随着国民消费能力及生活质量的提升,牛肉的消费量大幅增加,人们对牛肉品质的要求也不断提高。根据国家统计局发布的相关数据(https://data.stats.gov.cn),国内肉牛2021年出栏量为4 707.43万头,牛肉产量为697.51万吨,相比10年前增加了13.46%。我国本土牛品种繁多,根据粮农组织家畜多样性信息系统(https://www.fao.org/dad-is)统计数据显示,70多个牛种广泛分布于我国各个地区,其中50余种为我国本土品种[1-2]。根据牛种分布的地理位置可将我国牛种划分为3类:生活在中国北方的北方品种、黄河中下游的中部品种以及在南方分布的南方品种[3]。其中,南方品种牛体型较小,耐湿热能力及抗病能力强,具有优良的肌肉品质[4-5]。然而,相较于北方品种及中部品种,有关于南方品种牛肌生成及肉品质调控的研究较少,而南方地区尤其是华南地区有较大的牛肉消费市场,且对牛肉品质要求较高,因此,对华南地区固有品种牛进行肉用资源开发与利用具有重要意义。
屠宰率和胴体重是评估肉牛肉用价值的重要指标,而骨骼肌的发育直接影响肉牛的生产,因此使用各种手段调控骨骼肌肌纤维的生成与发育,对于提升肉牛肉用品质具有重要意义[6]。肌纤维的结构、组成以及生理生化活动可影响肉的嫩度、颜色以及pH下降速度等[7-9]。如肌纤维的直径与嫩度呈负相关[10-11],肌纤维中肌红蛋白的存在形式会直接影响肉色[12],肌纤维糖酵解能力会影响肌肉pH的下降速度[13-14]等。肌纤维的生长发育过程可以简单分为胚胎阶段以及出生后的肥大与转化阶段[8, 15]。肌肉在出生后的发育受到许多因素调控,如信号通路、肌源性调节因子以及非编码RNA(non-coding RNA, ncRNA)等[16-18]。长链非编码RNA (long noncoding RNA, lncRNA)是一种长度大于200个核苷酸的ncRNA,近年来不断有研究证明了其在转录过程中的调控功能,其中顺式(cis)调控和反式(trans)调控作用基因的搜寻是研究lncRNA靶标的常用方法之一。关于lncRNA顺式调控作用的机制一般被认为是lnc-RNA通过某些途径来影响其转录位点附近基因的表达,而反式调控作用则不局限于影响lncRNA的转录位点附近的基因[19-20]。例如,骨骼肌中lncMAAT的下调可以导致肌肉萎缩,而其过表达可以改善多种类型的肌肉萎缩,这是由于lncMAAT可以通过反式调控模块,利用SOX6来减少miR-29b的表达,同时通过顺式调控来增加Mbnl1基因的转录,从而减缓肌肉萎缩[21]。此外,lncRNA还可以作为miRNA的海绵,从而降低miRNA对转录本的抑制作用,以此为基础构建的竞争性内源RNA(competing endogenous RNAs,ceRNA)调控网络也是研究的热点[22]。如LncIRS1可以通过其海绵作用吸附miR-15家族,进而激活IGF1-PI3K/AKT途径以调控肌肉萎缩[23];再如,在成年和老年小鼠之间骨骼肌中新鉴定的名为MAR1的lncRNA可以通过吸附miR-487b来调节Wnt5a蛋白,从而促进骨骼肌分化与再生[24-25]。
上述研究表明,lncRNA在调控动物肌肉生成方面发挥着重要作用,现阶段有关挖掘调控牛肌生成及肌肉品质lncRNA的报道主要集中于牦牛[26-27]、秦川牛[28]以及山东黑牛[29]等华北肉用品种牛,而在华南地区地方品种牛的研究基本为空白。雷琼牛(LQN)和陆丰牛(LFN)主要分布在广东省及海南省,是华南地区特有品种黄牛。两种黄牛长期生活在亚热带气候环境,受人工引种影响较小,且饲养规模可观。雷琼牛与陆丰牛的亲缘关系较近,但其体貌外观以及肌纤维性状、肉品质差异较大[4, 30-31]。因此,以雷琼牛与陆丰牛作为对象,挖掘华南地区地方品种牛肌纤维lncRNA关键基因,对于进一步开发与高效利用华南地区地方品种肉用牛资源具有重要意义。
1 材料与方法 1.1 试验动物及样品采集本试验所涉及的雷琼牛、陆丰牛均来源于广东省梅州市保种场。所选牛均为4月龄,雌性,每个品种选择4头,饲养管理条件与营养条件均保持一致,健康无病且无人为杂交。选取的全部8头牛根据GB/T 19477—2018所述的畜禽屠宰操作规程进行屠宰后,迅速采集背最长肌组织样品于冻存管中,而后立刻放置于液氮冷冻,样品长期保存于-80 ℃冰箱。
1.2 总RNA提取和建库使用TRIzol(Thermo Fisher,上海,中国)法提取组织样品中总RNA,1%琼脂糖凝胶检测RNA降解情况。使用Agilent 2100生物分析仪和RNA 6000 Nano LabChip Kit(Agilent, Santa Clara, USA)测定RNA总量及完整性,质检符合测序要求的样品送至上海派森诺生物,使用Illumina HiSeq 2500平台进行测序。每个样品取1 μL总RNA用于文库的构建,根据NEB Next Ultra Directional RNA Library Prep Kit for Illumina(NEB,Ispawich,USA)说明书进行150 bp双末端测序。
small RNA测序单独建库,总RNA提取、质控方法与lncRNA、mRNA一致。RNA建库根据TruSeq Small RNA Sample Preparation Kits提供的方法进行,添加接头和index后,使用PCR富集文库,并利用聚丙烯酰胺凝胶对目的条带进行分离,质检合格的样品使用Illumina HiSeq 2500平台进行150 bp双末端测序。
1.3 数据质控、reads映射及转录本组装高通量测序过程中产生的原始reads(raw reads)储存为Fastq格式。使用Cutadapt对下机数据进行过滤,去除接头、低质量reads以及N-bases。过滤后的clean reads使用HiSAT2与牛(Bos taurus)参考基因组进行比对。在获得比较结果的bam文件后,使用StringTie软件比对到基因组上的reads。统计每个样品中转录本表达水平,并标准化为FPKM。
1.4 lncRNA的鉴定及顺式靶向调控基因的预测使用Stringtie软件,利用HiSAT2软件比对的结果进行转录本组装,在去除链方向不确定的转录本后,剩余的转录本用于lncRNA的筛选。筛选流程如下:Step1:筛选长度≥200 bp,以及外显子数目≥2的转录本;Step2:使用Cuffcompare软件筛选、去除与数据库已注释外显子区域有重叠的转录本;Step3:筛选出至少在一个样本中出现3次的lnc- RNA;Step4:使用PLEK、CNCI和PfamScan软件对lncRNA编码潜力进行预测,并对3个软件预测无编码功能的转录本取交集,作为高可信候选lnc-RNA。统计每个样品中lncRNA表达水平,并标准化为FPKM。
