2. 甘肃农业大学动物科学技术学院, 兰州 730050
2. College of Animal Science and Technology of Gansu Agricultural University, Lanzhou 730050, China
近年来,胚胎基因组选择(embryonic genome selection,EGS)技术可为牛繁殖育种工作提供诸多便利,逐渐成为牛繁殖育种研究的热点。EGS可以很容易地挑选出大量的全同胞胚胎和半同胞胚胎,只选择理想性别和基因型优良的胚胎进行移植;此外,可以淘汰携带隐性致死基因或其他染色体畸变的胚胎,降低致死基因的频率;还可以通过加大选择强度来加快育种进程,缩短世代间隔[1]。将基因组选择与生物技术相结合,能进一步加快牛的遗传改良[2]。近年来,随着胚胎生物技术的发展和牛育种工作的需求增加[3],EGS技术得到了长足的发展。本文综述了牛EGS技术的研究应用,并且分别概述了EGS技术常用的活检方法、扩增方法及该项技术当前存在的问题,旨在为相关领域的科研工作提供参考。
1 牛EGS常用活检方法胚胎活检指的是在胚胎移植之前,通过化学、机械和激光等方法在透明带打孔来获得胚胎细胞,了解胚胎的遗传特点,选择适宜的胚胎进行移植。牛EGS常用机械法进行活检,根据所用分割器械的不同又分为针法、抽吸法与微刀法。由于3种活检方法采取的措施不同,对胚胎造成的物理损伤也不同,对活检胚胎后续发育能力和移植妊娠率均有不同程度的影响。据报道,针法、抽吸法和微刀法活检胚胎冷冻保存解冻后的移植妊娠率分别为57%、43%和31%[4]。因此,选择适当的活检方法对于胚胎后续利用及冷冻保存具有重要意义。
1.1 针法针法通常更适合于早期的胚胎(卵裂期到桑葚胚期)[2]。如图 1A所示,该法的基本步骤是将胚胎置于含0.4%牛血清白蛋白的磷酸缓冲盐溶液(PBS)中,用吸液管固定胚胎,用细针将透明带(不损伤卵裂球)刺出一个小口,用活检移液管从桑椹胚中抽出2~3个细胞,或从囊胚滋养外胚层抽出4~5个细胞,将吸出的胚胎细胞转移到含有蛋白酶K的裂解缓冲液中[4-5]。该方法对胚胎透明带的损伤较小,后续冷冻保存后解冻的移植妊娠率可以高达57%。
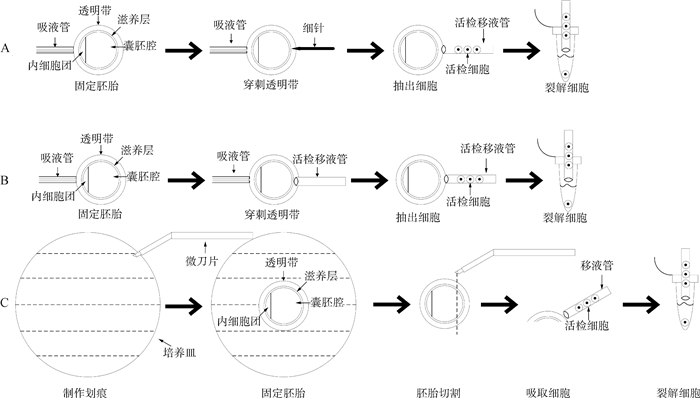
|
A.使用活检针进行穿刺透明带获取活检胚胎细胞;B.使用活检移液管进行穿刺透明带获取活检胚胎细胞;C.使用微刀片进行胚胎切割获取活检胚胎细胞 A.Biopsy needle was used to puncture the zona pellucida to obtain biopsied embryonic cells; B.Biopsy pipette was used to puncture the zona pellucida to obtain biopsied embryonic cells; C.Microblade was used for embryo cutting to obtain biopsied embryonic cells 图 1 牛EGS常用3种活检方法模式图 Fig. 1 Pattern diagram of 3 common biopsy methods for bovine EGS |
如图 1B所示,抽吸法的基本步骤是将胚胎置于含有0.4%牛血清白蛋白的PBS中,用吸液管固定,同时用活检移液管刺穿透明带,用活检移液管从桑椹胚中抽出2~3个细胞,或从囊胚滋养外胚层抽出4~5个细胞,将这些细胞转移到含有蛋白酶K的裂解缓冲液中。该方法对透明带的损伤适中,冷冻保存后解冻的移植妊娠率为43%[4-5],比针法低14%。
1.3 微刀法如图 1C所示,微刀法的基本步骤是将胚胎仅置于PBS缓冲液中,置于用微刀片划过底部的培养皿中,划痕可以固定胚胎[6],不再用手持移液管。将微刀片按压在胚胎的边缘上,缓慢地左右移动,直到胚胎的一小部分被切掉。然后,在培养皿中加入与先前PBS缓冲液等量的含0.8%牛血清蛋白的PBS,以防止细胞附着在刀片或培养皿底部。然后用吸液管取出活检细胞并将其放入含有蛋白酶K的裂解缓冲液中[4-5]。此方法易对胚胎后续发育造成不利影响,Tutt等[7]报道,采用该方法对牛高质量体内桑葚胚和囊胚进行活检后,妊娠率降低了5%~10%。以上3种牛EGS常用活检方法各有优缺点,具体如表 1所示。
|
|
表 1 牛EGS常用3种活检方法的优缺点 Table 1 Advantages and disadvantages of 3 biopsy methods for bovine EGS |
牛EGS的关键是要在不过度破坏胚胎后续发育能力的同时获得足够数量胚胎细胞DNA。然而,一方面,通过胚胎活检分割出的少量胚胎细胞所获取的DNA量不足以进行后续的全基因组测序、单核苷酸多态性(single-nucleotide polymorphism,SNP)基因分型等工作;另一方面,增加活检胚胎细胞数量虽然可以提高DNA的扩增成功率,但会严重降低活检胚胎后续发育能力。Lauri等[8]报道,取牛胚胎30个细胞进行活检后的胚胎冷冻存活率(62%)低于10个细胞的活检胚胎(89%)。由此可见,从少量胚胎细胞中提取足够的DNA是进行EGS的关键,为此,人们通常采用全基因组扩增(whole genomic amplification,WGA)技术对少量细胞的DNA进行扩增以获得足够的DNA。目前,胚胎基因组选择常用的扩增技术包括多重置换扩增(multiple displacement amplification, MDA)、多次退火环状循环扩增(multiple annealing and looping-based amplification cycles, MALBAC)和通过转座子插入的线性放大(linear amplification via transposon insertion, LIANTI)[9]。
