我国是一个养殖大国,近年来畜禽肉产品总产量保持稳中有升,基本能够满足消费市场需求。然而随着人们生活水平的提高,高端优质肉产品深受消费市场青睐,但目前高端优质肉的市场供应并不能满足消费者的需求。脂肪沉积是影响肉品质的重要因素,动物体脂分布在躯体皮下、腹腔、肌间和肌内等不同部位,其中肌内脂肪含量的高低是影响肉质的关键因素之一,其与风味、多汁性和嫩度等密切相关,对肉质性状的形成发挥着重要作用,因此探究脂肪沉积的分子机理对解析动物经济性状形成的规律具有重要意义。脂肪沉积受到动物品种、营养水平及表观遗传等多方面因素影响。近年来研究发现,DNA甲基化和非编码RNA调控等表观遗传修饰在动物脂肪沉积中发挥着关键作用[1-3],表观遗传修饰是调控脂肪沉积的关键因素,而m6A甲基化修饰是真核生物中含量最丰富的一种RNA表观遗传修饰,探究其调控作用与分子机制对脂肪生物学和动物性状改良具有重要作用[4-5]。因此,本文将综述有关m6A修饰、IGF2BP2及其对动物脂肪沉积影响的研究进展,并展望该领域未来的研究方向与发展趋势。
1 IGF2BP2介导的m6A修饰概述 1.1 RNA的m6A甲基化修饰过程RNA是实现遗传信息在蛋白质上表达、促进遗传信息向表型转化过程的桥梁。绝大多数生物体的RNA都可以接受多种化学修饰,其中m6A是真核生物mRNA中最丰富的化学修饰之一[6]。m6A是mRNA中腺嘌呤第6位N原子上的甲基化修饰,其修饰位点常存在于mRNA的CDS区和3′UTR区,主要发生在RRACH序列上,mRNA上可以含有一个或多个m6A修饰位点[7]。m6A修饰可以影响mRNA的运输、降解、翻译以及代谢等,在细胞增殖、分化、凋亡及免疫等多种生物学过程中发挥重要调控作用[8-10]。
RNA的m6A修饰涉及3种类型的蛋白质:甲基转移酶(writer)、去甲基酶(eraser)和m6A阅读蛋白(reader)。在细胞核内存在由多个亚基组成的writer复合体[11-12],此种复合体主要由METTL3、METTL14和Wilms肿瘤相关蛋白(WTAP)三个必不可少的亚基组成。METTL3充当重要的催化亚基[13-14],而METTL14则提供RNA结合支架,激活并增强METTL3的催化活性。WTAP与METTL3和METTL14相互作用,从而调节RNA的m6A修饰水平。writer复合体的各个亚基相互作用共同执行m6A修饰的写入功能[15]。m6A修饰处于动态调控之中,目前已知有两种不同的酶参与m6A去甲基化过程:FTO和ALKBH5[16-17],FTO是第一个被发现的m6A mRNA去甲基化酶,FTO被敲除后m6A峰升高[18-19],其调控脂肪相关基因的表达,并影响脂质代谢多个过程[20-21]。当mRNA进入细胞质后会与特定reader蛋白结合,不同的m6A reader蛋白有着不同的功能,如具有YTH结构域YTHDF1和YTHDF3通过协同作用提高mRNA的翻译效率,YTHDC1对于m6A介导的选择性剪接有一定的作用[22]。m6A reader蛋白除了参与脂肪细胞mRNA的稳定、翻译和降解等过程外(图 1),还参与调控肿瘤发生、血细胞生成、病毒复制和免疫应答等生物学过程[23-24]。
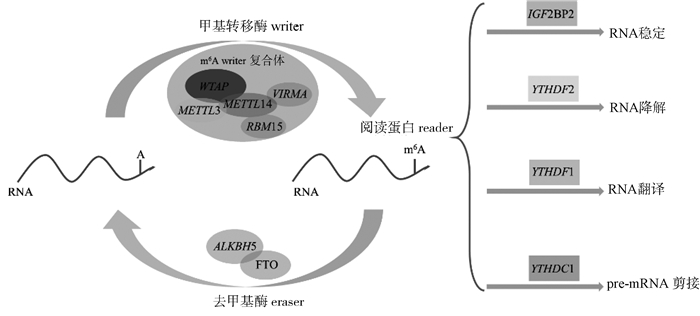
|
图 1 m6A甲基化修饰的作用机制 Fig. 1 The mechanism of m6A methylation modification |
IGF2BP蛋白(包括IGF2BP1/2/3)作为m6A阅读蛋白的一个新家族,通过识别一致的GG(m6A)C序列靶向数千个mRNA转录本[25]。与含有YTH结构域的蛋白家族促进mRNA衰变的功能相反,IGF2BPs蛋白成员在正常和应激条件下以m6A依赖的方式促进靶mRNA的稳定性和翻译,从而影响基因表达丰度[26]。IGF2BP1/2/3可以参与RNA生命周期调控,每个IGF2BP结合蛋白都能结合多种RNA,包括长链非编码RNA (lncRNA)和mRNA,以调控它们的剪接、运输、翻译和稳定性[27]。研究表明这三种IGF2BP家族基因在小鼠胚胎中协同表达,开始于胚胎发育第10.5天,峰值为第12.5天。IGF2BP1和IGF2BP3的表达在出生后基本消失,而IGF2BP2在出生后广泛表达,因此研究IGF2BP2对于性状表观遗传机制的调控尤为重要[28]。IGF2BP2是一种参与调节多种生物过程的RNA结合蛋白,且高度保守,由两个RNA识别基序结构域和4个hnRNP K同源结构域组成[29],其中K同源结构域是IGF2BP2识别m6A的必要结构域,通过与m6A修饰的相互识别可以提高m6A修饰mRNA的稳定性[30]。研究表明IGF2BP2影响糖尿病[31-32]、肥胖症和脂肪肝等多种疾病[33-34],且关于癌症的研究较多[35-37]。在肌肉生物学领域,有研究表明lncMyoD可以直接与IGF2BP2结合,负调控IGF2BP2介导的骨骼肌增殖基因N-Ras和c-Myc的翻译,表明IGF2BP2可以阻止肌细胞增殖以促进成肌分化[38],但对于IGF2BP2是否会依赖m6A或与其它lncRNAs及功能基因mRNA互作来调控脂肪沉积目前尚不清楚,有必要进行进一步研究。
2 m6A修饰对RNA的影响 2.1 m6A修饰影响RNA前体的剪接过程真核生物在转录过程中形成的RNA并不能直接翻译成蛋白,须经过一个剪接过程,剪接过程涉及对mRNA中内含子的识别、精准切除以及被切除后的外显子mRNA连接产生成熟mRNA,此过程中需要剪接因子参与。研究发现,WTAP作为m6A writer复合体成员之一,可以发挥剪接因子的作用[39],敲除WTAP或METTL3将会产生不同的mRNA亚型。在m6A去甲基化酶FTO敲除的前体脂肪细胞中,m6A修饰水平发生改变,导致剪接调控蛋白SRSF2对m6A mRNA进行可变剪接,产生不同的剪接产物,对脂肪沉积产生不同的影响[40]。因此,m6A修饰会影响mRNA的剪接过程。
