感器域、细胞质组氨酸激酶和反应调节域组成[11]。在低细胞密度下,LuxQ充当激酶,交叉磷酸化组氨酸激酶结构域内的组氨酸残基,经过一系列磷酸转移反应最终导致LuxO的磷酸化,使得磷酸化的LuxO间接抑制转录激活因子LuxR;在高细胞密度下,LuxQ从激酶转化为磷酸酶,与AI-2结合的LuxP相互作用[11]。LuxQ的去磷酸化导致磷酸基团通过AI-2信号转导途径逆行,从而抑制LuxR,LuxR的抑制会激活lux操纵子的转录,编码负责光产生的荧光素酶;因此,AI-2与LuxP的结合通过多条途径发挥作用,以刺激生物发光[11]。

|
虚线箭头表示低细胞密度下磷酰基的流动方向;在高细胞密度下,流动则是反向的 Dotted arrows indicate the phosphoryl group flow at low cell density; the flow is reversed at high cell density 图 2 哈维弧菌的QS信号转导[11] Fig. 2 Vibrio harveyi quorum-sensing signal transduction[11] |
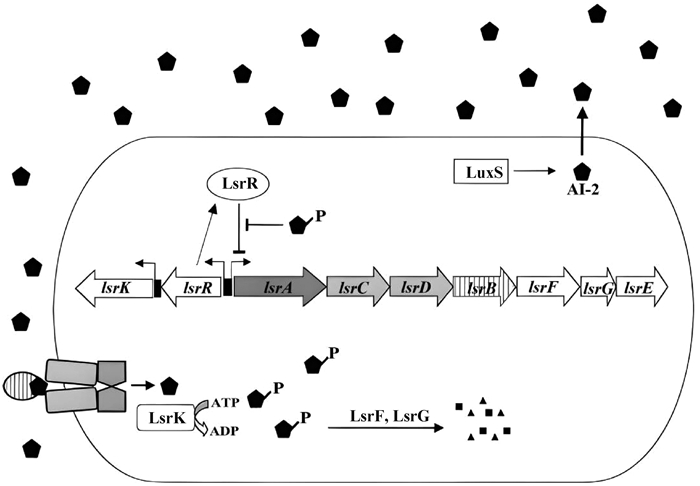
1.3.2 R-THMF的AI-2的信号转导 在鼠伤寒沙门菌中,R-THMF通过LsrATP结合ABC型转运蛋白进入细菌[27]。AI-2诱导lsrACDBFGE操纵子的转录,其中前4个编码Lsr转运体,LsrF和LsrG可能参与AI-2的磷酸化修饰,LsrE的功能尚不清楚[27]。LsrK是一种激酶,可以调节lsr操纵子和AI-2的转运,在进入细胞后磷酸化AI-2,AI-2的磷酸化导致其被隔离在细胞质中,进而诱导LsrR失活(LsrR是lsr操纵子的阻遏物),促进AI-2转运进入细胞(图 3)。
1.4 AI-2的终止在鼠伤寒沙门菌中,细胞外AI-2活性在指数生长后期累积到最大水平,随后AI-2活性从培养基中消失;然而也有证据显示,无细胞培养液中的AI-2可以长时间保持活性,这表明AI-2活性从细胞外环境中消失并不是由AI-2分子结构不稳定引起的[28]。从培养基中消除AI-2可能是细菌自身的行为,并且通过遗传分析已经确定鼠伤寒沙门菌中一组lsr基因和LsrR蛋白,lsr操纵子转录受AI-2信号分子的调控,而AI-2对lsr的调节受LsrR的影响;因此Lsr复合物的功能是将细胞外AI-2转运到细胞质中进行内化,随后内化的AI-2被其他的lsr操纵子编码的酶加工失活[27, 29]。
2 LuxS/AI-2群体感应对微生物的调控作用细菌生物膜是细菌为适应外界环境,通过分泌胞外聚合物(extracellular polymeric substances,EPS),使细菌包裹在EPS中从而形成的大量细菌聚集膜状物,是细菌的一种自我保护性生长方式[17]。微生物通过生物膜附着在物体表面来构建生物膜群落[30-31],这些附着的群落与医学、农业和环境息息相关[31-33]。
2.1 LuxS/AI-2群体感应对病原微生物的调控作用病原微生物群体感应是近年来微生物学领域的研究热点,这些研究主要关注微生物在人类和动植物体内形成QS并产生协同效应的机制及其对宿主的影响[34-35]。目前已发现多种病原微生物的群体感应信号分子和受体,这些信号分子能够影响病原微生物的发育、毒力和耐药性,目前确定的细菌及表型列于表 1。进一步研究群体感应机制可以更好地理解病原微生物与宿主的相互作用,为开发调控微生物菌群的策略提供新思路。
|
|
表 1 不同种类细菌中QS系统受体和表型功能 Table 1 QS system receptors and phenotypic functions in different types of bacteria |
细菌通过AI-2进行种间交流[45],AI-2作为DPD的衍生物,可以协调含有luxS基因的细菌在种间通讯中的行为。有研究发现,LuxS对生物膜形成的影响发生在初始黏附之后,在LuxS控制下的生物膜相关基因参与生物膜的早期形成,从而导致从表面黏附状态转变为新生生物膜状态[46]。Slater等[47]发现,LuxS/AI-2有可能通过诱导细菌差异代谢导致噬菌体介导宿主细胞裂解产生eDNA,eDNA进一步黏附细胞到生物膜内[48-51],进而开始生物膜的形成。Peng等[52]发现,在早期构建枯草芽孢杆菌3D生物膜结构过程中,eDNA与EPS具有协同作用,eDNA通过连接其他细菌的细胞质蛋白来逐渐稳定生物膜结构[53]。在植物乳杆菌中,添加外源AI-2可以增加EPS含量,并且抑制lamC和ftsH基因的表达[54]。在形成初级生物膜后可作为其他微生物的支架,生物膜形成后期EPS逐渐将细菌结合在一起发展为细胞聚集体,最终形成具有高细胞密度的多种类混合生物膜,这种生物膜表型使得单个细胞具有更强的环境耐受性和营养获取能力[52, 55-57]。此外,Sugimoto等[58]研究发现,在生物膜内的细胞外基质中微生物之间存在树枝状纳米管状网络,并且能观察到DNA和蛋白质网络,这表明生物膜中确实存在多细胞之间的通讯。
3 LuxS/AI-2群感应体在瘤胃内的研究进展随着检测技术的升级和普及,瘤胃微生物群体感应研究热度逐步提高,研究深度和广度也在进一步延伸和拓展。目前已证实AI-2介导的QS广泛存在于瘤胃微生物中[59],在多个瘤胃微生物菌属中均检测到luxS基因序列,其中包括许多裂解菌和饲料颗粒上生物膜内的微生物[60]。Xie等[61]通过宏基因组学发现,高剩余采食量(residual feed intake,RFI)的奶牛瘤胃内QS和DNA复制更加活跃,微生物多样性更为丰富,在代谢途径上更加多样且集中于丁酸盐和甲烷的代谢,而低RFI奶牛瘤胃内微生物则集中于丙酸的代谢。此外,Li等[62]的研究表明,丁酸盐和甲烷恰恰是半纤维素和果胶的代谢产物。低RFI奶牛表现为更高的饲料利用率,由此推测,CH4代谢可能是导致饲料利用率降低的主要原因,这使得AI-2介导的QS系统有可能成为调控瘤胃微生物生态和饲料利用率的一种方式[63-64],从而为反刍动物的高效生产提供新思路和新靶点。
