2. 朝阳市朝牧种畜场有限公司, 朝阳 122629
2. Chaoyang Chaomu Breeding Stock Farm Co. Ltd., Chaoyang 122629, China
布鲁氏菌病(以下简称布病)是一种由布鲁氏菌(以下简称布菌)感染引起的人畜共患病,具有高度传染性,在170多个国家和地区流行,每年感染人数逾50万[1]。根据布菌对宿主的偏好性,一般将其分为羊种(B. melitensis)、牛种(B. abortus)、猪种(B. suis)、鼠种(B. neotomae)、犬种(B. canis)和绵羊附睾种(B. ovis)等[2]。其中,B. melitensis(绵羊和山羊均易感)的毒力和感染性最强,也是人间布病的最主要病原,在我国占据主导地位[3]。此外,B. suis和B. ovis在绵羊群的感染状况也不容忽视[4]。布病的典型症状表现为波浪热、关节炎和繁殖障碍(公畜睾丸炎和附睾炎,母畜流产和不孕)。我国是绵羊生产大国,存栏量在2022年已达1.86亿只。但近年来,绵羊布病疫情在华北和西北等地呈蔓延趋势[5-6],不仅给我国绵羊养殖业带来巨大经济损失,还对公共卫生形成重大安全隐患。
布病是自然疫源性传染病,提高管理水平、接种疫苗和淘汰染疫动物是部分发达国家防控和净化布病的成功模式[3, 7],但在我国仍面临巨大挑战[8]。一方面,布菌是兼性胞内寄生菌,具有免疫逃避能力,布病临床表现为隐性感染和慢性经过,早期难发现,晚期难控制;另一方面,疫苗作用有限[9],加之我国羊群基数庞大,流动性强,尚未全面形成集约化养殖,疫苗难以完全覆盖。因此常规措施难以有效控制布病在我国的流行态势。从长远来看,开展绵羊抗布病新品种的选育工作,提高宿主动物抗病力,能根本上从动物源头切断布病的传播途径,真正落实“人病兽防,关口前移”。
1 布菌的致病机制布菌是兼性胞内寄生菌,能侵入多种细胞,介导凋亡与自噬进程,建立起复制生态位以逃避免疫杀伤,营造出微环境来维持慢性感染[10]。毒力因子的存在对于布菌生存繁殖的维系以及免疫逃避策略的执行不可或缺。如布菌的非典型脂多糖(lipopolysaccharide,LPS)协同外膜蛋白(outer membrane proteins,OMPs),不仅可抑制免疫激活,阻止补体沉积,还能限制溶酶体融合[11-13]。同时,布菌缺少诸如外毒素、菌毛、鞭毛和质粒等强效致病因子,因此其较弱的免疫原性往往不会引起宿主机体强烈的免疫应答[13]。
1.1 感染与侵袭布菌可经呼吸道、消化道、眼结膜和损伤皮肤感染宿主,侵入吞噬细胞(如巨噬细胞、树突状细胞和中性粒细胞)以及非吞噬细胞(如上皮细胞、成纤维细胞和滋养层细胞)[14]。脂筏(lipid raft)存在于质膜表面,富含胆固醇和鞘磷脂,是参与细胞膜内外信号传导和物质传递的专门膜微区[15],能介导巨噬细胞以内吞的方式摄取布菌[16]。布菌的环β-1,2-葡聚糖(cyclic β-1, 2-glucan,CβG)能扰乱脂筏中的胆固醇组分[17-18]。发动蛋白(dynamin)和网格蛋白(clathrin)共同作用于布菌内化过程中包被囊泡的再分配和重排[19]。发动蛋白具有GTP酶活性,可选择性地调节囊泡的组装[20];网格蛋白汇聚于脂筏,招募发动蛋白富集,挤压质膜内陷形成小窝,进而包被囊泡[21]。至此,布菌被质膜裹入细胞内部。
1.2 运输与定位巨噬细胞既是布菌侵入的主要靶细胞,又是先天免疫的重要功能细胞。90%的内化布菌在感染前期会被清除,余下部分则迁移到内质网处建立起复制生态位[22]。布菌内化后形成的囊泡结构统称布氏小体(Brucella containing vacuoles,BCVs),是布菌进行胞内运输与定位的载体。在转运初始阶段,由BCVs形成的初级内体(early endosome)短暂接触溶酶体,获得其表面标记分子而成为eBCVs(early BCVs)[23],布菌CβG可阻止eBCVs被溶酶体深度融合[17-18]。同时,这种有限融合产生的酸化环境提供了激活布菌IV型分泌系统(type IV secretion system,T4SS)的信号[24],T4SS分泌的效应子重定向eBCVs的胞内运输途径,以阻止其与溶酶体过分接触而被消化[13, 25]。少数(约10%)逃避降解的布菌脱离内体,驱动效应子截获由内质网出芽向高尔基体转运的囊泡[26]。BCVs与内质网融合形成rBCVs(replicative BCVs)区隔,并带有内质网的特异标记[22, 27]。另外,双组分调控系统(two-component regulatory system,TCS)时刻调控布菌对环境信号刺激的感知、传递和反应[28]。BCVs的胞内运输与定位伴随着分子标记的更新,内体特征的成熟标志着复制生态位的建立。
1.3 繁殖与扩散在感染后期,rBCVs参与巨噬细胞的自噬途径而转化为aBCVs(autophagy BCVs),布菌完成胞内循环后开始胞间传播[29]。巨噬细胞的裂解焦亡引发布菌的外流逸散,Hiyoshi等[30]发现布菌可利用T4SS穿透BCVs膜形成PIT(pore-induced intracellular trap)结构,诱使补体沉积其中,产生补体依赖的find-me信号,启动中性粒细胞对PIT的胞吞摄取,而留存于PIT中的布菌得以逃避呼吸爆发。
2 绵羊布病的抗病育种研究进展畜禽抗病育种,简言之就是通过定向选择或改变特定基因型来培育具有抗病能力的畜禽新品种,主要分为3个方向:家系群体选育、基因工程选育和标记辅助选育。
2.1 抗病性与遗传基础狭义的抗病性指畜禽抵抗寄生虫病和传染病的抗病力,可分为特殊抗病力和一般抗病力,二者遗传机制不同。抗病基因是抗病性的遗传基础,可分为单一主基因、微效多基因和独立多基因[31-32]。特殊抗病力指应对特定某种病原体的抗感染能力,主要被主基因所调控;一般抗病力指应对不限一种病原体的抗感染能力,总是由多基因所调控[31]。两种抗病力都受环境因素影响,其中受遗传因素影响的程度即遗传力,牛抗布病的遗传力为0.19[33],猪对布病的抗病性可以稳定遗传[34]。绵羊抗布病的遗传力未见相关报道。
2.2 家系群体选育群体选育指从畜禽种群挑选抗病力突出的个体作为亲本建立家系,再通过对子代逐代选择纯化目的基因,获得稳定遗传抗病性状的新品种。这种传统的育种方式实际上是对基因型的直接选择,耗时、费力和成本高的缺点突出,因此往往应用于世代间隔短、群体规模大的家禽研究中,如鸡抗沙门菌[35]和鸭抗肝炎病毒[36]等,而在绵羊的抗病研究中应用较少。
2.3 基因工程选育基因工程选育指利用锌指核酸酶(ZFNs)、转录激活因子样效应物核酸酶(TALENs)和成簇规律间隔短回文重复序列及其相关蛋白(CRISPR/Cas9)等基因编辑技术对畜禽基因组敲除受体基因或转入目的基因[37]。相较于其它基因编辑技术,CRISPR/Cas9因其高效、稳定和廉价的优点而被广泛应用于转基因动物生产中。
Toll样受体4(TLR4)能识别布菌的脂多糖成分,激活炎症反应和免疫反应。2012年,中国农业大学的研究团队对萨福克绵羊受精卵微量注射线性化的pTLR4-3S载体,借助胚胎移植技术,成功构建了过表达TLR4的转基因绵羊。进一步的攻毒试验结果显示,IL-6、IL-8和TNF-α的表达水平提高,炎症细胞的凋亡增加,机体的抗布菌感染能力明显增强[38]。在此基础上,该团队改进方法,在绵羊非繁殖季也能通过微量注射原核胚胎生产转基因绵羊,并且发现TLR4的过表达对绵羊的生长无不良影响[39]。
褪黑素(melatonin,MT)是由松果体分泌的神经类激素,作为一种抗氧化剂,可缓解氧化应激和促进免疫系统成熟,N-乙酰基-5-羟色胺-O-甲基转移酶(N-acetylserotonin O-methyltransferase,ASMT) 是其合成过程中的关键限速酶[40]。2017年,中国农业大学的研究团队选用杜泊绵羊作为供受体,利用CRISPR/Cas9、微量注射和胚胎移植技术成功构建出过表达ASMT的转基因绵羊[41]。布菌攻毒试验发现转基因绵羊均未患病,并且差异表达基因显著富集在自然杀伤细胞介导的相关通路,这表明转基因绵羊抗布病能力强于野生型[42]。