2.2 m6A修饰调控RNA的出核RNA经剪接后进入到细胞质,出核的过程受到m6A修饰的调控。研究表明细胞中存在一种TAP-P15复合体[41],而ASF/SF2的磷酸化水平决定这种复合体与mRNA的结合能力,当m6A去甲基化酶ALKBH5缺失时,m6A修饰水平增强使ASF/SF2去磷酸化程度上升,导致TAP-P15复合体与mRNA的结合能力上升,mRNA的核输出能力增强[42]。DEAD-box(DDX)解旋酶对RNA的识别和新陈代谢至关重要。DDX46通过其特有的螺旋结构域招募ALKBH5使与之结合的信号分子RNA去甲基化,导致其RNA不能出核[43]。因此,m6A修饰可以调控mRNA的出核过程。
2.3 m6A修饰调控RNA的稳定性和降解遗传信息通过表达蛋白实现其生物学功能,mRNA是用于翻译蛋白质的直接模板。大多数m6A修饰富集于外显子区和终止密码子区域[44-45]。在前体mRNA被剪接后形成成熟mRNA的过程中,m6A修饰被保留下来存在于成熟mRNA中。在调控mRNA稳定性方面,IGF2BP采用一种m6A依赖性方式来提高目标mRNA的稳定性[23]。HUR作为另外一种RNA结合蛋白,在各种组织中普遍表达[46],其包含核质穿梭序列(HNS),HNS的转录后修饰控制HUR在细胞核和细胞质之间的移位。HUR的功能依赖于动态的亚细胞定位,一旦HUR从细胞核穿梭到细胞质,导致HUR增加了目标RNA的稳定性和翻译[47-49]。当m6A与含有YTH结构域的YTHDF2结合后会导致m6A修饰的mRNA降解[50-51]。此外,有研究发现敲低胚胎干细胞中METTL3或METTL14后,其靶mRNA的稳定性降低[52-53]。因此,m6A修饰会影响m6A mRNA的稳定性与降解。
3 IGF2BP2介导的m6A修饰调控动物脂肪沉积 3.1 调控白色脂肪沉积白色脂肪是脂肪沉积的主要场所,其广泛分布在动物机体皮下组织和内脏周围,其中皮下脂肪含有较高的饱和脂肪酸,其摄入量过高会导致胆固醇、甘油三酯以及低密度脂蛋白升高,增加患各种心血管疾病的风险[54];而肌内脂肪含量与肉品质以及胰岛素敏感性等紧密相关[55]。因此, 研究白色脂肪沉积不仅对于动物肉质的改善有重要作用,且对人类健康也具有一定意义。在白色脂肪的成脂过程中,m6A表观遗传修饰发挥着不可或缺的作用。lncRAP2的表达具有很高的组织特异性,其能诱导白色脂肪细胞的分化,lncRAP2主要存在于细胞质中,不与核糖体和RNA直接结合,但可以与RNA结合蛋白IGF2BP2结合形成复合体,影响RNA稳定性或降解(图 2)。通过分析白色脂肪细胞中IGF2BP2靶向的mRNAs,发现其选择性地与编码成脂调节因子和能量消耗相关的mRNAs结合,如瘦素、脂联素及白介素等[56-58],表明这些成脂调控因子转录本上可能存在m6A修饰位点。此外,研究发现lncRNA Hilnc与IGF2BP2相互作用调节PPAR途径介导的肝脂质沉积[59](图 2)。全身敲除IGF2BP2抵抗高脂诱导的肥胖和脂肪肝发生,而在肝组织特异性过表达IGF2BP2会导致肝的脂肪变性[60]。肝细胞特异性IGF2BP2敲除小鼠导致PPARα、PPARγ和CPT1A等基因的表达水平的显著下调,使体内的甘油三酯的运转速率加快,抑制肝脂肪沉积。因此,IGF2BP2通过m6A依赖的方式识别成脂或脂代谢因子以调控靶mRNA的稳定性影响脂肪沉积[61-63]。

|
A. IGF2BP2与lncRNA lncRAP2结合通过招募Exosc6和Ddx47调控脂代谢基因表达;B. IGF2BP2与lncRNA Hilnc互作通过PPARγ调控肝脂质积累 A. IGF2BP2 regulates lipid metabolism by binding to lncRNA lncRAP2 and recruiting Exosc6 and Ddx47; B. The interaction between IGF2BP2 and lncRNA Hilnc regulates hepatic lipid accumulation through PPARγ 图 2 IGF2BP2与lncRNA互作调控脂质代谢的作用机制 Fig. 2 The mechanism of interaction between IGF2BP2 and lncRNA regulating lipid metabolism |
在猪脂肪沉积方面,发现IGF2BP2等RNA结合蛋白与选择性剪接事件区域内的核心剪接体相互作用,进行前体mRNA剪接,并对脂质代谢造成影响[64-65]。此外,研究表明IGF2BP2蛋白与IGF2具有较强的亲和力[66],可调控IGF2的生理作用,而IGF2基因可作为猪脂肪沉积的关键基因[67]。以上研究表明IGF2BP2可能对猪脂肪沉积发挥重要调控作用。在羊脂肪沉积方面的研究发现IGF2BP2在未成年山羊的肌肉和脾组织表达量最高,肾、卵巢、胃和大脑次之,肺、心和肝表达量相对较低;而在成年山羊中,IGF2BP2表达量最高的组织是脂肪,输卵管、卵巢、心和肺次之,肾、肌肉、脾和肝表达量相对较低,推测IGF2BP2在山羊个体不同发育时期发挥着重要的作用,特别是在脂肪沉积过程中[68]。
3.2 调控棕色脂肪产热棕色脂肪组织是动物体内呈棕色样的脂肪组织,其脂肪细胞体积较小,胞质中有多个较小的脂滴,并有较多的线粒体[69]。棕色脂肪的功能是促进适应性产热、ATP产生和底物氧化的解偶联,通过非颤抖产热的方式来维持体温变化。因此,棕色脂肪细胞能够通过氧化其自身储存的脂质,从而产生热量并增加代谢率[70]。UCP1是一个很重要的解偶联基因,激活该基因可显著抑制饮食诱导的肥胖。研究发现IGF2BP2对UCP1基因具有转录抑制作用,在小鼠上敲除IGF2BP2导致棕色脂肪的UCP1蛋白丰度增加,造成棕色脂肪产热能力增强,并对饮食诱导的肥胖和脂肪肝具有更强的抵抗力,以及更高的葡萄糖耐受性、胰岛素敏感性和更好的御寒能力[71]。因此,IGF2BP2介导的m6A修饰与棕色脂肪产热息息相关。