2.1 MDAMDA最早是由Dean等[10]改进传统的WGA技术而形成的,是一种全新的非PCR等温扩增技术[10-12]。如图 2A所示,该方法充分利用了phi29 DNA聚合酶强大的处理能力与高保真性[13],在恒温30 ℃的条件下,先是随机的6碱基引物在多个位点与模板DNA退火结合,再发生链置换扩增反应,被置换产生的单链产物又成为新的复制模板,再进行扩增,如此循环,最后产生大量片段大小>10 kb的扩增产物[14]。由于该方法属于恒温扩增,所以会产生非特异性扩增。为了克服MDA的缺陷,许多学者改良了MDA技术,如Sidore等[15]于2016年提出了数字液滴多重置换扩增(dd MDA);Zhou等[16]于2021年提出凝胶多重置换扩增(gel MDA)。
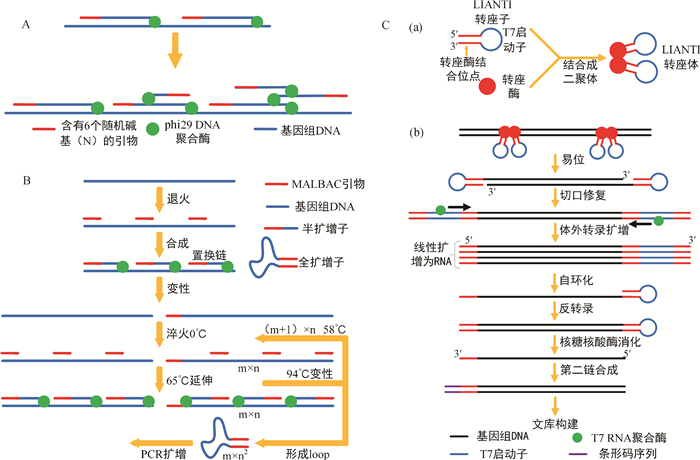
|
A.恒温条件下,含有6个随机碱基(N)的引物与基因组DNA随机退火,在phi29 DNA聚合酶的作用下发生链置换扩增反应;B.引物与模板随机结合,在65 ℃时,DNA链发生置换聚合反应,得到半扩增子;在94 ℃变性、0 ℃退火和65 ℃延伸循环后,半扩增子形成全扩增子后进行PCR扩增;C.(a)LIANTI转座体的合成;(b)LIANTI步骤:利用LIANTI转座子和转座酶,经过体外转录扩增,将基因组DNA片段线性扩增成基因组RNA,并反转录合成,构建基因组文库 A.Under the condition of constant temperature, the primers containing 6 random bases (N) were randomly degraded with genomic DNA and amplified by phi29 DNA polymerase; B.When the primers were randomly combined with the template, the replacement polymerization of DNA chain took place at 65 ℃, and the semi-amplification was obtained after denaturation at 94 ℃, degeneration at 0 ℃ and extension cycle at 65 ℃, and then PCR amplification was carried out after the semi-amplification was formed; C.(a) Synthesis of LIANTI transposon; (b) LIANTI step: using LIANTI transposon and transposase, genomic DNA fragments were linearly amplified into genomic RNA by in vitro transcription, and then reverse transcribed and synthesized to construct genomic library 图 2 牛EGS常用3种扩增技术原理示意图 Fig. 2 Schematic of the 3 commonly used amplification techniques for bovine EGS |
2012年,Lu等[17]应用MALBAC进行单细胞测序,Zong等[18]同时利用MALBAC方法成功地实现了单细胞水平的单核苷酸位点变异(single nucleo-tide variants,SNV)和拷贝数变异(copy number variation, CNV)的研究。如图 2B所示,该技术结合了PCR和MDA两种技术,同时兼顾了基因组扩增过程中的保真性和均一性,并实现了基因组的高覆盖率。该技术扩增的基本步骤先是预扩增阶段的部分随机引物退火;再通过具有链置换活性的DNA聚合酶实现链延伸;然后半扩增子产生3′和5′端互补的完整扩增子;全扩增子环化以防止全扩增子在第一时期PCR中进一步扩增和交叉杂交,而在第二时期PCR中作为靶标[19]。最后,产生长度为0.2~2.0 kb的扩增产物[20]。
2.3 LIANTI2017年,Chen等[21]报道了LIANTI技术,这项技术在整个基因组的扩增过程中都是线性扩增技术。如图 2C所示,该技术扩增的基本步骤首先是使用Tn5转座酶与T7启动子结合成二聚体LIANTI转座体,然后使用该转座体,随机插入单细胞基因组DNA,再使用T7 RNA聚合酶进行体外转录获得线性扩增的RNA转录本,之后再通过反转录和第二链合成得到线性扩增的双链DNA模板,这样就得到了足够进行正常建库的DNA样本[22]。LIANTI对于全基因组的覆盖率可以达到97%,等位基因缺失(allele dropout,ADO)率也只有17%,由于使用了更高效的DNA聚合酶,LIANTI技术的单碱基扩增错误率被降到了10-7以下[21]。