3.1 瘤胃微生物在饲料消化过程中的定殖模式瘤胃内纤维降解菌通常附着在摄入的植物颗粒上并与周围环境形成独特的微型生态系统,这种生态系统通过生物膜来降解和发酵纤维,发酵产物为宿主提供能量和营养[65-66]。Minato等[67]报道,附着于饲料颗粒的瘤胃细菌的内切葡聚糖酶占瘤胃总内切葡聚糖酶的80%。上述研究结果表明,附着于植物颗粒的瘤胃细菌在纤维降解中起主要作用。Cheng等[68]通过电子显微镜法观察到瘤胃细菌能迅速附着于植物细胞壁。Mosoni等[69]通过体外培养试验发现,在培养45 min后白色瘤胃球菌和黄色瘤胃球菌对纤维素的附着出现高峰。此外,Koike等[70]在瘤胃纤维降解菌对植物纤维附着的影响试验中发现,在瘤胃内培养5 min后,附着于干草的产琥珀酸丝状杆菌和黄色瘤胃球菌的数量分别达到105和104·g-1干物质(DM),培养10 min后,菌株的数量提高了10倍;此后数量持续上升,产琥珀酸丝状杆菌和黄色瘤胃球菌在培养24 h达到最大值(分别为109和107·g-1 DM),白色瘤胃球菌在培养48 h达到最大值(106·g-1 DM)。以上研究表明,瘤胃纤维降解细菌可迅速附着于进入瘤胃的饲料纤维,并且大部分的附着在10 min内完成。
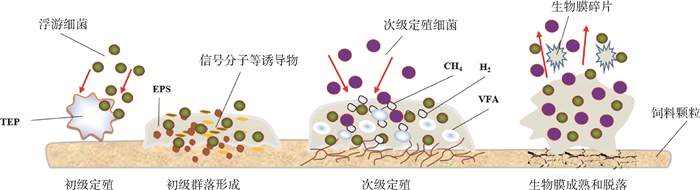
瘤胃微生物可在饲料消化过程中通过形成生物膜实现定殖,瘤胃内生物膜的形成主要有4个阶段(图 4):1)底物与细菌相互接近开始初级定殖。Bar-Zeev等[71]研究表明,大多数水生生态系统中都含有非常“黏”的浮游透明外聚合物颗粒(transparent exopolymer particles,TEP),虽然TEP尚未在瘤胃内发现,但它们很可能以动物反刍过程中从饲料颗粒表面释放的脱落生物膜碎片的形式存在,TEP可在饲料颗粒表面迅速形成生物膜微生物附着点[71-72];2)开始建立生物膜后,初级定殖者开始水解固体饲料中复杂结构的化合物,由此形成初级群落;3)次级定殖者逐渐被初级定殖者的代谢物或者其他细菌产生的信号分子等诱导物吸引[73],并将自己嵌入初始定殖者周围的细胞外聚合物中建立菌落,同化细菌水解的一些中间产物(主要是单糖、肽和氨基酸)来生长;随着生物膜的生长,更多需要特定底物的微生物也会进入生物膜;这些细菌可以利用最初定殖的微生物和植物颗粒内的真菌所产生的代谢产物作为生长所需的营养物质,生物膜随着它们的生长而成熟;通过糖酵解途径逐渐将碳水化合物降解为挥发性脂肪酸(volatile fatty acids,VFA);产生的H2大部分通过产甲烷菌合成CH4[74];4)最终细菌不可逆地附着在表面,形成具有三维结构的多糖体,生物膜完全成熟后就会脱落并在瘤胃内分散[53, 75]。生物膜形成过程中的每个阶段都取决于上一个阶段的完成程度,饲料消化过程中瘤胃内生物膜的形成类似于微生物黏附到固体基质上的一般模式,其可能的形成示意图如图 4。
|
图 4 饲料消化过程中瘤胃内生物膜形成示意图 Fig. 4 Schematic diagram of rumen biofilm formation during feed digestion |
瘤胃微生物LuxS/AI-2群体感应可以调控细菌生物膜的形成[76]。生物膜可以促进瘤胃微生物将纤维素转化为VFA[77]。从这个视角来看,调控这种感应可以促进细菌生物膜的形成,激活细菌的消化机制,进而维持瘤胃内微生物的生态平衡[78]。有研究发现,普雷沃氏菌作为初级定殖阶段的优势菌属,与QS相关的luxS基因表达最为丰富,且在QS中表现出“合作关系”,而其他优势菌属表现为“自私不合作”[79]。Ghali等[60]的研究同样表明,普雷沃氏菌中的luxS基因含量丰富、表达更为活跃,暗示该菌可能参与了纤维素的降解;此外在瘤胃内降解纤维的主要细菌是黄色瘤胃球菌、白色瘤胃球菌和产琥珀酸丝状杆菌,这些菌属中的luxS基因含量也较为丰富。
饲料消化与瘤胃微生物QS密切相关。在瘤胃微生物定殖初期,由于可溶性、易发酵的营养物质较多,变形菌门在降解初期大量增殖并在30 min后完成第一次的饲料降解,拟杆菌门和梭菌纲随后逐渐取代变形菌门,附着在饲料颗粒上形成的微生物群落可以在很长一段时间内得以保留[65]。这些初始定殖阶段微生物丰度的变化与Ghali等[60]的发现类似,在宏基因组数据库中检测到的luxS基因序列中,拟杆菌门(主要是普雷沃氏菌属)和厚壁菌门(主要是梭菌纲)占比超过96%。在次级阶段的定殖过程中,纤维杆菌作为主要的纤维素降解菌属,通过分泌一系列的纤维素分解酶来降解纤维[65]。然而也有研究发现,当把纤维素作为唯一碳源时纤维杆菌却无法生长[80],这可能是由于缺乏一些多糖的异构酶和转运蛋白[81]。退化的运动性和趋化性[82]暗示着纤维杆菌的作用仅仅是降解纤维素并为其他瘤胃微生物提供营养代谢产物,由此推测纤维杆菌是被动地通过初级定殖形成的生物膜基质黏附在植物纤维上,从而启动纤维降解[81]。然而,初级定殖的微生物群落如何识别纤维杆菌并与其“合作”的过程或纤维杆菌是否仅感知而不产生信号分子目前并未见报道[2]。在瘤胃中与纤维杆菌作用类似的密螺旋体不能以纤维素作为唯一碳源生长[65],目前已发现密螺旋体属是螺旋体门中luxS基因含量最丰富的属[60],但其是否与纤维杆菌类似仅作为单一的纤维素降解者还是兼具与其他微生物沟通的交流使命还有待研究。Huws等[79]在对附着群落进行基因相关性网络分析时发现,优势菌(除普雷沃氏菌外)几乎没有基因相关性。在代谢功能和微生物多样性丰富的瘤胃中,瘤胃微生物之间的交流频繁[61],表明群体感应这种交流机制同样存在于部分丰度较低的微生物群落中,这些丰度较低的菌属可能以某种方式对群落结构、稳定性和生态系统功能产生影响,并进一步影响宿主生产性能[83]。