2.4 标记辅助选育标记辅助选择(marker-assisted selection,MAS)指利用育种性状相关标记代替表型层次的选择,抗病性状标记辅助选择可分为免疫遗传学标记辅助选择和分子标记辅助选择[43]。其中,分子标记辅助选择指从分子水平利用与目标基因紧密连锁的遗传标记直接选择基因型,可在遗传本质上选育绵羊抗布病新品种。与家系群体选育相比,标记辅助选育可有效缩短世代间隔、增大选择强度、提高选种准确性和降低育种成本;与基因工程选育相比,标记辅助选育可避免生物安全、伦理道德和动物福利等方面的诸多限制。主效基因的挖掘是标记辅助选择的基础工作,以下从免疫和炎症反应的激活和效应两个阶段介绍布病相关基因和非编码RNA(non-coding RNAs,ncRNAs)。另外,如表 1和表 2所示,大多数研究选用B. abortus S2308经典菌株验证基因功能,不同菌株与宿主之间作用机制的比较还未见文献报道。
|
|
表 1 参与免疫和炎症反应激活阶段的基因或ncRNA Table 1 Genes or ncRNAs involved in the activation of immune and inflammatory responses |
|
|
表 2 参与免疫和炎症反应效应阶段的基因或ncRNA Table 2 Genes or ncRNAs involved in the effective stage of immune and inflammatory responses |
2.4.1 参与免疫和炎症反应激活阶段的基因或ncRNA 免疫和炎症反应的激活依赖于模式识别受体(pattern recognition receptors,PRRs)结合病原相关分子模式(pathogen-associated molecular patterns,PAMPs)或损伤相关分子模式(damage-associated molecular patterns,DAMPs)。能识别布菌的PRRs主要分为Toll样受体(toll-like receptors,TLRs)和NOD样受体(NOD-like receptors,NLRs)。
2.4.1.1 Toll样受体 TLRs是跨膜蛋白受体,其抗菌作用最早在1996年被发现于果蝇的真菌感染试验中[44]。迄今为止,在哺乳动物中共鉴定到13种TLRs,能监测多种PAMPs组分[45]。其中,TLR2驻留于质膜,能识别布菌的OMP[46];TLR3和TLR7驻留于内体膜,能识别布菌的RNA[47];TLR4驻留于质膜,能识别布菌的LPS[48];TLR9驻留于内体膜,能识别布菌的DNA[49]。除TLR3外,活化的TLRs均可募集髓样分化因子88(myeloid differentiation factor 88,MyD88)。MyD88是一种胞质可溶性的接头蛋白(adaptor),能将TLR信号向下游的IκB激酶(IKK)复合物和MAPK激酶激酶(MAP3K)转导,促进炎症介质和细胞因子的释放。若敲除小鼠的MyD88,则在感染布菌时,其巨噬细胞产生的TNF-α和IL-12显著减少[50],TNF-α和IL-12是首要的促炎症因子。由B. Melitensis分泌的TIR结构域蛋白(TIR domain-containing proteins,Tcps)称TcpB,TcpB可直接结合MyD88,阻碍TLR依赖于MyD88介导的NF-κB激活,还能诱导未折叠蛋白反应的靶基因表达[51-52]。
2.4.1.2 NOD样受体 NLRs家族是定位于胞浆的模式识别受体,其中,NOD1和NOD2可识别布菌胞壁的肽聚糖组分,通过结合接头蛋白RIP2激活IKK复合物和MAP3K[53]。此外,NOD1和NOD2还是诱导内质网应激的重要介质:布菌在侵入宿主细胞后会通过T4SS释放效应子VceC,触发由内质网应激传感器IRE1α介导的未折叠蛋白反应。其中,IRE1α是内质网跨膜蛋白,可募集TRAF2(TNF-receptor associated factor 2),依赖于NOD1和NOD2激活NF-κB受体[54]。布菌能通过IRE1α轴抑制宿主细胞的自噬[55]。NLRs家族的NLRP3受体蛋白在抗布菌感染中同样发挥重要作用。一般地,NF-κB信号的激活促进NLRP3的表达,NLRP3与凋亡相关斑点样蛋白(ASC)和半胱氨酸天冬氨酸蛋白酶1前体(pro-caspase-1)组装形成的NLRP3炎症小体,同源活化的caspase-1剪切促炎细胞因子IL-1β和IL-18的前体,使其成熟并释放到胞外,增强机体免疫能力[56]。最近的研究完善了该通路,在布菌感染时,IRE1α可诱导活性氧(ROS)依赖性的NLRP3易位到内质网与线粒体连接处,激活caspase-2和BID之间的信号轴,导致线粒体的损伤相关分子模式(DAMP)释放,刺激NLRP3炎症小体合成[57]。这一发现整合了布菌感染过程中内质网应激、线粒体损伤与先天免疫之间的联系。另外,黑色素瘤缺乏因子2(absent in melanoma 2,AIM2)是最近发现的胞内PRR,可识别布菌DNA,由其组装的AIM2炎症小体,同样能增进IL-1β和IL-18的产生[56, 58]。
2.4.1.3 其他模式识别受体 除TLRs和NLRs外,在2008年,首次发现了干扰素基因刺激蛋白(stimulator of interferon genes,STING)参与固有免疫应答的信号转导作用[59]。STING位于内质网膜,不仅能作为PRR行使区别于TLR9的“布菌DNA识别作用”,还能充当接头蛋白发挥二级受体结合cGAMP的“布菌DNA传感作用”,激活I型IFN的产生,调控促炎细胞因子通过NF-κB途径介导巨噬细胞的自噬,支持CD8+ T细胞的适应性免疫[59-60]。最近,Khan等[61]发现布菌可在感染早期抑制宿主细胞编码STING的Tmem173基因表达,并能诱导靶作用于STING的miR-24产生,增加免疫激活的成本,破坏胞浆监测的机制。在此基础上,Oliveira团队[62-63]的两项研究接连报道了STING通过缺氧诱导因子1α(HIF-1α)调控巨噬细胞在抗布菌感染中的代谢重编程,驱动其向促炎的M1型或抗炎的M2型极化,并指出该过程与IRE1α所介导的内质网应激紧密联系。有关HIF的发现已获得2019年的诺贝尔生理学和医学奖,但其在布病上的研究较少。
2.4.2 参与免疫和炎症反应效应阶段的基因或ncRNA2.4.2.1 免疫应答 主要组织相容性复合物(major histocompatibility complex,MHC)主要分为MHC I、MHC II和MHC III。经巨噬细胞处理的布菌肽段可由MHC II递呈给CD4+ T淋巴细胞中的辅助性T细胞(helper T cell,TH),促进特异性TH的增殖及其淋巴因子(如IFN-γ)的表达,IFN-γ能激活巨噬细胞的杀菌作用,在抗布菌的细胞因子中发挥首要作用;也可由MHC I递呈给CD8+ T淋巴细胞,使其活化成细胞毒性T淋巴细胞,杀伤感染布菌的靶细胞[64]。TH细胞包含TH1亚型和TH2亚型,它们分别是介导布菌感染前期细胞免疫和后期体液免疫的优势亚群[65-66]。而控制TH细胞分化方向的关键是cMAF。cMAF是促进CD4+ T细胞依赖IL-4向TH2亚型分化的特异性转录因子[67]。在2015年,Ranzani等[68]首次发现linc-MAF-4可抑制cMAF活性。最近,布病患者的血清学检测结果证实,过表达的linc-MAF-4调控CD4+ T细胞在感染早期趋向TH1亚型分化[69]。另外,miR-155最近也被证实可调控TH细胞的发育分化来应对布菌感染[70-71]。
2.4.2.2 NF-κB信号通路 由接头蛋白MyD88或RIP2激活的IKK复合物促使NF-κB与抑制因子IκB解聚,暴露出核定位序列,由胞浆转运至核内,启动细胞因子表达。若阻断NF-κB,则显著抑制肿瘤坏死因子α(tumor necrosis factor alpha,TNF-α)的表达和分泌。同时,TNF-α也是一种具有调节作用的促炎细胞因子,其与受体TNFR-1的结合能导致IκB泛素化,正反馈于NF-κB途径,减少抗炎细胞因子(如IL-10),增加促炎细胞因子(如活性氧和一氧化氮)[72]。但是,由TNF-α所诱导表达的一种泛素编辑酶A20,也称肿瘤坏死因子α诱导蛋白3(tumor necrosis factor alpha-induced protein 3,TNFAIP3),它能抑制NF-κB活化,该基因座的相关风险变异能增进IL-6和IL-1β产生,在应对布菌感染中发挥保护作用[73]。布菌通过影响巨噬细胞ncRNA的表达可抑制炎症反应的进行。比如,上调的Gm28309(竞争性内源RNA)能充当分子海绵富集miR-3068-5p,以阻止κB-Ras2蛋白的降解,抑制NF-κB的激活[74];miR-181a-5p和miR-21a-5p依赖于MyD88变化,前者上调,抑制TNF-α表达,后者下调,导致抗菌的鸟苷酸结合蛋白5(GBP5)减少和抗炎的IL-10增加[75]。