顺式作用是预测lncRNA靶标基因的主要方法之一。通过搜寻lncRNA基因上、下游100 kb以内的蛋白编码基因,认为搜索到的蛋白编码基因可能为其对应lncRNA的顺式调控靶基因。
1.5 miRNA的鉴定下机的原始数据删除ploy-N、低质量reads以获得高质量clean reads。根据片段大小,筛除小于18 nt以及大于30 nt的片段。miRNA的比对使用Bowtie软件,将干净的数据映射到参考序列,并使用miRDeep2来鉴定新的miRNA。估计每个样品的miRNA表达水平,并通过TPM算法进行标准化。使用DESeq2 R软件包(1.10.1)进行差异表达分析。DESeq2发现P < 0.05的miRNA被指定为候选差异表达miRNA。
将clean reads与Silva、GtRNAdb、Rfam和Repbase数据库进行比对,以过滤核糖体RNA(rRNA)、转运RNA(tRNA)、核小RNA(snRNA)以及胞质小RNA(scRNA)等。使用Bowtie软件将未注释的reads映射到参考基因组上,以确定已知miRNAs,同时使用miRBase(v21)和miRDeep 2(v2.0.1.2)预测的新miRNA。统计每个样品中miRNA表达水平,通过TPM算法进行标准化。
1.6 差异表达分析及富集分析根据各样品中lncRNA、miRNA以及mRNA的表达量,使用DESeq R包(1.18.0)进行差异表达分析,阈值为|log2 Fold Change|>1,显著性P < 0.05。为了解lncRNA靶基因功能,使用top GO进行基因本体(Gene Ontology,GO)富集分析,通过超几何分布方法计算显著富集的GO条目(显著富集的标准为P < 0.05)。使用cluster profiler软件进行京都基因与基因组百科全书(Kyoto Encyclopedia of Genes and Genomes,KEGG)通路富集分析,重点关注P < 0.05的通路。
1.7 lncRNA-miRNA-mRNA竞争调控网络的构建使用miRanda(v3.3a)和TargetScan(v7.0)对miRNA的靶基因进行预测,以斯皮尔曼相关系数大于0.8,P < 0.05作为条件进行筛选,结果使用Cytoscape软件(v3.9.1)进行可视化处理。
1.8 RT-qPCR验证分析从差异表达lncRNA、miRNA和mRNA中各随机选择3个基因,使用RT-qPCR进行转录组表达谱验证分析。反应体系20 μL:10 μL SYBR® Green Master Mix,上、下游引物各0.5 μL,1 μL cDNA和8 μL ddH2O。反应条件:95 ℃ 10 min;95 ℃ 5 s,60 ℃ 20 s,72 ℃ 1 min,40个循环。引物设计参考NCBI中牛(Bos taurus)的相关基因序列,GAPDH作为lncRNA和mRNA的内参基因,U6作为miRNA的内参基因,引物使用Primer 5.0软件设计,并送生工生物股份有限公司合成(表 1)。
|
|
表 1 引物序列 Table 1 Primer sequences |
RNA测序共获得870 359 532条reads,在去除接头以及低质量reads以后,758 859 982条高质量的有效reads可用于后续分析。有效reads占原始reads的(87.20±2.49)%。测序的质量分值(Q30)均在94%以上。各样本测序质量概况如表 2所示,所测各样本中lncRNA、miRNA和mRNA表达情况的相关性分析结果如图 1所示。
|
|
表 2 雷琼牛与陆丰牛背最长肌RNA测序概况 Table 2 RNA sequencing profile of longissimus dorsi from Leiqiong cattle and Lufeng cattle |

|
样本相关性采用皮尔逊相关性系数度量。a.样本中lncRNA表达相关性热图; b.样本中miRNA表达相关性热图;c.样本中mRNA表达相关性热图 The sample correlation is measured by Pearson correlation coefficient. a. Heatmap of lncRNA expression correlation in samples; b. Heatmap of miRNA expression correlation in samples; c. Heatmap of mRNA expression correlation in samples 图 1 雷琼牛与陆丰牛背最长肌基因表达样本相关性热图 Fig. 1 Correlation heatmap of gene expression for longissimus dorsi of Leiqiong cattle and Lufeng cattle |
在雷琼牛和陆丰牛背最长肌中共测得3 925个lncRNAs。以雷琼牛作为对照组,共有119条差异表达lncRNAs(differentially expressed lncRNAs, DE lncRNAs),其中下调表达的lncRNAs共46个,占总DE lncRNAs的38.7%;上调表达的lncRNAs共73个,占总DE lncRNAs的61.3%(图 2)。
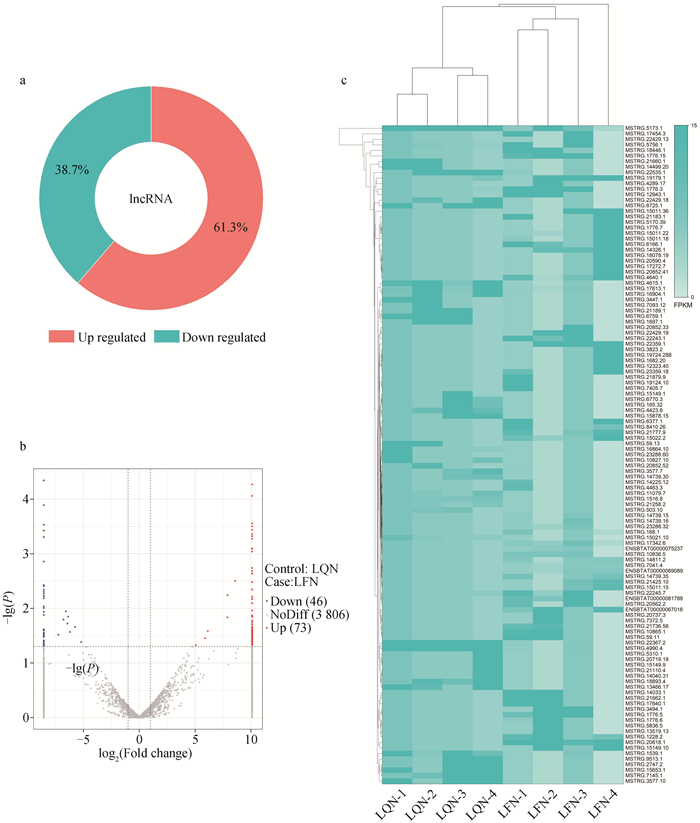
|
a.上/下调DE lncRNA占比;b. DE lncRNA火山图;c.DE lncRNA热图 a. Percentage of up/down-regulated DE lncRNAs; b. Volcano plot for DE lncRNA; c. Heatmap for DE lncRNA 图 2 雷琼牛与陆丰牛背最长肌DE lncRNA概况 Fig. 2 Overview of DE lncRNAs in the longissimus dorsi of Leiqiong cattle and Lufeng cattle |
图 3a展示了雷琼牛与陆丰牛背最长肌中miRNA表达的概况。在雷琼牛与陆丰牛背最长肌中共鉴定出来682个miRNAs。使用DEseq进行差异表达分析,共13条miRNAs在雷琼牛与陆丰牛背最长肌之间存在显著差异。以雷琼牛作为对照组,13条差异表达的miRNAs中7个为上调表达,6个为下调表达,分别占总差异miRNA的53.8%和46.2%。图 3b展示的结果为雷琼牛与陆丰牛背最长肌中mRNA表达的概况。测序并分析发现有17 313条mRNAs可以比对到参考基因组并被注释。差异表达分析结果显示,共599条mRNAs在雷琼牛与陆丰牛背最长肌之间存在显著差异,以雷琼牛作为对照组,155条mRNA为上调表达,444条mRNA为下调表达,分别占总差异mRNA的25.9%和74.1%。