4 总结与展望综上所述,IGF2BP2主要通过m6A依赖的方式影响脂肪沉积靶基因的稳定性,影响其RNA的丰度,进而调控动物脂肪沉积,但IGF2BP2介导的m6A修饰调控动物脂肪沉积的分子机制仍需进一步深入探究。因此,后续研究可借助多组学测序技术以及新的分子生物学方法挖掘IGF2BP2结合的m6A修饰下游靶基因,明确IGF2BP2介导的m6A修饰调控动物脂肪沉积的表观遗传机制,为畜禽遗传改良提供关键基因,并加快畜禽性状遗传改良进程,从而改善畜禽肉类品质,增加经济效益,满足国民对优质肉产品的需求。
| [1] |
SUN Y M, CHEN X C, QIN J, et al. Comparative analysis of long noncoding RNAs expressed during intramuscular adipocytes adipogenesis in fat-type and lean-type pigs[J]. J Agric Food Chem, 2018, 66(45): 12122-12130. DOI:10.1021/acs.jafc.8b04243 |
| [2] |
XU Z Y, YOU W J, CHEN W T, et al. Single-cell RNA sequencing and lipidomics reveal cell and lipid dynamics of fat infiltration in skeletal muscle[J]. J Cachexia Sarcopenia Muscle, 2021, 12(1): 109-129. DOI:10.1002/jcsm.12643 |
| [3] |
ZHANG Y F, ZHANG J J, GONG H F, et al. Genetic correlation of fatty acid composition with growth, carcass, fat deposition and meat quality traits based on GWAS data in six pig populations[J]. Meat Sci, 2019, 150: 47-55. DOI:10.1016/j.meatsci.2018.12.008 |
| [4] |
QIMUGE N, HE Z Z, QIN J, et al. Overexpression of DNMT3A promotes proliferation and inhibits differentiation of porcine intramuscular preadipocytes by methylating p21 and PPARg promoters[J]. Gene, 2019, 696: 54-62. DOI:10.1016/j.gene.2019.02.029 |
| [5] |
ZHAO C Z, WU H G, QIMUGE N, et al. MAT2A promotes porcine adipogenesis by mediating H3K27me3 at Wnt10b locus and repressing Wnt/β-catenin signaling[J]. Biochim Biophys Acta Mol Cell Biol Lipids, 2018, 1863(2): 132-142. |
| [6] |
SHI H L, WEI J B, HE C. Where, when, and how: context-dependent functions of RNA methylation writers, readers, and erasers[J]. Mol Cell, 2019, 74(4): 640-650. DOI:10.1016/j.molcel.2019.04.025 |
| [7] |
ZACCARA S, RIES R J, JAFFREY S R. Reading, writing and erasing mRNA methylation[J]. Nat Rev Mol Cell Biol, 2019, 20(10): 608-624. DOI:10.1038/s41580-019-0168-5 |
| [8] |
ROUNDTREE I A, EVANS M E, PAN T, et al. Dynamic RNA modifications in gene expression regulation[J]. Cell, 2017, 169(7): 1187-1200. DOI:10.1016/j.cell.2017.05.045 |
| [9] |
JIANG X L, LIU B Y, NIE Z, et al. The role of m6A modification in the biological functions and diseases[J]. Signal Transduct Target Ther, 2021, 6(1): 74. DOI:10.1038/s41392-020-00450-x |
| [10] |
张鑫芳, 乔成栋. mRNA m6A甲基化修饰在2型糖尿病发生发展中的作用机制研究进展[J]. 山东医药, 2022, 62(26): 85-88. ZHANG X F, QIAO C D. Research progress on the mechanism of mRNA m6A methylation modification in the occurrence and development of type 2 diabetes[J]. Shandong Medicine, 2022, 62(26): 85-88. DOI:10.3969/j.issn.1002-266X.2022.26.022 (in Chinese) |
| [11] |