4 展望LuxS/AI-2介导的QS作为存在于瘤胃内并调控瘤胃微生物生态的一种通讯方式,通过形成生物膜来帮助宿主提高纤维降解和养分利用能力,这种群体感应不仅调节了微生物自身的生理行为,对维持瘤胃内微生物多样性和群落稳定也起到了重要的作用。动物遭受应激后机体出现不同程度的免疫力降低和炎症反应,瘤胃微生物短时间内无法适应这种变化,进而无法有效降解日粮中的大部分营养素,导致采食量下降和各种应激综合征。理论上,动物应激后瘤胃微生物LuxS/AI-2群体感应系统会受到影响,使得微生物之间“通讯失联”,无法快速形成相应的细菌生物膜来抵抗这种应激,进而无法对剧烈变化的周围环境和体内代谢做出快速的应答。生物膜的形成受LuxS/AI-2群体感应调控,如果能在动物应激前就加入能增强瘤胃微生物LuxS/AI-2群体感应的物质,进而提前形成更多的生物膜来抵抗这种应激,是不是意味着可以缓解应激呢?AI-2作为细菌种间交流的一种通讯信号,进一步了解它的作用机制有助于人们更好地理解瘤胃微生物的交流模式,通过调控这种交流来加强或抑制生物膜的形成,进而调节各类营养物质的消化代谢、缓解应激,解决反刍动物生产上饲料转化效率低、环境应激等问题。
| [1] |
MORGAVI D P, KELLY W J, JANSSEN P H, et al. Rumen microbial (meta)genomics and its application to ruminant production[J]. Animal, 2013, 7(S1): 184-201. |
| [2] |
RANAVA D, BACKES C, KARTHIKEYAN G, et al. Metabolic exchange and energetic coupling between nutritionally stressed bacterial species: role of quorum-sensing molecules[J]. mBio, 2021, 12(1): e02758-20. |
| [3] |
MCNAB R, LAMONT R J. Microbial dinner-party conversations: the role of LuxS in interspecies communication[J]. J Med Microbiol, 2003, 52(Pt 7): 541-545. |
| [4] |
WANG Y S, BIAN Z R, WANG Y. Biofilm formation and inhibition mediated by bacterial quorum sensing[J]. Appl Microbiol Biotechnol, 2022, 106(19): 6365-6381. |
| [5] |
MITSUMORI M, XU L M, KAJIKAWA H, et al. Possible quorum sensing in the rumen microbial community: detection of quorum-sensing signal molecules from rumen bacteria[J]. FEMS Microbiol Lett, 2003, 219(1): 47-52. DOI:10.1016/S0378-1097(02)01192-8 |
| [6] |
WON M Y, OYAMA L B, COURTNEY S J, et al. Can rumen bacteria communicate to each other?[J]. Microbiome, 2020, 8(1): 23. DOI:10.1186/s40168-020-00796-y |
| [7] |
MILLER S T, XAVIER K B, CAMPAGNA S R, et al. Salmonella typhimurium recognizes a chemically distinct form of the bacterial quorum-sensing signal AI-2[J]. Mol Cell, 2004, 15(5): 677-687. DOI:10.1016/j.molcel.2004.07.020 |
| [8] |
MIRANDA V, TORCATO I M, XAVIER K B, et al. Synthesis of d-desthiobiotin-AI-2 as a novel chemical probe for autoinducer-2 quorum sensing receptors[J]. Bioorg Chem, 2019, 92: 103200. DOI:10.1016/j.bioorg.2019.103200 |
| [9] |