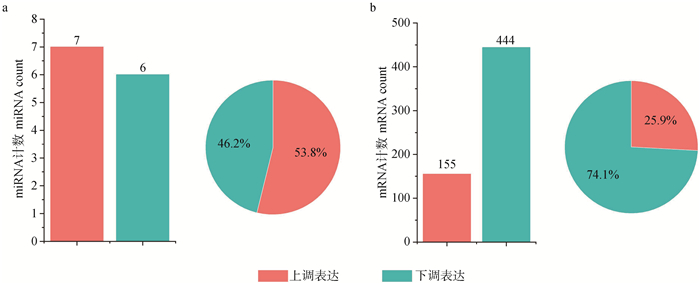
|
图 3 雷琼牛与陆丰牛背最长肌差异表达mRNA(A)和miRNA(B)的数量及占比 Fig. 3 Number and proportion of differentially expressed mRNAs (A) and miRNAs (B) in the longissimus dorsi of Leiqiong cattle and Lufeng cattle |
由于lncRNA可以通过顺式调控作用影响其转录位点临近处编码蛋白基因的表达,因此,本研究搜寻了已测得lncRNA基因上、下游100 kb以内的蛋白编码基因,用以分析可能受到lncRNA顺式作用调控的编码蛋白靶基因。共预测到98个基因可能受到lncRNA的顺式靶向调控作用,将预测到的靶基因进行富集分析,结果图 4如所示。KEGG富集分析结果表明(图 4a),预测的lncRNA顺式调控靶基因共富集到128个通路,其中显著富集的通路为29个(P<0.05),包括MAPK信号传导途径、局部粘附等;GO富集分析共产生了476个显著富集的GO条目(P<0.05),其中有378个GO条目类别为生物过程(biological process, BP)。GO富集分析中根据显著性排序前20的GO条目如图 4b所示,主要包括肌肉结构的发展、肌肉器官的发育、横纹肌组织发育的调节、细胞分化、肌肉器官发育的调节、肌肉组织发育的调节、横纹肌组织的发育以及肌肉组织的发育等,其中HDAC4(histone deacetylase 4)、SOX6(SRY-box transcription factor 6)以及KEL(Kell metallo-endopeptidase)为富集的主要基因。
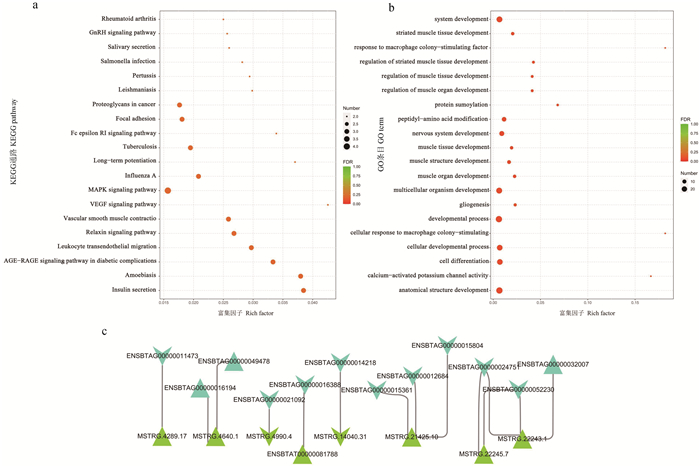
|
a.lncRNA顺式靶向调控基因KEGG富集分析气泡图;b. lncRNA顺式靶向调控基因GO富集分析气泡图;c. 雷琼牛与陆丰牛背最长肌lncRNA顺势靶向调控网络图 a. Bubble diagram of KEGG enrichment analysis of lncRNA cis-targeted regulatory genes; b. Bubble diagram of GO enrichment analysis of lncRNA cis-targeted regulatory genes; c. lncRNA cis-targeted regulatory network in longissimus dorsi of Leiqiong cattle and Lufeng cattle 图 4 差异lncRNA靶基因功能富集分析及与差异mRNA共表达网络 Fig. 4 Functional enrichment analysis of differential lncRNA target genes and its co-expression network with differential mRNAs |
此外,对预测到的lncRNA顺式调控靶基因与雷琼牛与陆丰牛背最长肌差异表达mRNA取交集,用于预测存在于雷琼牛与陆丰牛背最长肌中,且可能受到lncRNA的顺势调控作用的基因。所筛选出的mRNA与lncRNA作用关系如图 4c所示,共有12个存在于两种牛背最长肌中的差异表达mRNAs被筛选出来,8个lncRNAs可能通过顺式调控作用参与其表达的调控。
2.5 lncRNA相关ceRNA网络与功能富集分析lncRNA可以作为miRNA的海绵,通过富集miRNA以降低其对mRNA表达的抑制作用。根据差异表达的lncRNA、miRNA和mRNA序列,分析miRNA在lncRNA和mRNA上的结合集合位点,从而构建lncRNA相关的ceRNA网络。结果显示,13个miRNAs分别与77个lncRNAs和138个mRNAs有结合位点,网络关系如图 5a所示。在靶基因的KEGG富集分析中,Rap1信号传导途径、MAPK信号传导途径、PI3K-Akt信号通路被显著富集(图 5b);靶基因的GO富集分析中,磷酸化以及膜结构等相关功能被显著富集(图 5c)。
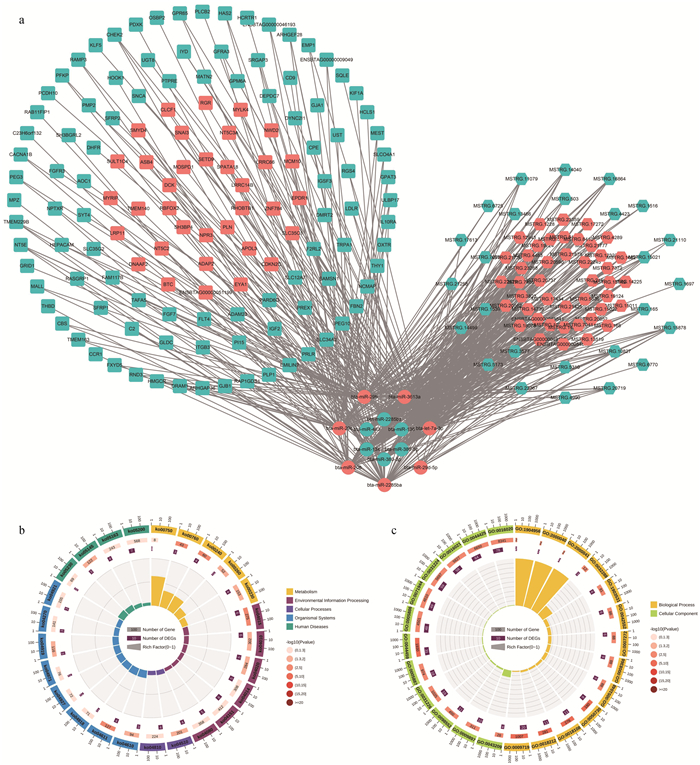
|
a.lncRNA-miRNA-mRNA竞争性调控网络;b. ceRNA网络中靶基因KEGG富集分析;c. ceRNA网络中靶基因GO富集分析 a. lncRNA-miRNA-mRNA competitive regulatory network; b. KEGG enrichment analysis of target genes in ceRNA network; c. GO enrichment analysis of target genes in ceRNA network 图 5 lncRNA-miRNA-mRNA竞争性调控网络及富集分析 Fig. 5 lncRNA-miRNA-mRNA competitive regulatory network and enrichment analysis |
如图 6所示,通过RT-qPCR对9个差异表达lncRNAs、miRNAs和mRNAs进行验证,结果与测序结果趋势相似,证明雷琼牛与陆丰牛背最长肌转录组测序结果可信,可用于后续分析与讨论。

|
图 6 差异表达基因RT-qPCR验证结果 Fig. 6 Results of RT-qPCR validation of differentially expressed genes |
lncRNA是一种长度大于200 nt的ncRNA,具有广泛的基因表达调控功能,且一般不具有编码蛋白的功能[32-33]。lncRNA曾被认为是基因表达过程的“噪音”,而高通量技术(如下一代测序)的发展使得能够以前所未有的分辨率以及规模对lncRNA进行深入研究,lncRNA如今已被证实作为调控基因功能和表达水平的重要参与者[34-35]。此外,lncRNA还被证明参与许多如蛋白修饰、细胞增殖分化等生物进程[19, 36-38]。lncRNA的顺式调控作用一般是指lncRNA可以通过某些途径影响其转录位点附近基因表达,如lncRNA可以通过招募调节因子至基因座,进而调节周边基因;lncRNA的反式调控功能则与lncRNA转录的位点无关,这些lnc-RNA通过影响染色体与核结构,或与蛋白质及其他RNA分子相互作用,从而调节细胞生物学功能[19]。肌肉生成是一个生理过程,从胎儿时期开始就受到各种内部或外部因素的影响,过去的研究证明,lnc-RNA能够有效调节骨骼肌的肌生成过程[39-40]。