MI S Y, SHI Y J, DARI G, et al. Function of m6A and its regulation of domesticated animals ' complex traits[J]. J Anim Sci, 2022, 100(3): skac034. DOI:10.1093/jas/skac034 |
| [12] |
OERUM S, MEYNIER V, CATALA M, et al. A comprehensive review of m6A/m6Am RNA methyltransferase structures[J]. Nucleic Acids Res, 2021, 49(13): 7239-7255. DOI:10.1093/nar/gkab378 |
| [13] |
BATISTA P J, MOLINIE B, WANG J K, et al. m6A RNA modification controls cell fate transition in mammalian embryonic stem cells[J]. Cell Stem Cell, 2014, 15(6): 707-719. DOI:10.1016/j.stem.2014.09.019 |
| [14] |
LIU J Z, YUE Y N, HAN D L, et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation[J]. Nat Chem Biol, 2014, 10(2): 93-95. DOI:10.1038/nchembio.1432 |
| [15] |
XU W Q, LI J H, HE C X, et al. METTL3 regulates heterochromatin in mouse embryonic stem cells[J]. Nature, 2021, 591(7849): 317-321. DOI:10.1038/s41586-021-03210-1 |
| [16] |
JIA G F, FU Y, ZHAO X, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO[J]. Nat Chem Biol, 2011, 7(12): 885-887. DOI:10.1038/nchembio.687 |
| [17] |
HUANG Y, YAN J L, LI Q, et al. Meclofenamic acid selectively inhibits FTO demethylation of m6A over ALKBH5[J]. Nucleic Acids Res, 2015, 43(1): 373-384. DOI:10.1093/nar/gku1276 |
| [18] |
潘明敏, 王启阳, 杨丽萍. FTO介导RNA的m6A修饰与发育的研究进展[J]. 基础医学与临床, 2022, 42(10): 1591-1595. PAN M M, WANG Q Y, YANG L P. Progress in research on m6A modification of FTO mediated-RNA and development[J]. Basic and Clinical Medicine, 2022, 42(10): 1591-1595. (in Chinese) |
| [19] |
CAO G C, LI H B, YIN Z N, et al. Recent advances in dynamic m6A RNA modification[J]. Open Biol, 2016, 6(4): 160003. DOI:10.1098/rsob.160003 |
| [20] |
JIA G F, FU Y, ZHAO X, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO[J]. Nat Chem Biol, 2011, 7(12): 885-887. DOI:10.1038/nchembio.687 |
| [21] |
宋兴亚, 彭巍, 刘贤, 等. m6A甲基化修饰及其影响动物脂肪生成的分子机制研究进展[J]. 中国畜牧杂志, 2022, 58(10): 53-58. SONG X Y, PENG W, LIU X, et al. The Research progress in methylation modification of m a and its molecular mechanism affecting the Adipogenesis of animals[J]. Chinese Journal of Animal Science, 2022, 58(10): 53-58. (in Chinese) |
| [22] |
XIAO W, ADHIKARI S, DAHAL U, et al. Nuclear m6A reader YTHDC1 regulates mRNA splicing[J]. Mol Cell, 2016, 61(4): 507-519. DOI:10.1016/j.molcel.2016.01.012 |
| [23] |
ZHAO Y C, SHI Y F, SHEN H F, et al. m6A-binding proteins: the emerging crucial performers in epigenetics[J]. J Hematol Oncol, 2020, 13(1): 35. DOI:10.1186/s13045-020-00872-8 |
| [24] |
王静, 闫爽. m6A修饰调控成脂分化的研究进展[J]. 东南大学学报(医学版), 2022, 41(6): 894-898. WANG J, YAN S. Research progress on the regulation of lipid differentiation by m6A modification[J]. Journal of Southeast University (Medical Science Edition), 2022, 41(6): 894-898. DOI:10.3969/j.issn.1671-6264.2022.06.025 (in Chinese) |