2.4.2.3 MAPK信号通路 丝裂原活化蛋白激酶(mitogen-activated protein kinase,MAPK)信号通路包括MAPK激酶激酶(MAP kinase kinase kinase,MAP3K)、MAPK激酶(MAP kinase kinase,MAP2K)和MAPK。由接头蛋白MyD88或RIP2激活的MAP3K逐级激活MAP2K和MAPK,MAPK家族成员包括ERK1/2、JNK和p38。MAPK级联系统调控免疫和炎症反应,或还影响BCVs的转运与定位。早先,在小鼠巨噬细胞中发现布菌LPS通过限制MAPK信号通路(p38、ERK1/2和JNK)的激活,抑制TNF-α等细胞因子的合成[76]。随后的研究指出,由TLR2诱导的MAPK信号通路(p38和ERK1/2)激活能抑制由TLR9介导的IL-12产生[77]。布菌在人单核细胞内的维持与复制反而依赖于由TLR2介导的MAPK信号通路(p38和JNK)延迟激活[78]。MAPK信号通路可激活转录因子激活蛋白-1(activator protein-1,AP-1),有效促进炎症反应清除布菌。AP-1是由原癌基因c-Fos和c-Jun所编码的蛋白二聚体,其中,c-Fos可通过抑制促炎因子IL-10的产生来限制布菌的生存,但也会负调控TNF-α、IL-6和IL-10等抗炎因子,并且c-Fos还负反馈于TLR4[79]。综上,在响应布菌刺激时,NF-κB与MAPK通路之间密切联系,信号串扰(crosstalk)。
2.4.2.4 其他细胞因子 在布菌胞内寄生的各环节,某些炎症因子可作用于囊泡、溶酶体和内质网来控制感染。例如:1)溶酶体相关细胞器生物发生复合物-1亚基1(biogenesis of lysosome-related organelles complex-1 subunit 1,BLOS1)在线粒体和溶酶体的功能行使以及囊泡和内体的胞内运输中承担重要功能[80]。Wells等[81]发现BLOS1是防御布菌感染的新型免疫因子,而布菌能通过内质网应激途径的IRE1α-RIDD轴定向降解翻译BLOS1的mRNA。2)干扰素刺激基因家族(interferon-stimulated genes,ISGs)表达的干扰素诱导跨膜蛋白(IFITM3)是先天免疫的关键效应器[82],可介导溶酶体pH降低,有效抑制布菌等多种胞内病原体[83]。经布菌刺激的小鼠巨噬细胞能分泌包含IFITM3的外泌体,广泛地增强微环境中未感染细胞的免疫能力[84]。3)天然抗性相关巨噬蛋白1(natural resistance associated macrophage protein 1,NRAMP1)通过竞争性消耗布菌[85-86]和分枝杆菌[87]等胞内寄生菌繁殖所必需的铁、镁等二价金属离子而发挥抗菌作用。
3 绵羊抗布病育种研究存在的问题目前,绵羊抗布病育种工作还处于研究阶段,正面临许多问题,如:1)遗传机制复杂,易受环境因素影响,目前绵羊抗布病性状的遗传力及其与生产性状的相关作用有待评估;2)筛选指标不明确,目前抗布病性状主要用免疫指标表征,如血清抗体滴度,但将其视作阈性状抑或数量性状还有待研究;3)主效基因稀缺,目前国内外挖掘出的抗布病候选基因均非主效基因,不足以支撑分子标记辅助选择工作的展开;4)功能验证困难,布病为二类动物疫病,布菌攻毒试验要求在三级及以上生物安全实验室进行,增加了候选基因下游验证的难度。
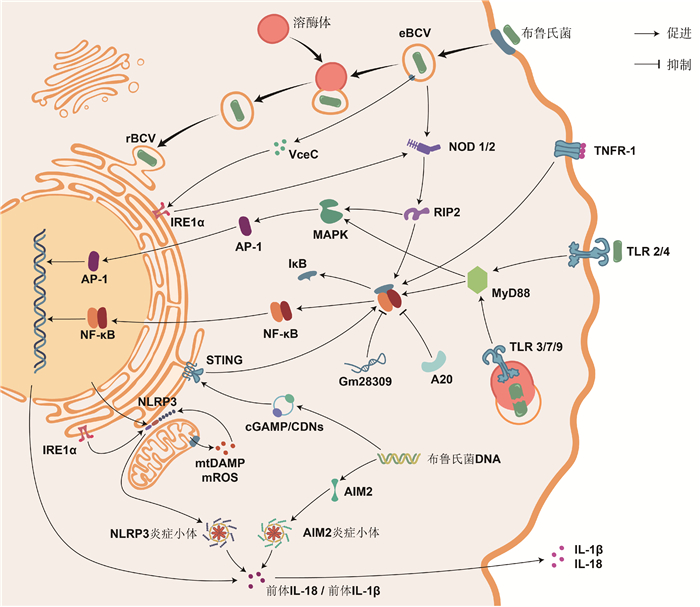
|
图 1 参与免疫和炎症反应的基因或ncRNA及其通路 Fig. 1 Genes or ncRNAs involved in immune and inflammatory responses and their pathways |
机器学习(mechine learning,ML)是人工智能领域近年新兴的分支,适用于数据驱动的科学,能有效处理基因组高维数据信息,在生物育种研究中展现出巨大潜力,为基因挖掘工作提供了全新视角。基于监督机器学习算法,例如决策树(decision tree,DT)、随机森林(random forest,RF)、支持向量机(support vector machines,SVM)和神经网络(neural network,NN)等[90],输入训练集、验证集和测试集数据的基因型特征与抗布病表型标签,建立结合年龄、性别和品种等效应的“基因多态性-抗布病性能”预测模型,以达到根据绵羊基因分型信息预测其抗布病能力表型的目的,实现依据特征值贡献比例筛选有效抗布病突变位点的目标。总之,随着布病研究的深入,布菌的毒力因子作用和免疫逃避机制日渐明晰,越来越多的研究者加入抗布病位点基因的挖掘和验证工作中,相信终将培育出兼顾优良经济性状的抗布病新品种绵羊。
| [1] |
PAPPAS G, PAPADIMITRIOU P, AKRITIDIS N, et al. The new global map of human brucellosis[J]. Lancet Infect Dis, 2006, 6(2): 91-99. DOI:10.1016/S1473-3099(06)70382-6 |
| [2] |
MORENO E. The one hundred year journey of the genus Brucella (Meyer and Shaw 1920)[J]. FEMS Microbiol Rev, 2021, 45(1): fuaa045. DOI:10.1093/femsre/fuaa045 |
| [3] |
高彦辉, 赵丽军, 孙殿军, 等. 布鲁氏菌病防治基础研究现状与展望[J]. 中国科学: 生命科学, 2014, 44(6): 628-635. GAO Y H, ZHAO L J, SUN D J, et al. Status and perspective of basic research related to the prevention and control of brucellosis[J]. Scientia Sinica Vitae, 2014, 44(6): 628-635. (in Chinese) |
| [4] |
MA J Y, WANG H, ZHANG X F, et al. MLVA and MLST typing of Brucella from Qinghai, China[J]. Infect Dis Poverty, 2016, 5: 26. DOI:10.1186/s40249-016-0123-z |
| [5] |
CAO X A, LI S E, LI Z C, et al. Enzootic situation and molecular epidemiology of Brucella in livestock from 2011 to 2015 in Qingyang, China[J]. Emerg Microbes Infect, 2018, 7(1): 1-8. |
| [6] |
YANG X W, PIAO D, MAO L L, et al. Whole-genome sequencing of rough Brucella melitensis in China provides insights into its genetic features[J]. Emerg Microbes Infect, 2020, 9(1): 2147-2156. DOI:10.1080/22221751.2020.1824549 |
| [7] |
汪洁英, 宁博, 景伟, 等. 布鲁氏菌病及其在我国的防控现状与建议[J]. 中国兽医科学, 2022, 52(12): 1578-1585. WANG J Y, NING B, JING W, et al. Research progress and suggestions regarding on the prevention and control of brucellosis in China: a review[J]. Chinese Veterinary Science, 2022, 52(12): 1578-1585. (in Chinese) |
| [8] |
JIANG H, O'CALLAGHAN D, DING J B. Brucellosis in China: history, progress and challenge[J]. Infect Dis Poverty, 2020, 9(1): 55. DOI:10.1186/s40249-020-00673-8 |