上述3种牛EGS常用扩增方法各有优缺点,具体如表 2所示。
|
|
表 2 牛EGS常用3种扩增方法的优缺点 Table 2 Advantages and disadvantages of 3 commonly used amplification methods for bovine EGS |
目前,在马[31]、山羊[32]、绵羊[33]等家畜主要应用植入前遗传学诊断(preimplantation genetic diagnosis)来检测性别决定基因或其他致病基因,很少涉及到全基因组层面的分析。根据现有EGS技术的报道,该技术主要应用在日本黑牛上,主要目的是为了获取牛的基因型数据、选育提高牛的胴体性状、对牛的基因组估计育种值进行估计。
2017年,Fujii等[34]利用刀片法切割从透明带孵化出来的日本黑牛和安格斯牛延长概念的胚胎细胞中提取DNA,经MDA扩增出足够量的DNA后,再通过密度为30K的SNP芯片分析来检测基因分型的准确性,以开发一个结合延长概念移植技术的牛胚胎植入前基因组选择系统,获得准确的基因分型数据,从而通过该技术估计基因组育种值。
此外,2019年,Fujii等[35]对日本黑牛屠体性状进行了基因组选择,通过分别对体内囊胚分割产生的约5个活检细胞、约15个活检细胞提取的DNA经MDA后进行密度为50K的SNP芯片分析,并且活检后玻璃化囊胚的胚胎移植40日妊娠率达到41.9%。首次证明了在日本黑牛胴体性状中使用胚胎附植前基因组选择系统的可行性,表明胚胎附植前基因组选择在日本黑牛胴体性状的实际应用中具有潜力。
2021年,Fujii等[36]还报道了卵裂球分离技术为牛体外受精胚胎的移植前基因组选择提供了一种简单的方法,他在2~8细胞阶段分离体外受精胚胎的卵裂球来产生单卵双胚胎。其中一个单卵半胚胎用于估计育种值进行基因组选择,而另一个用于胚胎移植,并且卵裂球分离技术不会降低胚胎基因组选择的效率。他还评估了第8天分离的卵裂球产生的半囊胚DNA经WGA后进行SNP基因分型的准确性,均得到了良好的检出率。他发现的卵裂球分离技术产生的第8天半囊胚进行准确的SNP基因分型和基因组估计育种值的计算是可能的。表 3汇总了以上3篇报道中应用SNP芯片分析胚胎DNA的检出率。
|
|
表 3 不同密度SNP芯片对胚胎DNA检出率的影响 Table 3 Effect of SNP microarray with different density on the detection rates of embryo DNA |
虽然EGS技术已被成功用于日本黑牛,但仍存在一些问题亟待解决。首先,胚胎活检是胚胎基因组选择的前提,但活检的显微操作需要昂贵的设备和复杂的技术支持[36],并且活检本身会损害胚胎,影响妊娠率[33]。其次,WGA容易出现扩增偏置、污染和覆盖度较差等问题[37]。最后,活检的胚胎需要经过一段时间的冷冻保存后才会选出所需胚胎进行移植,活检胚胎的冷冻存活率低也是制约EGS发展的重要问题[38]。
4.1 胚胎活检中存在的问题影响胚胎活检后剩余胚胎质量的因素有很多。首先就是胚胎活检后的细胞数量会减少[39],这可能会影响胚胎的生存能力[40-41],从而降低妊娠率[42-44]。目前,从4细胞期到桑葚胚期活检会导致早期胚胎停止发育[45-47]、囊胚率较低[48-51]等,同时囊胚期活检对发育中的胚胎的危害小于卵裂球活检[52],所以囊胚期的胚胎活检仍是目前首选的方法[53-54]。与此同时,较少的细胞活检对囊胚的破坏较小,但会导致扩增失败的风险升高,而增加活检细胞的数量会使扩增失败的风险降低,但会增加囊胚破裂的风险[55]。Mara等[56]也提到胚胎活检考虑的主要因素是:1)采集的胚胎细胞数量应该尽量不影响活检胚胎正常发育;2)获得足够数量的DNA以进行分析;3)对胚胎的损害最小。所以目前,囊胚滋养外胚层(trophectoderm,TE)活检所采集的细胞数量一般为5~10个[57]。其次就是囊胚由TE和内细胞团(inner cell mass,ICM)组成,根据Lawrenz等[58]的研究,TE活检的可靠性比ICM活检相对更高。
4.2 WGA中存在的问题成功的全基因组扩增是当代胚胎基因组选择的基石[19]。Del等[59]已经证明下一代测序技术(next-generation sequencing,NGS)对鉴定胚胎活检中突变有用,但现代测序平台要求下一代测序(NGS)技术至少需要1 ng DNA;第三代测序至少需要1 μg DNA;读长测序技术也需要高摩尔重量的DNA作为模板,通常DNA片段需要是>10 kb[60]。因此,为了满足测序的要求,需要WGA具备以下条件: 1)使用具有高加工效率和高保真度的酶,即具有校对活性,以确保较低的错误率和较长的扩增子;2)通过使用通用引物和调整退火或再退火条件(即循环条件)来增加启动次数;3)在扩增(例如通过片段化)之前降低基因组的复杂性[19]。然而,目前的扩增方法在扩增成本、扩增均匀性、基因组覆盖度等方面还存在不足,还有待继续改进。