本研究发现,雷琼牛与陆丰牛背最长肌中98个基因可能受到DE lncRNAs的顺式调控作用,通过富集分析发现,RBFOX1、HDAC4以及SOX6等基因富集到了多个与肌生成相关的GO条目。RBFOX1是一种RNA结合蛋白,该家族蛋白高度保守,Pedrotti等[41]的试验结果表明RBFOX1基因敲除小鼠表现出显著的肌肉发力减少,并证实RBFOX1调节着一个维持肌肉生理学多个方面所需的选择性剪切事件网络。HDAC4是一种IIa类组蛋白去乙酰化酶,该酶在骨骼肌中上调表达可以响应去神经支配诱发的萎缩[42]。有研究发现,lncMir22hg衍生的miR-22-3p可以通过抑制HDAC4的表达来促进骨骼肌分化和再生[43]。但在HDAC4对鸡骨骼肌卫星细胞影响的研究中发现,HDAC4在胚胎骨骼肌中富集,并且在胚胎肌组织中比在产后肌组织中表达量更高,HDAC4基因的敲低可以显着抑制鸡骨骼肌卫星细胞的增殖和分化,但对骨骼肌卫星细胞的凋亡似乎没有显著影响[44]。SOX6是肌肉慢纤维特异性基因的关键抑制因子,有报道证明组蛋白脱乙酰酶Sirt6可以通过增加CREB的转录来下调SOX6基因的表达,进而导致纤维类型发生转化[45]。在骨骼肌中缺乏SOX6的小鼠肌肉中发现慢纤维数量增多,而快肌纤维基因程序下调,肌肉耐力明显增强[45]。在雷琼牛与陆丰牛背最长肌的差异mRNA中,预测到了可能受到DE lncRNA顺式调控作用的基因共12个,在这其中DKK1和SIGMAR1基因可能参与肌肉生成的调控。Wnt信号是广泛存在于胚胎时期的信号通路,但也调节成年动物机体的生理过程,已被证明在成年动物骨骼发育和稳态等方面发挥重要作用[46-47]。DKK1是Wnt信号传导的蛋白质抑制剂,其过表达可以明显抑制Wnt信号通路[48],但DKK1是否能够通过抑制Wnt通路来影响牛骨骼肌的发育尚不明确。SIGMAR1是一种广泛表达的多任务分子伴侣蛋白,在多个细胞过程中起着功能作用,SIGMAR1敲除小鼠的骨骼肌肌病与骨骼肌中央核数量增加,且耐力与运动能力均明显下降[49]。本研究发现,RBFOX1、HDAC4以及SOX6基因位点分别位于MSTRG.13466、MSTRG.5308以及ENSBTAG00000054834的转录位点位附近,这些lnc-RNA可能参与肌生成的调控。
除了上述调控方式,lncRNA还可以充当miRNA的海绵,吸附miRNA以降低其对基因表达的负调控作用[38, 50]。受到DE lncRNAs间接调控作用,靶基因显著富集(P<0.05)于PI3K-AKT信号通路、MAPK信号传导途径等功能上。PI3K-AKT通路已被广泛证明与肌肉肥大和萎缩有关[51-54]。最近的研究也证明了一些ncRNA可以通过内源竞争的方式来调节PI3K/AKT通路,例如,环状RNA circRILPL1可以作为miR-145的海绵,调控IGF1R的表达,从而减少miR-145对PI3K/AKT信号通路的抑制作用,促进成肌细胞生长[55]。MAPK信号通路是驱动骨骼肌对运动代谢适应的信号通路之一,MAPK信号通路在运动过程中会增加胰岛素依赖的葡萄糖摄取和氧化代谢,以及肌肉中线粒体的氧化磷酸化[56-57]。此外,MAPK信号通路还被证明可能与成肌细胞的细胞周期停滞有关,这对成肌细胞启动肌肉分化至关重要[58];在山羊骨骼肌的研究中,发现p38-MAPK途径可以被GADD45b激活从而促进肌源性分化。本研究表明,MSTRG.13466、MSTRG.5308、ENSBTAG00000054834等或可参与华南地区特有黄牛品种肌生成的调节,可作为进一步提升华南地区黄牛肉用价值的候选lncRNAs。
4 结论本研究通过RNA测序,获得了雷琼牛与陆丰牛背最长肌中119个差异表达lncRNAs、13个差异表达miRNAs以及599个差异表达mRNAs。差异表达基因进行了lncRNA-miRNA-mRNA竞争性调控网络的构建以及lncRNA顺式靶向调控作用靶基因的搜寻。对上述靶基因进行GO和KEGG富集分析,筛选出了多个与牛骨骼肌肌生成相关,且可能受到lncRNA调控的关键基因。该结果为进一步了解华南地区特有黄牛品种基因组表达特点以及肉用价值进行合理开发与利用提供了理论支持。
| [1] |
ZHANG W G, GAO X, ZHANG Y, et al. Genome-wide assessment of genetic diversity and population structure insights into admixture and introgression in Chinese indigenous cattle[J]. BMC Genet, 2018, 19(1): 114. DOI:10.1186/s12863-018-0705-9 |
| [2] |
QIU H, JU Z Y, CHANG Z J. A survey of cattle production in China[J]. World Animal Review, 1993(76): 12-18. |
| [3] |
ZENG L, CHEN N, NING Q, et al. PRLH and SOD1 gene variations associated with heat tolerance in Chinese cattle[J]. Anim Genet, 2018, 49(5): 447-451. DOI:10.1111/age.12702 |
| [4] |
LIU Y Q, XU L Y, YANG L, et al. Discovery of genomic characteristics and selection signatures in southern Chinese local cattle[J]. Front Genet, 2020, 11: 533052. DOI:10.3389/fgene.2020.533052 |
| [5] |
LU X B, ARBAB A A I, ZHANG Z P, et al. Comparative transcriptomic analysis of the pituitary gland between cattle breeds differing in growth: Yunling cattle and leiqiong cattle[J]. Animals (Basel), 2020, 10(8): 1271. |
| [6] |