| [9] |
OLSEN S C, STOFFREGEN W S. Essential role of vaccines in brucellosis control and eradication programs for livestock[J]. Expert Rev Vaccines, 2005, 4(6): 915-928. DOI:10.1586/14760584.4.6.915 |
| [10] |
JIAO H W, ZHOU Z X, LI B W, et al. The mechanism of facultative intracellular parasitism of Brucella[J]. Int J Mol Sci, 2021, 22(7): 3673. DOI:10.3390/ijms22073673 |
| [11] |
CONDE-ÁLVAREZ R, ARCE-GORVEL V, IRIARTE M, et al. The lipopolysaccharide core of Brucella abortus acts as a shield against innate immunity recognition[J]. PLoS Pathog, 2012, 8(5): e1002675. DOI:10.1371/journal.ppat.1002675 |
| [12] |
MANCILLA M. Smooth to Rough dissociation in Brucella: the missing link to virulence[J]. Front Cell Infect Microbiol, 2016, 5: 98. |
| [13] |
ROOP Ⅱ R M, BARTON I S, HOPERSBERGER D, et al. Uncovering the hidden credentials of Brucella virulence[J]. Microbiol Mol Biol Rev, 2021, 85(1): e00021-19. |
| [14] |
COPIN R, VITRY M A, HANOT MAMBRES D, et al. In situ microscopy analysis reveals local innate immune response developed around Brucella infected cells in resistant and susceptible mice[J]. PLoS Pathog, 2012, 8(3): e1002575. DOI:10.1371/journal.ppat.1002575 |
| [15] |
LINGWOOD D, SIMONS K. Lipid rafts as a membrane-organizing principle[J]. Science, 2010, 327(5961): 46-50. DOI:10.1126/science.1174621 |
| [16] |
WATARAI M, MAKINO S I, FUJⅡ Y, et al. Modulation of Brucella-induced macropinocytosis by lipid rafts mediates intracellular replication[J]. Cell Microbiol, 2002, 4(6): 341-355. DOI:10.1046/j.1462-5822.2002.00195.x |
| [17] |
ARELLANO-REYNOSO B, LAPAQUE N, SALCEDO S, et al. Cyclic β-1, 2-glucan is a brucella virulence factor required for intracellular survival[J]. Nat Immunol, 2005, 6(6): 618-625. DOI:10.1038/ni1202 |
| [18] |
GUIDOLIN L S, ARCE-GORVEL V, CIOCCHINI A E, et al. Cyclic β-glucans at the bacteria-host cells interphase: one sugar ring to rule them all[J]. Cell Microbiol, 2018, 20(6): e12850. DOI:10.1111/cmi.12850 |
| [19] |
LEE J J, KIM D G, KIM D H, et al. Interplay between clathrin and Rab5 controls the early phagocytic trafficking and intracellular survival of Brucella abortus within HeLa cells[J]. J Biol Chem, 2013, 288(39): 28049-28057. DOI:10.1074/jbc.M113.491555 |
| [20] |
METTLEN M, CHEN P H, SRINIVASAN S, et al. Regulation of clathrin-mediated endocytosis[J]. Annu Rev Biochem, 2018, 87: 871-896. DOI:10.1146/annurev-biochem-062917-012644 |
| [21] |
EHRLICH M, BOLL W, VAN OIJEN A, et al. Endocytosis by random initiation and stabilization of clathrin-coated pits[J]. Cell, 2004, 118(5): 591-605. DOI:10.1016/j.cell.2004.08.017 |
| [22] |
CELLI J, DE CHASTELLIER C, FRANCHINI D M, et al. Brucella evades macrophage killing via VirB-dependent sustained interactions with the endoplasmic reticulum[J]. J Exp Med, 2003, 198(4): 545-556. DOI:10.1084/jem.20030088 |
| [23] |
VON BARGEN K, GORVEL J P, SALCEDO S P. Internal affairs: investigating the Brucella intracellular lifestyle[J]. FEMS Microbiol Rev, 2012, 36(3): 533-562. DOI:10.1111/j.1574-6976.2012.00334.x |
| [24] |
BOSCHIROLI M L, OUAHRANI-BETTACHE S, FOULONGNE V, et al. The Brucella suis virB operon is induced intracellularly in macrophages[J]. Proc Natl Acad Sci U S A, 2002, 99(3): 1544-1549. DOI:10.1073/pnas.032514299 |
| [25] |
MILLER C N, SMITH E P, CUNDIFF J A, et al. A Brucella type IV effector targets the COG tethering complex to remodel host secretory traffic and promote intracellular replication[J]. Cell Host Microbe, 2017, 22(3): 317-329. e7. DOI:10.1016/j.chom.2017.07.017 |
| [26] |