4.3 活检胚胎冷冻中存在的问题胚胎冷冻保存会降低活检胚胎的活力,进而影响EGS的应用。首先,胎冷冻保存的方法主要包括缓慢冷冻法和玻璃化冷冻法[61],但在牛上玻璃化胚胎比慢速冷冻胚胎具有更高的存活率[62]。Miki等[63]使用小鼠模型检查了从TE活检到玻璃化的不同时间间隔与囊胚存活率和囊胚生长能力的相关性,表明了囊胚玻璃化应在TE活检后1 h或3 h以上进行,在再扩张过程中对活检囊胚进行玻璃化处理可能会降低植入后发育的能力,从而降低妊娠率。此外,根据Wilsher等[64]的报道,存活率还跟活检囊胚的大小有关,根据大小细分胚胎表明,较小胚胎(≤550 μm)玻璃化冷冻的妊娠率更低。
5 小结与展望胚胎基因组选择技术体系在畜牧业中具有重要应用价值,然而为保障胚胎发育潜力和移植妊娠率,现有的胚胎活检体系只能切割微量细胞用于后续的全基因组扩增,微量细胞数的限制无法实现扩增后基因组的覆盖度和准确性。因此需要开发微量细胞的高效扩增体系,目前研究进展中,由于现有单细胞基因组扩增方法还不足以获得所有基因组位点信息,致使胚胎基因组选择技术的应用受限。随着扩增体系的进一步发展,研究人员需要改良单细胞扩增体系,改造高效的高保真聚合酶以及扩增方法等,进一步提高WGA准确性、并需要完善育种SNP芯片以及胚胎活检细胞数量和后期冻存体系,使得移植妊娠率与鲜胚无异,从而保障EGS可以达到产业化应用的地步。
| [1] |
MULLAART E, WELLS D. Embryo biopsies for genomic selection[M]//NIEMANN H, WRENZYCKI C. Animal Biotechnology 2: Emerging Breeding Technologies. Cham: Springer, 2018: 81-94.
|
| [2] |
HOUSTON R D, BEAN T P, MACQUEEN D J, et al. Harnessing genomics to fast-track genetic improvement in aquaculture[J]. Nat Rev Genet, 2020, 21(7): 389-409. DOI:10.1038/s41576-020-0227-y |
| [3] |
胡智辉, 王欢, 衡诺, 等. 高通量SNP芯片在牛体外早期胚胎染色体质量鉴定中的初步应用[J]. 畜牧兽医学报, 2022, 53(11): 3866-3879. HU Z H, WANG H, HENG N, et al. Preliminary application of high throughput SNP chip in chromosome quality identification of bovine early in vitro embryos[J]. Acta Veterinaria et Zootechnica Sinica, 2022, 53(11): 3866-3879. (in Chinese) |
| [4] |
CENARIU M, PALL E, CERNEA C, et al. Evaluation of bovine embryo biopsy techniques according to their ability to preserve embryo viability[J]. J Biomed Biotechnol, 2012, 2012: 541384. |
| [5] |
PONSART C, LE BOURHIS D, KNIJN H, et al. Reproductive technologies and genomic selection in dairy cattle[J]. Reprod Fertil Dev, 2013, 26(1): 12-21. |
| [6] |
GONZÁLEZ-RODRÍGUEZ N, MARTÍNEZ-RODERO I, SCHERZER J, et al. Vitrification and in-straw warming do not affect pregnancy rates of biopsied bovine embryos[J]. Theriogenology, 2022, 191: 221-230. DOI:10.1016/j.theriogenology.2022.07.021 |
| [7] |
TUTT D A R, PASSARO C, WHITWORTH D J, et al. Laser assisted blastomere extrusion biopsy of in vitro produced cattle embryos-A potential high throughput, minimally invasive approach for sampling pre-morula and morula stage embryos[J]. Anim Reprod Sci, 2020, 219: 106546. DOI:10.1016/j.anireprosci.2020.106546 |
| [8] |