| [25] |
HUANG H L, WENG H Y, SUN W J, et al. Recognition of RNA N6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation[J]. Nat Cell Biol, 2018, 20(3): 285-295. DOI:10.1038/s41556-018-0045-z |
| [26] |
DAI N, RAPLEY J, ANGEL M, et al. mTOR phosphorylates IMP2 to promote IGF2 mRNA translation by internal ribosomal entry[J]. Genes Dev, 2011, 25(11): 1159-1172. DOI:10.1101/gad.2042311 |
| [27] |
HUANG H L, WENG H Y, SUN W J, et al. Publisher correction: recognition of RNA N6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation[J]. Nat Cell Biol, 2020, 22(10): 1288. DOI:10.1038/s41556-020-00580-y |
| [28] |
DAI N, ZHAO L P, WRIGHTING D, et al. IGF2BP2/IMP2-deficient mice resist obesity through enhanced translation of Ucp1 mRNA and other mRNAs encoding mitochondrial proteins[J]. Cell Metab, 2015, 21(4): 609-621. DOI:10.1016/j.cmet.2015.03.006 |
| [29] |
ZHANG P P, WU W Y, MA C F, et al. RNA-binding proteins in the regulation of adipogenesis and adipose function[J]. Cells, 2022, 11(15): 2357. DOI:10.3390/cells11152357 |
| [30] |
ZHANG X, YIN H L, ZHANG X F, et al. N6-methyladenosine modification governs liver glycogenesis by stabilizing the glycogen synthase 2 mRNA[J]. Nat Commun, 2022, 13(1): 7038. DOI:10.1038/s41467-022-34808-2 |
| [31] |
陈颖. 高胰岛素和高血糖对糖尿病患者IGF2BP2基因表达的影响[J]. 实用糖尿病杂志, 2020, 16(3): 80-81. CHEN Y. Effect of high insulin and high blood glucose on the IGF2BP2 gene expression of diabetes patients[J]. Journal of Practical Diabetology, 2020, 16(3): 80-81. (in Chinese) |
| [32] |
PENG T W, LIU M C, HU L, et al. LncRNA Airn alleviates diabetic cardiac fibrosis by inhibiting activation of cardiac fibroblasts via a m6A-IMP2-p53 axis[J]. Biol Direct, 2022, 17(1): 32. DOI:10.1186/s13062-022-00346-6 |
| [33] |
XU Z J, QIN Y, LV B B, et al. Intermittent fasting improves high-fat diet-induced obesity cardiomyopathy via alleviating lipid deposition and apoptosis and decreasing m6A methylation in the heart[J]. Nutrients, 2022, 14(2): 251. DOI:10.3390/nu14020251 |
| [34] |
HU X G, PENG W X, ZHOU H X, et al. IGF2BP2 regulates DANCR by serving as an N6-methyladenosine reader[J]. Cell Death Differ, 2020, 27(6): 1782-1794. DOI:10.1038/s41418-019-0461-z |
| [35] |
WANG Y, LU J H, WU Q N, et al. LncRNA LINRIS stabilizes IGF2BP2 and promotes the aerobic glycolysis in colorectal cancer[J]. Mol Cancer, 2019, 18(1): 174. DOI:10.1186/s12943-019-1105-0 |
| [36] |