FUGIER E, SALCEDO S P, DE CHASTELLIER C, et al. The glyceraldehyde-3-phosphate dehydrogenase and the small GTPase Rab 2 are crucial for Brucella replication[J]. PLoS Pathog, 2009, 5(6): e1000487. DOI:10.1371/journal.ppat.1000487 |
| [27] |
SEDZICKI J, TSCHON T, LOW S H, et al. 3D correlative electron microscopy reveals continuity of Brucella-containing vacuoles with the endoplasmic reticulum[J]. J Cell Sci, 2018, 131(4): jcs210799. |
| [28] |
RAMBOW-LARSEN A A, PETERSEN E M, GOURLEY C R, et al. Brucella regulators: self-control in a hostile environment[J]. Trends Microbiol, 2009, 17(8): 371-377. DOI:10.1016/j.tim.2009.05.006 |
| [29] |
STARR T, CHILD R, WEHRLY T D, et al. Selective subversion of autophagy complexes facilitates completion of the Brucella intracellular cycle[J]. Cell Host Microbe, 2012, 11(1): 33-45. DOI:10.1016/j.chom.2011.12.002 |
| [30] |
HIYOSHI H, ENGLISH B C, DIAZ-OCHOA V E, et al. Virulence factors perforate the pathogen-containing vacuole to signal efferocytosis[J]. Cell Host Microbe, 2022, 30(2): 163-170. e6. DOI:10.1016/j.chom.2021.12.001 |
| [31] |
张世栋, 金维江. 动物抗病育种研究进展[J]. 中国畜牧杂志, 1999, 35(4): 55-57. ZHANG S D, JIN W J. Research progress in animal disease resistance breeding[J]. Chinese Journal of Animal Science, 1999, 35(4): 55-57. DOI:10.3969/j.issn.0258-7033.1999.04.033 (in Chinese) |
| [32] |
朱猛进, 吴珍芳, 赵书红. 猪抗病育种研究进展及对几个认识问题的讨论[J]. 中国畜牧兽医, 2007, 34(4): 63-67. ZHU M J, WU Z F, ZHAO S H. Research progress on pig disease resistance breeding and discussion of several cognitive issues[J]. China Animal Husbandry & Veterinary Medicine, 2007, 34(4): 63-67. DOI:10.3969/j.issn.1671-7236.2007.04.019 (in Chinese) |
| [33] |
施启顺. 畜禽抗病育种研究进展[J]. 中国畜牧杂志, 1995, 31(6): 48-51. SHI Q S. Research progress in disease resistance breeding of livestock and poultry[J]. Chinese Journal of Animal Science, 1995, 31(6): 48-51. (in Chinese) |
| [34] |
CAMERON H S, HUGHES E H, GREGORY P W. Genetic resistance to brucellosis in swine[J]. J Anim Sci, 1942, 1(2): 106-110. DOI:10.2527/jas1942.12106x |
| [35] |
朱波. 鸡H/L选育系选择效果分析及功能基因筛选[D]. 北京: 中国农业科学院, 2019. ZHU B. Chicken H/L selection effect analysis and functional gene screening[D]. Beijing: Chinese Academy of Agricultural Sciences, 2019. (in Chinese) |
| [36] |
王笑言. 抗DHAV-3北京鸭专门化品系的选育及G2代群体的抗病力差异机制[D]. 北京: 中国农业科学院, 2017. WANG X Y. Study on the resistant breeding of Pekin duck and the differences in the second generation resistance to DHAV-3[D]. Beijing: Chinese Academy of Agricultural Sciences, 2017. (in Chinese) |
| [37] |
谢晓刚, 薛嘉, 康健, 等. 基因编辑技术发展及其在家畜上的应用[J]. 农业生物技术学报, 2019, 27(1): 139-149. XIE X G, XUE J, KANG J, et al. Development of gene editing techniques and its application in livestock[J]. Journal of Agricultural Biotechnology, 2019, 27(1): 139-149. (in Chinese) |
| [38] |
DENG S L, WU Q, YU K, et al. Changes in the relative inflammatory responses in sheep cells overexpressing of toll-like receptor 4 when stimulated with LPS[J]. PLoS One, 2012, 7(10): e47118. DOI:10.1371/journal.pone.0047118 |
| [39] |
LI Y, LIAN D, DENG S L, et al. Efficient production of pronuclear embryos in breeding and nonbreeding season for generating transgenic sheep overexpressing TLR4[J]. J Anim Sci Biotechnol, 2016, 7: 38. DOI:10.1186/s40104-016-0096-6 |
| [40] |
杨爱玲, 李广栋, 吴昊, 等. 褪黑素合成酶AANAT/ASMT基因过表达绵羊生物安全评价研究[J]. 畜牧兽医学报, 2020, 51(7): 1563-1572. YANG A L, LI G D, WU H, et al. Study of biosafety evaluation on melatonin synthase AANAT/ASMT overexpressed sheep[J]. Acta Veterinaria et Zootechnica Sinica, 2020, 51(7): 1563-1572. (in Chinese) |
| [41] |
MA T, TAO J L, YANG M H, et al. An AANAT/ASMT transgenic animal model constructed with CRISPR/Cas9 system serving as the mammary gland bioreactor to produce melatonin-enriched milk in sheep[J]. J Pineal Res, 2017, 63(1): e12406. DOI:10.1111/jpi.12406 |