LAURI A, LAZZARI G, GALLI C, et al. Assessment of MDA efficiency for genotyping using cloned embryo biopsies[J]. Genomics, 2013, 101(1): 24-29. DOI:10.1016/j.ygeno.2012.09.002 |
| [9] |
姚雅馨, 喇永富, 狄冉, 等. 不同单细胞全基因组扩增方法的比较及MALBAC在辅助生殖中的应用[J]. 遗传, 2018, 40(8): 620-631. YAO Y X, LA Y F, DI R, et al. Comparison of different single cell whole genome amplification methods and MALBAC applications in assisted reproduction[J]. Hereditas (Beijing), 2018, 40(8): 620-631. DOI:10.16288/j.yczz.18-091 (in Chinese) |
| [10] |
DEAN F B, HOSONO S, FANG L H, et al. Comprehensive human genome amplification using multiple displacement amplification[J]. Proc Natl Acad Sci U S A, 2002, 99(8): 5261-5266. DOI:10.1073/pnas.082089499 |
| [11] |
ORDÓÑEZ C D, REDREJO-RODRÍGUEZ M. DNA polymerases for whole genome amplification: considerations and future directions[J]. Int J Mol Sci, 2023, 24(11): 9331. DOI:10.3390/ijms24119331 |
| [12] |
HUTCHISON Ⅲ C A, SMITH H O, PFANNKOCH C, et al. Cell-free cloning using φ29 DNA polymerase[J]. Proc Natl Acad Sci U S A, 2005, 102(48): 17332-17336. DOI:10.1073/pnas.0508809102 |
| [13] |
ZHANG J, SU X L, WANG Y F, et al. Improved single-cell genome amplification by a high-efficiency phi29 DNA polymerase[J]. Front Bioeng Biotechnol, 2023, 11: 1233856. DOI:10.3389/fbioe.2023.1233856 |
| [14] |
PATRO S C, NIYONGABO A, MALDARELLI F, et al. New approaches to multi-parametric HIV-1 genetics using multiple displacement amplification: determining the what, how, and where of the HIV-1 reservoir[J]. Viruses, 2021, 13(12): 2475. DOI:10.3390/v13122475 |
| [15] |
SIDORE A M, LAN F, LIM S W, et al. Enhanced sequencing coverage with digital droplet multiple displacement amplification[J]. Nucleic Acids Res, 2016, 44(7): e66. DOI:10.1093/nar/gkv1493 |
| [16] |
ZHOU Y, JIA E T, QIAO Y, et al. Low bias multiple displacement amplification with confinement effect based on agarose gel[J]. Anal Bioanal Chem, 2021, 413(17): 4397-4405. DOI:10.1007/s00216-021-03415-3 |
| [17] |
LU S J, ZONG C H, FAN W, et al. Probing meiotic recombination and aneuploidy of single sperm cells by whole-genome sequencing[J]. Science, 2012, 338(6114): 1627-1630. DOI:10.1126/science.1229112 |
| [18] |
ZONG C H, LU S J, CHAPMAN A R, et al. Genome-wide detection of single-nucleotide and copy-number variations of a single human cell[J]. Science, 2012, 338(6114): 1622-1626. DOI:10.1126/science.1229164 |