PENG F, XU J, CUI B, et al. Oncogenic AURKA-enhanced N6-methyladenosine modification increases DROSHA mRNA stability to transactivate STC1 in breast cancer stem-like cells[J]. Cell Res, 2021, 31(3): 345-361. DOI:10.1038/s41422-020-00397-2 |
| [37] |
LI B T, ZHU L L, LU C L, et al. circNDUFB2 inhibits non-small cell lung cancer progression via destabilizing IGF2BPs and activating anti-tumor immunity[J]. Nat Commun, 2021, 12(1): 295. DOI:10.1038/s41467-020-20527-z |
| [38] |
GONG C G, LI Z Z, RAMANUJAN K, et al. A long non-coding RNA, LncMyoD, regulates skeletal muscle differentiation by blocking IMP2-mediated mRNA translation[J]. Dev Cell, 2015, 34(2): 181-191. DOI:10.1016/j.devcel.2015.05.009 |
| [39] |
PING X L, SUN B F, WANG L, et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase[J]. Cell Res, 2014, 24(2): 177-189. DOI:10.1038/cr.2014.3 |
| [40] |
ZHAO X, YANG Y, SUN B F, et al. FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis[J]. Cell Res, 2014, 24(12): 1403-1419. DOI:10.1038/cr.2014.151 |
| [41] |
TANIGUCHI I, OHNO M. ATP-dependent recruitment of export factor Aly/REF onto intronless mRNAs by RNA helicase UAP56[J]. Mol Cell Biol, 2008, 28(2): 601-608. DOI:10.1128/MCB.01341-07 |
| [42] |
ZHENG G Q, DAHL J A, NIU Y M, et al. Sprouts of RNA epigenetics: the discovery of mammalian RNA demethylases[J]. RNA Biol, 2013, 10(6): 915-918. DOI:10.4161/rna.24711 |
| [43] |
ZHENG Q L, HOU J, ZHOU Y, et al. The RNA helicase DDX46 inhibits innate immunity by entrapping m6A-demethylated antiviral transcripts in the nucleus[J]. Nat Immunol, 2017, 18(10): 1094-1103. DOI:10.1038/ni.3830 |
| [44] |
DOMINISSINI D, MOSHITCH-MOSHKOVITZ S, SCHWARTZ S, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq[J]. Nature, 2012, 485(7397): 201-206. DOI:10.1038/nature11112 |
| [45] |
LUO G Z, MACQUEEN A, ZHENG G Q, et al. Unique features of the m6A methylome in Arabidopsis thaliana[J]. Nat Commun, 2014, 5: 5630. DOI:10.1038/ncomms6630 |
| [46] |
HINMAN M N, LOU H. Diverse molecular functions of Hu proteins[J]. Cell Mol Life Sci, 2008, 65(20): 3168-3181. DOI:10.1007/s00018-008-8252-6 |
| [47] |
VISVANATHAN A, PATIL V, ARORA A, et al. Essential role of METTL3-mediated m6A modification in glioma stem-like cells maintenance and radioresistance[J]. Oncogene, 2018, 37(4): 522-533. DOI:10.1038/onc.2017.351 |
| [48] |
SIANG D T C, LIM Y C, KYAW A M M, et al. The RNA-binding protein HuR is a negative regulator in adipogenesis[J]. Nat Commun, 2020, 11(1): 213. DOI:10.1038/s41467-019-14001-8 |
| [49] |
SCHULTZ C, PREET R, DHIR T, et al. Understanding and targeting the disease-related RNA binding protein human antigen R (HuR)[J]. Wiley Interdiscip Rev RNA, 2020, 11(3): e1581. DOI:10.1002/wrna.1581 |