| [42] |
LI G D, LV D Y, YAO Y J, et al. Overexpression of ASMT likely enhances the resistance of transgenic sheep to brucellosis by influencing immune-related signaling pathways and gut microbiota[J]. FASEB J, 2021, 35(9): e21783. DOI:10.1096/fj.202100651R |
| [43] |
李林召, 张龙超. 国内外抗病育种技术研究进展[J]. 中国畜牧兽医, 2009, 36(9): 104-106. LI L Z, ZHANG L C. Research progress of disease resistance breeding technology at home and abroad[J]. China Animal Husbandry & Veterinary Medicine, 2009, 36(9): 104-106. (in Chinese) |
| [44] |
LEMAITRE B, NICOLAS E, MICHAUT L, et al. The dorsoventral regulatory gene cassette spätzle/Toll/cactus controls the potent antifungal response in drosophila adults[J]. Cell, 1996, 86(6): 973-983. DOI:10.1016/S0092-8674(00)80172-5 |
| [45] |
KAWAI T, AKIRA S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity[J]. Immunity, 2011, 34(5): 637-650. DOI:10.1016/j.immuni.2011.05.006 |
| [46] |
GIAMBARTOLOMEI G H, ZWERDLING A, CASSATARO J, et al. Lipoproteins, not lipopolysaccharide, are the key mediators of the proinflammatory response elicited by heat-killed Brucella abortus[J]. J Immunol, 2004, 173(7): 4635-4642. DOI:10.4049/jimmunol.173.7.4635 |
| [47] |
CAMPOS P C, GOMES M T R, GUIMARÃES E S, et al. TLR7 and TLR3 sense Brucella abortus RNA to induce proinflammatory cytokine production but they are dispensable for host control of infection[J]. Front Immunol, 2017, 8: 28. |
| [48] |
CAMPOS M A, ROSINHA G M S, ALMEIDA I C, et al. Role of toll-like receptor 4 in induction of cell-mediated immunity and resistance to Brucella abortus infection in mice[J]. Infect Immun, 2004, 72(1): 176-186. DOI:10.1128/IAI.72.1.176-186.2004 |
| [49] |
GOMES M T, CAMPOS P C, PEREIRA G D S, et al. TLR9 is required for MAPK/NF-κB activation but does not cooperate with TLR2 or TLR6 to induce host resistance to Brucella abortus[J]. J Leukoc Biol, 2016, 99(5): 771-780. DOI:10.1189/jlb.4A0815-346R |
| [50] |
MACEDO G C, MAGNANI D M, CARVALHO N B, et al. Central role of MyD88-dependent dendritic cell maturation and proinflammatory cytokine production to control Brucella abortus infection[J]. J Immunol, 2008, 180(2): 1080-1087. DOI:10.4049/jimmunol.180.2.1080 |
| [51] |
CIRL C, WIESER A, YADAV M, et al. Subversion of Toll-like receptor signaling by a unique family of bacterial Toll/interleukin-1 receptor domain-containing proteins[J]. Nat Med, 2008, 14(4): 399-406. DOI:10.1038/nm1734 |
| [52] |
SMITH J A, KHAN M, MAGNANI D D, et al. Brucella induces an unfolded protein response via TcpB that supports intracellular replication in macrophages[J]. PLoS Pathog, 2013, 9(12): e1003785. DOI:10.1371/journal.ppat.1003785 |
| [53] |
GOMES M T R, CAMPOS P C, DE ALMEIDA L A, et al. The role of innate immune signals in immunity to Brucella abortus[J]. Front Cell Infect Microbiol, 2012, 2: 130. |
| [54] |
KEESTRA-GOUNDER A M, BYNDLOSS M X, SEYFFERT N, et al. NOD1 and NOD2 signalling links ER stress with inflammation[J]. Nature, 2016, 532(7599): 394-397. DOI:10.1038/nature17631 |
| [55] |
QIN Q M, PEI J W, ANCONA V, et al. RNAi screen of endoplasmic reticulum-associated host factors reveals a role for IRE1α in supporting Brucella replication[J]. PLoS Pathog, 2008, 4(7): e1000110. DOI:10.1371/journal.ppat.1000110 |
| [56] |
GOMES M T R, CAMPOS P C, OLIVEIRA F S, et al. Critical role of ASC inflammasomes and bacterial type IV secretion system in caspase-1 activation and host innate resistance to Brucella abortus infection[J]. J Immunol, 2013, 190(7): 3629-3638. DOI:10.4049/jimmunol.1202817 |
| [57] |
BRONNER D N, ABUAITA B H, CHEN X Y, et al. Endoplasmic reticulum stress activates the inflammasome via NLRP3-and caspase-2-driven mitochondrial damage[J]. Immunity, 2015, 43(3): 451-462. DOI:10.1016/j.immuni.2015.08.008 |