BEAK S H, PARK S J, FASSAH D M, et al. Relationships among carcass traits, auction price, and image analysis traits of marbling characteristics in Korean cattle beef[J]. Meat Sci, 2021, 171: 108268. DOI:10.1016/j.meatsci.2020.108268 |
| [7] |
PICARD B, GAGAOUA M. Muscle fiber properties in cattle and their relationships with meat qualities: an overview[J]. J Agric Food Chem, 2020, 68(22): 6021-6039. DOI:10.1021/acs.jafc.0c02086 |
| [8] |
ZHUANG X N, LIN Z K, XIE F, et al. Identification of circRNA-associated ceRNA networks using longissimus thoracis of pigs of different breeds and growth stages[J]. BMC Genomics, 2022, 23(1): 294. DOI:10.1186/s12864-022-08515-7 |
| [9] |
LEE S H, JOO S T, RYU Y C. Skeletal muscle fiber type and myofibrillar proteins in relation to meat quality[J]. Meat Sci, 2010, 86(1): 166-170. DOI:10.1016/j.meatsci.2010.04.040 |
| [10] |
WHITE A, O'SULLIVAN A, TROY D J, et al. Manipulation of the pre-rigor glycolytic behaviour of bovine M. longissimus dorsi in order to identify causes of inconsistencies in tenderness[J]. Meat Sci, 2006, 73(1): 151-156. DOI:10.1016/j.meatsci.2005.11.021 |
| [11] |
GAGAOUA M, PICARD B, MONTEILS V. Assessment of cattle inter-individual cluster variability: the potential of continuum data from the farm-to-fork for ultimate beef tenderness management[J]. J Sci Food Agric, 2019, 99(8): 4129-4141. DOI:10.1002/jsfa.9643 |
| [12] |
WICKS J, BELINE M, GOMEZ J F M, et al. Muscle energy metabolism, growth, and meat quality in beef cattle[J]. Agriculture (Basel), 2019, 9(9): 195. DOI:10.3390/agriculture9090195 |
| [13] |
HUGHES J M, CLARKE F M, PURSLOW P P, et al. Meat color is determined not only by chromatic heme pigments but also by the physical structure and achromatic light scattering properties of the muscle[J]. Compr Rev Food Sci Food Saf, 2020, 19(1): 44-63. DOI:10.1111/1541-4337.12509 |
| [14] |
PURSLOW P P, WARNER R D, CLARKE F M, et al. Variations in meat colour due to factors other than myoglobin chemistry; a synthesis of recent findings (invited review)[J]. Meat Sci, 2020, 159: 107941. DOI:10.1016/j.meatsci.2019.107941 |
| [15] |
LEFAUCHEUR L. Myofiber typing and pig meat production[J]. Slov Vet Zb, 2001, 38(1): 5-33. |
| [16] |
HE S Q, FU T T, YU Y, et al. IRE1α regulates skeletal muscle regeneration through Myostatin mRNA decay[J]. J Clin Invest, 2021, 131(17): e143737. DOI:10.1172/JCI143737 |
| [17] |
COUDERT L, OSSENI A, GANGLOFF Y G, et al. The ESCRT-0 subcomplex component Hrs/Hgs is a master regulator of myogenesis via modulation of signaling and degradation pathways[J]. BMC Biol, 2021, 19(1): 153. DOI:10.1186/s12915-021-01091-4 |
| [18] |
ARCHACKA K, CIEMERYCH M A, FLORKOWSKA A, et al. Non-coding RNAs as regulators of myogenesis and postexercise muscle regeneration[J]. Int J Mol Sci, 2021, 22(21): 11568. DOI:10.3390/ijms222111568 |
| [19] |