SCHAUDER S, SHOKAT K, SURETTE M G, et al. The LuxS family of bacterial autoinducers: biosynthesis of a novel quorum-sensing signal molecule[J]. Mol Microbiol, 2001, 41(2): 463-476. DOI:10.1046/j.1365-2958.2001.02532.x |
| [10] |
TORCATO I M, KASAL M R, BRITO P H, et al. Identification of novel autoinducer-2 receptors in Clostridia reveals plasticity in the binding site of the LsrB receptor family[J]. J Biol Chem, 2019, 294(12): 4450-4463. DOI:10.1074/jbc.RA118.006938 |
| [11] |
NEIDITCH M B, FEDERLE M J, MILLER S T, et al. Regulation of LuxPQ receptor activity by the quorum-sensing signal autoinducer-2[J]. Mol Cell, 2005, 18(5): 507-518. DOI:10.1016/j.molcel.2005.04.020 |
| [12] |
CHEN X, SCHAUDER S, POTIER N, et al. Structural identification of a bacterial quorum-sensing signal containing boron[J]. Nature, 2002, 415(6871): 545-549. DOI:10.1038/415545a |
| [13] |
RAJAMANI S, ZHU J G, PEI D H, et al. A LuxP-FRET-based reporter for the detection and quantification of AI-2 bacterial quorum-sensing signal compounds[J]. Biochemistry, 2007, 46(13): 3990-3997. DOI:10.1021/bi602479e |
| [14] |
LAGANENKA L, LEE J W, MALFERTHEINER L, et al. Chemotaxis and autoinducer-2 signalling mediate colonization and contribute to co-existence of Escherichia coli strains in the murine gut[J]. Nat Microbiol, 2023, 8(2): 204-217. DOI:10.1038/s41564-022-01286-7 |
| [15] |
KEIZERS M, DOBRINDT U, BERGER M. A simple biosensor-based assay for quantitative autoinducer-2 analysis[J]. ACS Synth Biol, 2022, 11(2): 747-759. DOI:10.1021/acssynbio.1c00459 |
| [16] |
KUMAR S, KUMAR C B, RAJENDRAN V, et al. Delineating virulence of Vibrio campbellii: a predominant luminescent bacterial pathogen in Indian shrimp hatcheries[J]. Sci Rep, 2021, 11(1): 15831. DOI:10.1038/s41598-021-94961-4 |
| [17] |
BISWAS S, MUKHERJEE P, MANNA T, et al. Quorum sensing autoinducer(s) and flagellum independently mediate EPS signaling in Vibrio cholerae through LuxO-independent mechanism[J]. Microb Ecol, 2019, 77(3): 616-630. DOI:10.1007/s00248-018-1262-5 |
| [18] |
TALÀ A, SIDE D D, BUCCOLIERI G, et al. Exposure to static magnetic field stimulates quorum sensing circuit in luminescent Vibrio strains of the Harveyi clade[J]. PLoS One, 2014, 9(6): e100825. DOI:10.1371/journal.pone.0100825 |
| [19] |
GU Y, LI B, TIAN J J, et al. The response of LuxS/AI-2 quorum sensing in Lactobacillus fermentum 2-1 to changes in environmental growth conditions[J]. Ann Microbiol, 2018, 68(5): 287-294. DOI:10.1007/s13213-018-1337-z |