| [58] |
MARIM F M, FRANCO M M C, GOMES M T R, et al. The role of NLRP3 and AIM2 in inflammasome activation during Brucella abortus infection[J]. Semin Immunopathol, 2017, 39(2): 215-223. DOI:10.1007/s00281-016-0581-1 |
| [59] |
ISHIKAWA H, BARBER G N. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling[J]. Nature, 2008, 455(7213): 674-678. DOI:10.1038/nature07317 |
| [60] |
BARBER G N. STING: infection, inflammation and cancer[J]. Nat Rev Immunol, 2015, 15(12): 760-770. DOI:10.1038/nri3921 |
| [61] |
KHAN M, HARMS J S, LIU Y P, et al. Brucella suppress STING expression via miR-24 to enhance infection[J]. PLoS Pathog, 2020, 16(10): e1009020. DOI:10.1371/journal.ppat.1009020 |
| [62] |
GOMES M T R, GUIMARÃES E S, MARINHO F V, et al. STING regulates metabolic reprogramming in macrophages via HIF-1α during Brucella infection[J]. PLoS Pathog, 2021, 17(5): e1009597. DOI:10.1371/journal.ppat.1009597 |
| [63] |
GUIMARÃES E S, GOMES M T R, SANCHES R C O, et al. The endoplasmic reticulum stress sensor IRE1α modulates macrophage metabolic function during Brucella abortus infection[J]. Front Immunol, 2022, 13: 1063221. |
| [64] |
BARRIONUEVO P, GIAMBARTOLOMEI G H. Inhibition of antigen presentation by Brucella: many more than many ways[J]. Microbes Infect, 2019, 21(3-4): 136-142. DOI:10.1016/j.micinf.2018.12.004 |
| [65] |
RAFIEI A, ARDESTANI S K, KARIMINIA A, et al. Dominant Th1 cytokine production in early onset of human brucellosis followed by switching towards Th2 along prolongation of disease[J]. J Infect, 2006, 53(5): 315-324. DOI:10.1016/j.jinf.2005.11.024 |
| [66] |
ZHENG R J, XIE S S, ZHANG Q, et al. Circulating Th1, Th2, Th17, Treg, and PD-1 levels in patients with brucellosis[J]. J Immunol Res, 2019, 2019: 3783209. |
| [67] |
HWANG E S, WHITE I A, HO I C. An IL-4-independent and CD25-mediated function of c-maf in promoting the production of Th2 cytokines[J]. Proc Natl Acad Sci U S A, 2002, 99(20): 13026-13030. DOI:10.1073/pnas.202474499 |
| [68] |
RANZANI V, ROSSETTI G, PANZERI I, et al. The long intergenic noncoding RNA landscape of human lymphocytes highlights the regulation of T cell differentiation by linc-MAF-4[J]. Nat Immunol, 2015, 16(3): 318-325. DOI:10.1038/ni.3093 |
| [69] |
GHEITASI R, KERAMAT F, SOLGI G, et al. Investigation of Linc-MAF-4 expression as an effective marker in brucellosis[J]. Mol Immunol, 2020, 123: 60-63. DOI:10.1016/j.molimm.2020.04.022 |
| [70] |
THAI T H, CALADO D P, CASOLA S, et al. Regulation of the germinal center response by microRNA-155[J]. Science, 2007, 316(5824): 604-608. DOI:10.1126/science.1141229 |
| [71] |
ZHANG X, CHEN J J, CHENG H M, et al. MicroRNA-155 expression with Brucella infection in vitro and in vivo and decreased serum levels of MicroRNA-155 in patients with brucellosis[J]. Sci Rep, 2022, 12(1): 4181. DOI:10.1038/s41598-022-08180-6 |
| [72] |
HOP H T, REYES A W B, HUY T X N, et al. Activation of NF-κB-mediated TNF-induced antimicrobial immunity is required for the efficient Brucella abortus clearance in RAW 264.7 cells[J]. Front Cell Infect Microbiol, 2017, 7: 437. DOI:10.3389/fcimb.2017.00437 |
| [73] |
LOU L X, BAO W G, LIU X J, et al. An autoimmune disease-associated risk variant in the TNFAIP3 gene plays a protective role in brucellosis that is mediated by the NF-κB signaling pathway[J]. J Clin Microbiol, 2018, 56(4): e01363-17. |
| [74] |
DENG X M, GUO J, SUN Z H, et al. Brucella-induced downregulation of lncRNA Gm28309 triggers macrophages inflammatory response through the miR-3068-5p/NF-κB pathway[J]. Front Immunol, 2020, 11: 581517. DOI:10.3389/fimmu.2020.581517 |