| [50] |
WANG L F, MCFADDEN J W, YANG G Q, et al. Effect of melatonin on visceral fat deposition, lipid metabolism and hepatic lipo-metabolic gene expression in male rats[J]. J Anim Physiol Anim Nutr (Berl), 2021, 105(4): 787-796. DOI:10.1111/jpn.13497 |
| [51] |
WANG J Y, LU A Q. The biological function of m6A reader YTHDF2 and its role in human disease[J]. Cancer Cell Int, 2021, 21(1): 109. DOI:10.1186/s12935-021-01807-0 |
| [52] |
GEULA S, MOSHITCH-MOSHKOVITZ S, DOMINISSINI D, et al. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation[J]. Science, 2015, 347(6225): 1002-1006. DOI:10.1126/science.1261417 |
| [53] |
WANG Y, LI Y, TOTH J I, et al. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells[J]. Nat Cell Biol, 2014, 16(2): 191-198. DOI:10.1038/ncb2902 |
| [54] |
宋丹丹, 宋延彬, 张富洋, 等. 白色脂肪组织来源细胞外囊泡在代谢性心血管疾病发病中的作用机制研究进展[J]. 山东医药, 2021, 61(32): 89-93. SONG D D, SONG Y B, ZHANG F Y, et al. Research progress on the Mechanism of White Adipose tissue-derived extracellular vesicles in the pathogenesis of Metabolic cardiovascular disease[J]. Shandong Med J, 2021, 61(32): 89-93. DOI:10.3969/j.issn.1002-266X.2021.32.023 (in Chinese) |
| [55] |
LIU F, CAI Z X, YANG Y, et al. The adipocyte-enriched secretory protein tetranectin exacerbates type 2 diabetes by inhibiting insulin secretion from β cells[J]. Sci Adv, 2022, 8(38): eabq1799. DOI:10.1126/sciadv.abq1799 |
| [56] |
唐妮, 王书瑶, 齐锦雯, 等. 脂联素调控脂质代谢的研究进展[J]. 畜牧兽医学报, 2018, 49(12): 2550-2557. TANG N, WANG S Y, QI J W, et al. Research progress on adiponectin regulating lipid metabolism[J]. Acta Veterinaria et Zootechnica Sinica, 2018, 49(12): 2550-2557. DOI:10.11843/j.issn.0366-6964.2018.12.003 (in Chinese) |
| [57] |
ALVAREZ-DOMINGUEZ J R, WINTHER S, HANSEN J B, et al. An adipose lncRAP2-Igf2bp2 complex enhances adipogenesis and energy expenditure by stabilizing target mRNAs[J]. iScience, 2021, 25(1): 103680. |
| [58] |
孟珊, 杨阳, 李睿霄, 等. lncRNA-6617调控猪肌内前体脂肪细胞分化的筛选与功能研究[J]. 畜牧兽医学报, 2022, 53(6): 1712-1722. MENG S, YANG Y, LI R X, et al. Screening and functional study of lncRNA-6617 regulating porcine intramuscular preadipocytes differentiation[J]. Acta Veterinaria et Zootechnica Sinica, 2022, 53(6): 1712-1722. (in Chinese) |
| [59] |
JIANG Y, PENG J Y, SONG J W, et al. Loss of Hilnc prevents diet-induced hepatic steatosis through binding of IGF2BP2[J]. Nat Metab, 2021, 3(11): 1569-1584. DOI:10.1038/s42255-021-00488-3 |
| [60] |