| [19] |
VOLOZONOKA L, MISKOVA A, GAILITE L. Whole genome amplification in preimplantation genetic testing in the era of massively parallel sequencing[J]. Int J Mol Sci, 2022, 23(9): 4819. DOI:10.3390/ijms23094819 |
| [20] |
LI N, WANG L, WANG H, et al. The performance of whole genome amplification methods and next-generation sequencing for pre-implantation genetic diagnosis of chromosomal abnormalities[J]. J Genet Genomics, 2015, 42(4): 151-159. DOI:10.1016/j.jgg.2015.03.001 |
| [21] |
CHEN C Y, XING D, TAN L Z, et al. Single-cell whole-genome analyses by Linear Amplification via Transposon Insertion (LIANTI)[J]. Science, 2017, 356(6334): 189-194. DOI:10.1126/science.aak9787 |
| [22] |
LI N N, JIN K R, BAI Y M, et al. Tn5 transposase applied in genomics research[J]. Int J Mol Sci, 2020, 21(21): 8329. DOI:10.3390/ijms21218329 |
| [23] |
ZHOU X X, XU Y, ZHU L B, et al. Comparison of multiple displacement amplification (MDA) and multiple annealing and looping-based amplification cycles (MALBAC) in limited DNA sequencing based on tube and droplet[J]. Micromachines (Basel), 2020, 11(7): 645. DOI:10.3390/mi11070645 |
| [24] |
FU Y, SHEN X T, WU H T, et al. Preimplantation genetic testing for monogenic disease of spinal muscular atrophy by multiple displacement amplification: 11 unaffected livebirths[J]. Int J Med Sci, 2019, 16(9): 1313-1319. DOI:10.7150/ijms.32319 |
| [25] |
GAWAD C, KOH W, QUAKE S R. Single-cell genome sequencing: current state of the science[J]. Nat Rev Genet, 2016, 17(3): 175-188. DOI:10.1038/nrg.2015.16 |
| [26] |
SOBOL M S, KASTER A K. Back to basics: a simplified improvement to multiple displacement amplification for microbial single-cell genomics[J]. Int J Mol Sci, 2023, 24(5): 4270. DOI:10.3390/ijms24054270 |
| [27] |
YAO K, GONZÁLEZ-ESCALONA N, HOFFMANN M. Multiple displacement amplification as a solution for low copy number plasmid sequencing[J]. Front Microbiol, 2021, 12: 617487. DOI:10.3389/fmicb.2021.617487 |
| [28] |
RUAN Q Y, RUAN W D, LIN X Y, et al. Digital-WGS: automated, highly efficient whole-genome sequencing of single cells by digital microfluidics[J]. Sci Adv, 2020, 6(50): eabd6454. DOI:10.1126/sciadv.abd6454 |