| [75] |
CORSETTI P P, DE ALMEIDA L A, GONÇALVES A N A, et al. miR-181a-5p regulates TNF-α and miR-21a-5p influences gualynate-binding protein 5 and IL-10 expression in macrophages affecting host control of brucella abortus infection[J]. Front Immunol, 2018, 9: 1331. DOI:10.3389/fimmu.2018.01331 |
| [76] |
JIMÉNEZ DE BAGVÉS M P, GROSS A, TERRAZA A, et al. Regulation of the mitogen-activated protein kinases by Brucella spp. expressing a smooth and rough phenotype: relationship to pathogen invasiveness[J]. Infect Immun, 2005, 73(5): 3178-3183. DOI:10.1128/IAI.73.5.3178-3183.2005 |
| [77] |
ZHANG C Y, BAI N, ZHANG Z H, et al. TLR2 signaling subpathways regulate TLR9 signaling for the effective induction of IL-12 upon stimulation by heat-killed Brucella abortus[J]. Cell Mol Immunol, 2012, 9(4): 324-333. DOI:10.1038/cmi.2012.11 |
| [78] |
DIMITRAKOPOULOS O, LIOPETA K, DIMITRACOPOULOS G, et al. Replication of Brucella melitensis inside primary human monocytes depends on mitogen activated protein kinase signaling[J]. Microbes Infect, 2013, 15(6-7): 450-460. DOI:10.1016/j.micinf.2013.04.007 |
| [79] |
HOP H T, ARAYAN L T, HUY T X N, et al. The key role of c-fos for immune regulation and bacterial dissemination in Brucella infected macrophage[J]. Front Cell Infect Microbiol, 2018, 8: 287. DOI:10.3389/fcimb.2018.00287 |
| [80] |
SCOTT I, WANG L D, WU K Y, et al. GCN5L1/BLOS1 links acetylation, organelle remodeling, and metabolism[J]. Trends Cell Biol, 2018, 28(5): 346-355. DOI:10.1016/j.tcb.2018.01.007 |
| [81] |
WELLS K M, HE K, PANDEY A, et al. Brucella activates the host RIDD pathway to subvert BLOS1-directed immune defense[J]. Elife, 2022, 11: e73625. DOI:10.7554/eLife.73625 |
| [82] |
WONG M T, CHEN S S L. Emerging roles of interferon-stimulated genes in the innate immune response to hepatitis C virus infection[J]. Cell Mol Immunol, 2016, 13(1): 11-35. DOI:10.1038/cmi.2014.127 |
| [83] |
RANJBAR S, HARIDAS V, JASENOSKY L D, et al. A role for IFITM proteins in restriction of Mycobacterium tuberculosis infection[J]. Cell Rep, 2015, 13(5): 874-883. DOI:10.1016/j.celrep.2015.09.048 |
| [84] |
YI J H, WANG Y L, ZHANG H, et al. Interferon-inducible transmembrane protein 3-containing exosome as a new carrier for the cell-to-cell transmission of anti-Brucella activity[J]. Front Vet Sci, 2021, 8: 642968. DOI:10.3389/fvets.2021.642968 |
| [85] |
SATHIYASEELAN J, JIANG X, BALDWIN C L. Growth of Brucella abortus in macrophages from resistant and susceptible mouse strains[J]. Clin Exp Immunol, 2000, 121(2): 289-294. |
| [86] |
BORRIELLO G, CAPPARELLI R, BIANCO M, et al. Genetic resistance to Brucella abortus in the water buffalo (Bubalus bubalis)[J]. Infect Immun, 2006, 74(4): 2115-2120. DOI:10.1128/IAI.74.4.2115-2120.2006 |
| [87] |
ANTELO G T, VILA A J, GIEDROC D P, et al. Molecular evolution of transition metal bioavailability at the host-pathogen interface[J]. Trends Microbiol, 2021, 29(5): 441-457. DOI:10.1016/j.tim.2020.08.001 |
| [88] |
TALTY A, DEEGAN S, LJUJIC M, et al. Inhibition of IRE1α RNase activity reduces NLRP3 inflammasome assembly and processing of pro-IL1β[J]. Cell Death Dis, 2019, 10(9): 622. DOI:10.1038/s41419-019-1847-z |
| [89] |
COSTA FRANCO M M S, MARIM F M, ALVES-SILVA J, et al. AIM2 senses Brucella abortus DNA in dendritic cells to induce IL-1β secretion, pyroptosis and resistance to bacterial infection in mice[J]. Microbes Infect, 2019, 21(2): 85-93. DOI:10.1016/j.micinf.2018.09.001 |
| [90] |
ENOMA D O, BISHUNG J, ABIODUN T, et al. Machine learning approaches to genome-wide association studies[J]. J King Saud Univ Sci, 2022, 34(4): 101847. DOI:10.1016/j.jksus.2022.101847 |
(编辑 郭云雁)