| [20] |
CHAI Y M, MA Q W, NONG X, et al. Dissecting LuxS/AI-2 quorum sensing system-mediated phenyllactic acid production mechanisms of Lactiplantibacillus plantarum L3[J]. Food Res Int, 2023, 166: 112582. DOI:10.1016/j.foodres.2023.112582 |
| [21] |
AQAWI M, SIONOV R V, FRIEDMAN M, et al. The antibacterial effect of cannabigerol toward Streptococcus mutans is influenced by the autoinducers 21-CSP and AI-2[J]. Biomedicines, 2023, 11(3): 668. DOI:10.3390/biomedicines11030668 |
| [22] |
MENG F Q, ZHAO M W, LU Z X. The LuxS/AI-2 system regulates the probiotic activities of lactic acid bacteria[J]. Trends Food Sci Technol, 2022, 127: 272-279. DOI:10.1016/j.tifs.2022.05.014 |
| [23] |
HERZOG R, PESCHEK N, FRÖHLICH K S, et al. Three autoinducer molecules act in concert to control virulence gene expression in Vibrio cholerae[J]. Nucleic Acids Res, 2019, 47(6): 3171-3183. DOI:10.1093/nar/gky1320 |
| [24] |
BRIDGES A A, BASSLER B L. The intragenus and interspecies quorum-sensing autoinducers exert distinct control over Vibrio cholerae biofilm formation and dispersal[J]. PLoS Biol, 2019, 17(11): e3000429. DOI:10.1371/journal.pbio.3000429 |
| [25] |
BOYACI H, SHAH T, HURLEY A, et al. Structure, regulation, and inhibition of the quorum-sensing signal integrator LuxO[J]. PLoS Biol, 2016, 14(5): e1002464. DOI:10.1371/journal.pbio.1002464 |
| [26] |
RAYCHAUDHURI S, JAIN V, DONGRE M. Identification of a constitutively active variant of LuxO that affects production of HA/protease and biofilm development in a non-O1, non-O139 Vibrio cholerae O110[J]. Gene, 2006, 369: 126-133. DOI:10.1016/j.gene.2005.10.031 |
| [27] |
TAGA M E, MILLER S T, BASSLER B L. Lsr-mediated transport and processing of AI-2 in Salmonella typhimurium[J]. Mol Microbiol, 2003, 50(4): 1411-1427. DOI:10.1046/j.1365-2958.2003.03781.x |
| [28] |
TAGA M E, SEMMELHACK J L, BASSLER B L. The LuxS-dependent autoinducer AI-2 controls the expression of an ABC transporter that functions in AI-2 uptake in Salmonella typhimurium[J]. Mol Microbiol, 2001, 42(3): 777-793. DOI:10.1046/j.1365-2958.2001.02669.x |
| [29] |
TAGA M E, BASSLER B L. Chemical communication among bacteria[J]. Proc Natl Acad Sci U S A, 2003, 100(S2): 14549-14554. |
| [30] |
TOLKER-NIELSEN T. Biofilm development[M]//GHANNOUM M, PARSEK M, WHITELEY M, et al. Microbial Biofilms. 2nd ed. Washington: American Society for Microbiology, 2015: 51-66.