| [29] |
ARAKAWA K. Ultralow-input genome library preparation for nanopore sequencing with droplet MDA[M]//ARAKAWA K. Nanopore Sequencing. New York: Humana, 2023: 91-100.
|
| [30] |
SHOJAEI SAADI H A, VIGNEAULT C, SARGOLZAEI M, et al. Impact of whole-genome amplification on the reliability of pre-transfer cattle embryo breeding value estimates[J]. BMC Genomics, 2014, 15(1): 889. DOI:10.1186/1471-2164-15-889 |
| [31] |
GUIGNOT F, REIGNER F, PERREAU C, et al. Preimplantation genetic diagnosis in Welsh pony embryos after biopsy and cryopreservation[J]. J Anim Sci, 2015, 93(11): 5222-5231. DOI:10.2527/jas.2015-9469 |
| [32] |
GUIGNOT F, PERREAU C, CAVARROC C, et al. Sex and PRNP genotype determination in preimplantation caprine embryos[J]. Reprod Domest Anim, 2011, 46(4): 656-663. DOI:10.1111/j.1439-0531.2010.01724.x |
| [33] |
GUIGNOT F, BARIL G, DUPONT F, et al. Determination of sex and scrapie resistance genotype in preimplantation ovine embryos[J]. Mol Reprod Dev, 2009, 76(2): 183-190. DOI:10.1002/mrd.20940 |
| [34] |
FUJII T, HIRAYAMA H, NAITO A, et al. Production of calves by the transfer of cryopreserved bovine elongating conceptuses and possible application for preimplantation genomic selection[J]. J Reprod Dev, 2017, 63(5): 497-504. DOI:10.1262/jrd.2017-025 |
| [35] |
FUJII T, NAITO A, HIRAYAMA H, et al. Potential of preimplantation genomic selection for carcass traits in Japanese Black cattle[J]. J Reprod Dev, 2019, 65(3): 251-258. DOI:10.1262/jrd.2019-009 |
| [36] |
FUJII T, NAITO A, MORIYASU S, et al. Potential of preimplantation genomic selection using the blastomere separation technique in bovine in vitro fertilized embryos[J]. J Reprod Dev, 2021, 67(2): 155-159. DOI:10.1262/jrd.2020-153 |
| [37] |
LEUNG K, KLAUS A, LIN B K, et al. Robust high-performance nanoliter-volume single-cell multiple displacement amplification on planar substrates[J]. Proc Natl Acad Sci U S A, 2016, 113(30): 8484-8489. DOI:10.1073/pnas.1520964113 |
| [38] |
PÉREZ-MARÍN C C, VIZUETE G, VAZQUEZ-MARTINEZ R, et al. Comparison of different cryopreservation methods for horse and donkey embryos[J]. Equine Vet J, 2018, 50(3): 398-404. DOI:10.1111/evj.12777 |
| [39] |