KOPP F, MENDELL J T. Functional classification and experimental dissection of long noncoding RNAs[J]. Cell, 2018, 172(3): 393-407. DOI:10.1016/j.cell.2018.01.011 |
| [20] |
李艳艳, 顾志良. 长链非编码RNA在肌肉发育中的作用研究进展[J]. 中国畜牧兽医, 2014, 41(11): 232-236. LI Y Y, GU Z L. Research progress of long noncoding RNA in muscle development[J]. China Animal Husbandry & Veterinary Medicine, 2014, 41(11): 232-236. (in Chinese) |
| [21] |
LI J, YANG T T, TANG H F, et al. Inhibition of lncRNA MAAT controls multiple types of muscle atrophy by cis- and trans-regulatory actions[J]. Mol Ther, 2021, 29(3): 1102-1119. DOI:10.1016/j.ymthe.2020.12.002 |
| [22] |
ZHANG P P, CHAO Z, ZHANG R, et al. Circular RNA regulation of myogenesis[J]. Cells, 2019, 8(8): 885. DOI:10.3390/cells8080885 |
| [23] |
LI Z H, CAI B L, ABDALLA B A, et al. LncIRS1 controls muscle atrophy via sponging miR-15 family to activate IGF1-PI3K/AKT pathway[J]. J Cachexia Sarcopenia Muscle, 2019, 10(2): 391-410. DOI:10.1002/jcsm.12374 |
| [24] |
ZHANG Z K, LI J, GUAN D G, et al. A newly identified lncRNA MAR1 acts as a miR-487b sponge to promote skeletal muscle differentiation and regeneration[J]. J Cachexia Sarcopenia Muscle, 2018, 9(3): 613-626. DOI:10.1002/jcsm.12281 |
| [25] |
HERMAN A B, TSITSIPATIS D, GOROSPE M. Integrated lncRNA function upon genomic and epigenomic regulation[J]. Mol Cell, 2022, 82(12): 2252-2266. DOI:10.1016/j.molcel.2022.05.027 |
| [26] |
HUANG C, DAI R F, MENG G Y, et al. Transcriptome-wide study of mRNAs and lncRNAs modified by m6A RNA methylation in the Longissimus dorsi muscle development of cattle-yak[J]. Cells, 2022, 11(22): 3654. DOI:10.3390/cells11223654 |
| [27] |
HUANG C, GE F, MA X M, et al. Comprehensive analysis of mRNA, lncRNA, circRNA, and miRNA expression profiles and their ceRNA networks in the Longissimus dorsi muscle of cattle-yak and yak[J]. Front Genet, 2021, 12: 772557. DOI:10.3389/fgene.2021.772557 |
| [28] |
JIANG R, LI H, HUANG Y Z, et al. Transcriptome profiling of lncRNA related to fat tissues of Qinchuan cattle[J]. Gene, 2020, 742: 144587. DOI:10.1016/j.gene.2020.144587 |
| [29] |
LIU R L, HAN M X, LIU X X, et al. Genome-wide identification and characterization of long non-coding RNAs in longissimus dorsi skeletal muscle of shandong black cattle and luxi cattle[J]. Front Genet, 2022, 13: 849399. DOI:10.3389/fgene.2022.849399 |
| [30] |
黄定庆, 王亮星. 雷琼黄牛品种改良中存在的问题和对策[J]. 世界热带农业信息, 2023(4): 69-71. HUANG D Q, WANG L X. Problems and countermeasures in the improvement of Leiqiong cattle breed[J]. World Tropical Agriculture Information, 2023(4): 69-71. (in Chinese) |
| [31] |
LIU Y Q, ZHAO G Y, LIN X J, et al. Genomic inbreeding and runs of homozygosity analysis of indigenous cattle populations in southern China[J]. PLoS One, 2022, 17(8): e0271718. DOI:10.1371/journal.pone.0271718 |
| [32] |
ANDERSON D M, ANDERSON K M, CHANG C L, et al. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance[J]. Cell, 2015, 160(4): 595-606. DOI:10.1016/j.cell.2015.01.009 |