|
| [31] |
KOLTER R, GREENBERG E P. Microbial sciences: the superficial life of microbes[J]. Nature, 2006, 441(7091): 300-302. DOI:10.1038/441300a |
| [32] |
CHU P L, FENG Y M, LONG Z Q, et al. Novel benzothiazole derivatives as potential anti-quorum sensing agents for managing plant bacterial diseases: synthesis, antibacterial activity assessment, and SAR study[J]. J Agric Food Chem, 2023, 71(17): 6525-6540. DOI:10.1021/acs.jafc.2c07810 |
| [33] |
RYBTKE M, HULTQVIST L D, GIVSKOV M, et al. Pseudomonas aeruginosa biofilm infections: community structure, antimicrobial tolerance and immune response[J]. J Mol Biol, 2015, 427(23): 3628-3645. DOI:10.1016/j.jmb.2015.08.016 |
| [34] |
HÄUSSLER S, BECKER T. The pseudomonas quinolone signal (PQS) balances life and death in Pseudomonas aeruginosa populations[J]. PLoS Pathog, 2008, 4(9): e1000166. DOI:10.1371/journal.ppat.1000166 |
| [35] |
VENDEVILLE A, WINZER K, HEURLIER K, et al. Making 'sense' of metabolism: autoinducer-2, LuxS and pathogenic bacteria[J]. Nat Rev Microbiol, 2005, 3(5): 383-396. DOI:10.1038/nrmicro1146 |
| [36] |
WANG Y, WANG Y X, SUN L Y, et al. The LuxS/AI-2 system of Streptococcus suis[J]. Appl Microbiol Biotechnol, 2018, 102(17): 7231-7238. DOI:10.1007/s00253-018-9170-7 |
| [37] |
WANG Y, YI L, ZHANG Z C, et al. Overexpression of luxS cannot increase autoinducer-2 production, only affect the growth and biofilm formation in Streptococcus suis[J]. Sci World J, 2013, 2013: 924276. |
| [38] |
LAGANENKA L, COLIN R, SOURJIK V. Chemotaxis towards autoinducer 2 mediates autoaggregation in Escherichia coli[J]. Nat Commun, 2016, 7(1): 12984. DOI:10.1038/ncomms12984 |
| [39] |
HEGDE M, ENGLERT D L, SCHROCK S, et al. Chemotaxis to the quorum-sensing signal AI-2 requires the Tsr chemoreceptor and the periplasmic LsrB AI-2-binding protein[J]. J Bacteriol, 2011, 193(3): 768-773. DOI:10.1128/JB.01196-10 |
| [40] |
WEN Y C, HUANG H M, TANG T C, et al. AI-2 represses CagA expression and bacterial adhesion, attenuating the Helicobacter pylori-induced inflammatory response of gastric epithelial cells[J]. Helicobacter, 2021, 26(2): e12778. DOI:10.1111/hel.12778 |
| [41] |
ANDERSON J K, HUANG J Y, WREDEN C, et al. Chemorepulsion from the quorum signal autoinducer-2 promotes Helicobacter pylori biofilm dispersal[J]. mBio, 2015, 6(4): e00379. |
| [42] |
CLUZEL M E, ZANELLA-CLE?ON I, COZZONE A J, et al. The Staphylococcus aureus autoinducer-2 synthase LuxS is regulated by Ser/Thr phosphorylation[J]. J Bacteriol, 2010, 192(23): 6295-6301. DOI:10.1128/JB.00853-10 |
| [43] |
ZHANG L J, SHEN Y, QIU L L, et al. The suppression effect of SCH-79797 on Streptococcus mutans biofilm formation[J]. J Oral Microbiol, 2022, 14(1): 2061113. DOI:10.1080/20002297.2022.2061113 |
| [44] |
SZTAJER H, LEMME A, VILCHEZ R, et al. Autoinducer-2-regulated genes in Streptococcus mutans UA159 and global metabolic effect of the luxS mutation[J]. J Bacteriol, 2008, 190(1): 401-415. DOI:10.1128/JB.01086-07 |
| [45] |
RODRIGUES M V, KIS P, XAVIER K B, et al. Synthesis and potential of Autoinducer-2 and analogs to manipulate inter-species quorum sensing[J]. ISR J Chem, 2023, 63(5-6): e202200091. DOI:10.1002/ijch.202200091 |
| [46] |
MAYER C, BORGES A, FLAMENT-SIMON S C, et al. Quorum sensing architecture network in Escherichia coli virulence and pathogenesis[J]. FEMS Microbiol Rev, 2023, 47(4): fuad031. DOI:10.1093/femsre/fuad031 |
| [47] |
SLATER R T, FROST L R, JOSSI S E, et al. Clostridioides difficile LuxS mediates inter-bacterial interactions within biofilms[J]. Sci Rep, 2019, 9: 9903. DOI:10.1038/s41598-019-46143-6 |
| [48] |
ARENAS J, TOMMASSEN J. Meningococcal biofilm formation: Let's stick together[J]. Trends Microbiol, 2017, 25(2): 113-124. DOI:10.1016/j.tim.2016.09.005 |
| [49] |