REGUÉ L, MINICHIELLO L, AVRUCH J, et al. Liver-specific deletion of IGF2 mRNA binding protein-2/IMP2 reduces hepatic fatty acid oxidation and increases hepatic triglyceride accumulation[J]. J Biol Chem, 2019, 294(31): 11944-11951. DOI:10.1074/jbc.RA119.008778 |
| [61] |
TYBL E, SHI F D, KESSLER S M, et al. Overexpression of the IGF2-mRNA binding protein p62 in transgenic mice induces a steatotic phenotype[J]. J Hepatol, 2011, 54(5): 994-1001. DOI:10.1016/j.jhep.2010.08.034 |
| [62] |
LAGGAI S, KESSLER S M, BOETTCHER S, et al. The IGF2 mRNA binding protein p62/IGF2BP2-2 induces fatty acid elongation as a critical feature of steatosis[J]. J Lipid Res, 2014, 55(6): 1087-1097. DOI:10.1194/jlr.M045500 |
| [63] |
KESSLER S M, LAGGAI S, VAN WONTERGHEM E, et al. Transient hepatic overexpression of insulin-like growth factor 2 induces free cholesterol and lipid droplet formation[J]. Front Physiol, 2016, 7: 328. |
| [64] |
NIKOLAOU K C, VATANDASLAR H, MEYER C, et al. The RNA-Binding protein A1CF regulates hepatic fructose and glycerol metabolism via alternative RNA splicing[J]. Cell Rep, 2019, 29(2): 283-300. DOI:10.1016/j.celrep.2019.08.100 |
| [65] |
XIAO P, GOODARZI P, PEZESHKI A, et al. RNA-seq reveals insights into molecular mechanisms of metabolic restoration via tryptophan supplementation in low birth weight piglet model[J]. J Anim Sci, 2022, 100(5): skac156. DOI:10.1093/jas/skac156 |
| [66] |
CAI Z X, SARUP P, OSTERSEN T, et al. Genomic diversity revealed by whole-genome sequencing in three Danish commercial pig breeds[J]. J Anim Sci, 2020, 98(7): skaa229. DOI:10.1093/jas/skaa229 |
| [67] |
YOUNIS S, SCHÖNKE M, MASSART J, et al. The ZBED6-IGF2 axis has a major effect on growth of skeletal muscle and internal organs in placental mammals[J]. Proc Natl Acad Sci USA, 2018, 115(9): E2048-E2057. |
| [68] |
辛东芸. 陕北白绒山羊IGF2BP2和IGF2BP3基因mRNA表达、变异位点检测及遗传效应研究[D]. 杨凌: 西北农林科技大学, 2022. XIN D Y. mRNA expression, mutation locus detection and genetic effects of IGF2BP2 and IGF2BP3 genes in Shaanbei White Cashmere goat[D]. Yangling: Northwest A & F University, 2022. (in Chinese) |
| [69] |
WANG W S, SEALE P. Control of brown and beige fat development[J]. Nat Rev Mol Cell Biol, 2016, 17(11): 691-702. DOI:10.1038/nrm.2016.96 |
| [70] |
MARLATT K L, RAVUSSIN E. Brown adipose tissue: an update on recent findings[J]. Curr Obes Rep, 2017, 6(4): 389-396. DOI:10.1007/s13679-017-0283-6 |
| [71] |
DAI N, ZHAO L P, WRIGHTING D, et al. IGF2BP2/IMP2-deficient mice resist obesity through enhanced translation of Ucp1 mRNA and other mRNAs encoding mitochondrial proteins[J]. Cell Metab, 2015, 21(4): 609-621. DOI:10.1016/j.cmet.2015.03.006 |
(编辑 郭云雁)