| [33] |
BRIDGES M C, DAULAGALA A C, KOURTIDIS A. LNCcation: lncRNA localization and function[J]. J Cell Biol, 2021, 220(2): e202009045. DOI:10.1083/jcb.202009045 |
| [34] |
QIAN X Y, ZHAO J Y, YEUNG P Y, et al. Revealing lncRNA structures and interactions by sequencing-based approaches[J]. Trends Biochem Sci, 2019, 44(1): 33-52. DOI:10.1016/j.tibs.2018.09.012 |
| [35] |
NOJIMA T, PROUDFOOT N J. Mechanisms of lncRNA biogenesis as revealed by nascent transcriptomics[J]. Nat Rev Mol Cell Biol, 2022, 23(6): 389-406. DOI:10.1038/s41580-021-00447-6 |
| [36] |
ZHOU T, DING J W, WANG X A, et al. Long noncoding RNAs and atherosclerosis[J]. Atherosclerosis, 2016, 248: 51-61. DOI:10.1016/j.atherosclerosis.2016.02.025 |
| [37] |
任灵通, 刘凌斌, 李佳璐, 等. 肌肉发生相关长链非编码RNA研究进展[J]. 中国畜牧兽医, 2021, 48(8): 2957-2965. REN L T, LIU L F, LI J L, et al. Research progress on long non-coding RNA associated with myogenesis[J]. China Animal Husbandry & Veterinary Medicine, 2021, 48(8): 2957-2965. DOI:10.16431/j.cnki.1671-7236.2021.08.028 (in Chinese) |
| [38] |
郭志峰. 长链非编码RNA作为竞争性内源RNA在Ⅰ期肺腺癌中的分析研究[D]. 福州: 福建医科大学, 2021. GUO Z F. Comprehensive analysis of dysregulated LncRNAs and their competing endogenous RNA network in stage I lung adenocarcinoma[D]. Fuzhou: Fujian Medical University, 2021. (in Chinese) |
| [39] |
叶峻宁, 邓铭, 薛慧雯, 等. 影响山羊胎儿肌肉发育mRNA和lncRNA的鉴定与分析[J]. 畜牧兽医学报, 2023, 54(3): 989-1002. YE J N, DENG M, XUE H W, et al. Identification and analysis of mRNA and lncRNA affecting goat fetal muscle development[J]. Acta Veterinaria et Zootechnica Sinica, 2023, 54(3): 989-1002. (in Chinese) |
| [40] |
李文雅, 牛欣然, 任团辉, 等. 鸡骨骼肌中天然反义lncRNA VGLL2-AS的鉴定及其与VGLL2的关系研究[J]. 畜牧兽医学报, 2023, 54(1): 122-132. LI W Y, NIU X R, REN T H, et al. Identification of natural antisense lncRNA VGLL2-AS in chicken skeletal muscle and its relationship with VGLL2[J]. Acta Veterinaria et Zootechnica Sinica, 2023, 54(1): 122-132. (in Chinese) |
| [41] |
PEDROTTI S, GIUDICE J, DAGNINO-ACOSTA A, et al. The RNA-binding protein Rbfox1 regulates splicing required for skeletal muscle structure and function[J]. Hum Mol Genet, 2015, 24(8): 2360-2374. DOI:10.1093/hmg/ddv003 |
| [42] |
LUO L Q, MARTIN S C, PARKINGTON J, et al. HDAC4 controls muscle homeostasis through deacetylation of myosin heavy chain, PGC-1α, and Hsc70[J]. Cell Rep, 2019, 29(3): 749-763.e12. DOI:10.1016/j.celrep.2019.09.023 |
| [43] |
LI R, LI B, CAO Y, et al. Long non-coding RNA Mir22hg-derived miR-22-3p promotes skeletal muscle differentiation and regeneration by inhibiting HDAC4[J]. Mol Ther Nucleic Acids, 2021, 24: 200-211. DOI:10.1016/j.omtn.2021.02.025 |
| [44] |
ZHAO J, SHEN X X, CAO X A, et al. HDAC4 regulates the proliferation, differentiation and apoptosis of chicken skeletal muscle satellite cells[J]. Animals (Basel), 2020, 10(1): 84. |
| [45] |