ROUSSEL-JAZÉDÉ V, GRIJPSTRA J, VAN DAM V, et al. Lipidation of the autotransporter NalP of Neisseria meningitidis is required for its function in the release of cell-surface-exposed proteins[J]. Microbiology (Reading), 2013, 159(Pt 2): 286-295. |
| [50] |
ROUSSEL-JAZÉDÉ V, JONGERIUS I, BOS M P, et al. NalP-mediated proteolytic release of lactoferrin-binding protein B from the meningococcal cell surface[J]. Infect Immun, 2010, 78(7): 3083-3089. DOI:10.1128/IAI.01193-09 |
| [51] |
VAN ULSEN P, VAN ALPHEN L, HOVE J T, et al. A Neisserial autotransporter NalP modulating the processing of other autotransporters[J]. Mol Microbiol, 2003, 50(3): 1017-1030. DOI:10.1046/j.1365-2958.2003.03773.x |
| [52] |
PENG N, CAI P, MORTIMER M, et al. The exopolysaccharide-eDNA interaction modulates 3D architecture of Bacillus subtilis biofilm[J]. BMC Microbiol, 2020, 20: 115. DOI:10.1186/s12866-020-01789-5 |
| [53] |
HOBLEY L, HARKINS C, MACPHEE C E, et al. Giving structure to the biofilm matrix: an overview of individual strategies and emerging common themes[J]. FEMS Microbiol Rev, 2015, 39(5): 649-669. DOI:10.1093/femsre/fuv015 |
| [54] |
GU Y, TIAN J J, ZHANG Y, et al. Dissecting signal molecule AI-2 mediated biofilm formation and environmental tolerance in Lactobacillus plantarum[J]. J Biosci Bioeng, 2021, 131(2): 153-160. DOI:10.1016/j.jbiosc.2020.09.015 |
| [55] |
MUHAMMAD M H, IDRIS A L, FAN X, et al. Beyond risk: bacterial biofilms and their regulating approaches[J]. Front Microbiol, 2020, 11: 928. DOI:10.3389/fmicb.2020.00928 |
| [56] |
KRAGH K N, HUTCHISON J B, MELAUGH G, et al. Role of multicellular aggregates in biofilm formation[J]. mBio, 2016, 7(2): e00237. |
| [57] |
NUNAN N, WU K J, YOUNG I M, et al. Spatial distribution of bacterial communities and their relationships with the micro-architecture of soil[J]. FEMS Microbiol Ecol, 2003, 44(2): 203-215. DOI:10.1016/S0168-6496(03)00027-8 |
| [58] |
SUGIMOTO S, OKUDA K I, MIYAKAWA R, et al. Imaging of bacterial multicellular behaviour in biofilms in liquid by atmospheric scanning electron microscopy[J]. Sci Rep, 2016, 6: 25889. DOI:10.1038/srep25889 |
| [59] |
LIU X Z, LIU Q M, SUN S H, et al. Exploring AI-2-mediated interspecies communications within rumen microbial communities[J]. Microbiome, 2022, 10: 167. DOI:10.1186/s40168-022-01367-z |
| [60] |
GHALI I, SHINKAI T, MITSUMORI M. Mining of luxS genes from rumen microbial consortia by metagenomic and metatranscriptomic approaches[J]. Anim Sci J, 2016, 87(5): 666-673. DOI:10.1111/asj.12476 |
| [61] |
XIE Y Y, SUN H Z, XUE M Y, et al. Metagenomics reveals differences in microbial composition and metabolic functions in the rumen of dairy cows with different residual feed intake[J]. Anim Microbiome, 2022, 4(1): 19. DOI:10.1186/s42523-022-00170-3 |
| [62] |
LI Q S, WANG R, MA Z Y, et al. Dietary selection of metabolically distinct microorganisms drives hydrogen metabolism in ruminants[J]. ISME J, 2022, 16(11): 2535-2546. DOI:10.1038/s41396-022-01294-9 |
| [63] |
郭海康, 万发春, 沈维军, 等. 畜禽消化道细菌群体感应及相关调控技术研究进展[J]. 畜牧兽医学报, 2022, 53(6): 1678-1688. GUO H K, WAN F C, SHEN W J, et al. Research progress and related regulation technology on bacterial quorum sensing in the gastro-intestinal tract of livestock and poultry[J]. Acta Veterinaria et Zootechnica Sinica, 2022, 53(6): 1678-1688. (in Chinese) |
| [64] |
冉涛, 谭支良. 反刍家畜瘤胃微生物群体感应[J]. 动物营养学报, 2012, 24(7): 1207-1215. RAN T, TAN Z L. Rumen microbial quorum-sensing of ruminant livestock[J]. Chinese Journal of Animal Nutrition, 2012, 24(7): 1207-1215. (in Chinese) |
| [65] |
VAHIDI M F, GHARECHAHI J, BEHMANESH M, et al. Diversity of microbes colonizing forages of varying lignocellulose properties in the sheep rumen[J]. PeerJ, 2021, 9: e10463. DOI:10.7717/peerj.10463 |
| [66] |
COMTET-MARRE S, PARISOT N, LEPERCQ P, et al. Metatranscriptomics reveals the active bacterial and eukaryotic fibrolytic communities in the rumen of dairy cow fed a mixed diet[J]. Front Microbiol, 2017, 8: 67. |