QUIAT D, VOELKER K A, PEI J M, et al. Concerted regulation of myofiber-specific gene expression and muscle performance by the transcriptional repressor Sox6[J]. Proc Natl Acad Sci U S A, 2011, 108(25): 10196-10201. DOI:10.1073/pnas.1107413108 |
| [46] |
REGARD J B, ZHONG Z, WILLIAMS B O, et al. Wnt signaling in bone development and disease: making stronger bone with Wnts[J]. Cold Spring Harb Perspect Biol, 2012, 4(12): a007997. |
| [47] |
RUDNICKI M A, WILLIAMS B O. Wnt signaling in bone and muscle[J]. Bone, 2015, 80: 60-66. DOI:10.1016/j.bone.2015.02.009 |
| [48] |
HE Y, CHEN Y, ZHAO Q, et al. Roles of brain and muscle ARNT-like 1 and Wnt antagonist Dkk1 during osteogenesis of bone marrow stromal cells[J]. Cell Prolif, 2013, 46(6): 644-653. DOI:10.1111/cpr.12075 |
| [49] |
AISHWARYA R, ABDULLAH C S, REMEX N S, et al. Molecular characterization of skeletal muscle dysfunction in sigma 1 receptor (Sigmar1) knockout mice[J]. Am J Pathol, 2022, 192(1): 160-177. DOI:10.1016/j.ajpath.2021.10.003 |
| [50] |
YE W D, DUAN Y, ZHANG W T, et al. Comprehensive analysis of hub mRNA, lncRNA and miRNA, and associated ceRNA networks implicated in grass carp (Ctenopharyngodon idella) growth traits[J]. Genomics, 2021, 113(6): 4004-4014. DOI:10.1016/j.ygeno.2021.10.001 |
| [51] |
GLASS D J. Signalling pathways that mediate skeletal muscle hypertrophy and atrophy[J]. Nat Cell Biol, 2003, 5(2): 87-90. DOI:10.1038/ncb0203-87 |
| [52] |
YU M L, WANG H, XU Y L, et al. Insulin-like growth factor-1 (IGF-1) promotes myoblast proliferation and skeletal muscle growth of embryonic chickens via the PI3K/Akt signalling pathway[J]. Cell Biol Int, 2015, 39(8): 910-922. |
| [53] |
STITT T N, DRUJAN D, CLARKE B A, et al. The IGF-1/PI3K/Akt pathway prevents expression of muscle atrophy-induced ubiquitin ligases by inhibiting FOXO transcription factors[J]. Mol Cell, 2004, 14(3): 395-403. |
| [54] |
KUMAR A, XIE L T, TA C M, et al. SWELL1 regulates skeletal muscle cell size, intracellular signaling, adiposity and glucose metabolism[J]. Elife, 2020, 9: e58941. |
| [55] |
SHEN X M, TANG J, JIANG R, et al. CircRILPL1 promotes muscle proliferation and differentiation via binding miR-145 to activate IGF1R/PI3K/AKT pathway[J]. Cell Death Dis, 2021, 12(2): 142. |
| [56] |
BENGAL E, AVIRAM S, HAYEK T. p38 MAPK in glucose metabolism of skeletal muscle: beneficial or harmful?[J]. Int J Mol Sci, 2020, 21(18): 6480. |
| [57] |
SOMWAR R, KOTERSKI S, SWEENEY G, et al. A dominant-negative p38 MAPK mutant and novel selective inhibitors of p38 MAPK reduce insulin-stimulated glucose uptake in 3T3-L1 adipocytes without affecting GLUT4 translocation[J]. J Biol Chem, 2002, 277(52): 50386-50395. |
| [58] |
LEE J, HONG F, KWON S, et al. Activation of p38 MAPK induces cell cycle arrest via inhibition of Raf/ERK pathway during muscle differentiation[J]. Biochem Biophys Res Commun, 2002, 298(5): 765-771. |
(编辑 郭云雁)