| [67] |
MINATO H, ENDO A, OOTOMO Y, et al. Ecological treatise on the rumen fermentation: Ⅱ.The amylolytic and cellulolytic activities of the fractionated bacterial portions attached to the rumen solids[J]. J Gen Appl Microbiol, 1966, 12(1): 53-69. DOI:10.2323/jgam.12.53 |
| [68] |
CHENG K J, STEWART C S, DINSDALE D, et al. Electron microscopy of bacteria involved in the digestion of plant cell walls[J]. Anim Feed Sci Technol, 1984, 10(2-3): 93-120. DOI:10.1016/0377-8401(84)90002-6 |
| [69] |
MOSONI P, FONTY G, GOUET P. Competition between ruminal cellulolytic bacteria for adhesion to cellulose[J]. Ann Zootech, 1996, 45(S1): 298. |
| [70] |
KOIKE S, PAN J, KOBAYASHI Y, et al. Kinetics of in sacco fiber-attachment of representative ruminal cellulolytic bacteria monitored by competitive PCR[J]. J Dairy Sci, 2003, 86(4): 1429-1435. DOI:10.3168/jds.S0022-0302(03)73726-6 |
| [71] |
BAR-ZEEV E, BERMAN-FRANK I, GIRSHEVITZ O, et al. Revised paradigm of aquatic biofilm formation facilitated by microgel transparent exopolymer particles[J]. Proc Natl Acad Sci U S A, 2012, 109(23): 9119-9124. DOI:10.1073/pnas.1203708109 |
| [72] |
LENG R A. The rumen- a fermentation vat or a series of organized structured microbial consortia: implications for the mitigation of enteric methane production by feed additives[J]. Livest Res Rural Dev, 2011, 23(12): 258. |
| [73] |
SPEZIALE P, PIETROCOLA G, FOSTER T J, et al. Protein-based biofilm matrices in Staphylococci[J]. Front Cell Infect Microbiol, 2014, 4: 171. |
| [74] |
LENG R A. Interactions between microbial consortia in biofilms: a paradigm shift in rumen microbial ecology and enteric methane mitigation[J]. Anim Prod Sci, 2014, 54(5): 519-543. DOI:10.1071/AN13381 |
| [75] |
MCCALL A, EDGERTON M. Real-time approach to flow cell imaging of Candida albicans biofilm development[J]. J Fungi, 2017, 3(1): 13. DOI:10.3390/jof3010013 |
| [76] |
HUWS S, MAYORGA O L, KIM E J, et al. Microbial colonization and subsequent biofilm formation by ruminal microorganisms on fresh perennial ryegrass[C]//2007 Conference on Gastrointestinal Function (CGIF). Chicago, 2007.
|
| [77] |
XIROS C, SHAHAB R L, STUDER M H P. A cellulolytic fungal biofilm enhances the consolidated bioconversion of cellulose to short chain fatty acids by the rumen microbiome[J]. Appl Microbiol Biotechnol, 2019, 103(8): 3355-3365. DOI:10.1007/s00253-019-09706-1 |
| [78] |
DOBRETSOV S, TEPLITSKI M, PAUL V. Mini-review: quorum sensing in the marine environment and its relationship to biofouling[J]. Biofouling, 2009, 25(5): 413-427. DOI:10.1080/08927010902853516 |
| [79] |
HUWS S A, EDWARDS J E, LIN W C, et al. Microbiomes attached to fresh perennial ryegrass are temporally resilient and adapt to changing ecological niches[J]. Microbiome, 2021, 9(1): 143. DOI:10.1186/s40168-021-01087-w |
| [80] |
SOROKIN D Y, GUMEROV V M, RAKITIN A L, et al. Genome analysis of Chitinivibrio alkaliphilus gen.nov., sp.nov., a novel extremely haloalkaliphilic anaerobic chitinolytic bacterium from the candidate phylum Termite Group 3[J]. Environ Microbiol, 2014, 16(6): 1549-1565. DOI:10.1111/1462-2920.12284 |
| [81] |
SUEN G, WEIMER P J, STEVENSON D M, et al. The complete genome sequence of Fibrobacter succinogenes S85 reveals a cellulolytic and metabolic specialist[J]. PLoS One, 2011, 6(4): e18814. DOI:10.1371/journal.pone.0018814 |
| [82] |
RAHMAN N A, PARKS D H, VANWONTERGHEM I, et al. A phylogenomic analysis of the bacterial phylum Fibrobacteres[J]. Front Microbiol, 2016, 6: 1469. |
| [83] |
SHABAT S K B, SASSON G, DORON-FAIGENBOIM A, et al. Specific microbiome-dependent mechanisms underlie the energy harvest efficiency of ruminants[J]. ISME J, 2016, 10(12): 2958-2972. DOI:10.1038/ismej.2016.62 |
(编辑 范子娟)