LOS F J, VAN OPSTAL D, VAN DEN BERG C. The development of cytogenetically normal, abnormal and mosaic embryos: a theoretical model[J]. Hum Reprod Update, 2004, 10(1): 79-94. DOI:10.1093/humupd/dmh005 |
| [40] |
SCOTT R T Jr, FERRY K, SU J, et al. Comprehensive chromosome screening is highly predictive of the reproductive potential of human embryos: a prospective, blinded, nonselection study[J]. Fertil Steril, 2012, 97(4): 870-875. DOI:10.1016/j.fertnstert.2012.01.104 |
| [41] |
BRODIE D, BEYER C E, OSBORNE E, et al. Preimplantation genetic diagnosis for chromosome rearrangements- one blastomere biopsy versus two blastomere biopsy[J]. J Assist Reprod Genet, 2012, 29(8): 821-827. DOI:10.1007/s10815-012-9782-2 |
| [42] |
ADLER A, LEE H L, MCCULLOH D H, et al. Blastocyst culture selects for euploid embryos: comparison of blastomere and trophectoderm biopsies[J]. Reprod Biomed Online, 2014, 28(4): 485-491. DOI:10.1016/j.rbmo.2013.11.018 |
| [43] |
SCOTT R T Jr, UPHAM K M, FORMAN E J, et al. Cleavage-stage biopsy significantly impairs human embryonic implantation potential while blastocyst biopsy does not: a randomized and paired clinical trial[J]. Fertil Steril, 2013, 100(3): 624-630. DOI:10.1016/j.fertnstert.2013.04.039 |
| [44] |
LEAVER M, WELLS D. Non-invasive preimplantation genetic testing (niPGT): the next revolution in reproductive genetics?[J]. Hum Reprod Update, 2020, 26(1): 16-42. DOI:10.1093/humupd/dmz033 |
| [45] |
MASTENBROEK S, TWISK M, VAN ECHTEN-ARENDS J, et al. In vitro fertilization with preimplantation genetic screening[J]. N Engl J Med, 2007, 357(1): 9-17. DOI:10.1056/NEJMoa067744 |
| [46] |
ZAKHAROVA E E, ZALETOVA V V, KRIVOKHARCHENKO A S. Biopsy of human morula-stage embryos: outcome of 215 IVF/ICSI cycles with PGS[J]. PLoS One, 2014, 9(9): e106433. DOI:10.1371/journal.pone.0106433 |
| [47] |
MUNNÉ S. Chromosome abnormalities and their relationship to morphology and development of human embryos[J]. Reprod Biomed Online, 2006, 12(2): 234-253. DOI:10.1016/S1472-6483(10)60866-8 |
| [48] |
STAESSEN C, VERPOEST W, DONOSO P, et al. Preimplantation genetic screening does not improve delivery rate in women under the age of 36 following single-embryo transfer[J]. Hum Reprod, 2008, 23(12): 2818-2825. DOI:10.1093/humrep/den367 |
| [49] |
DEBROCK S, MELOTTE C, SPIESSENS C, et al. Preimplantation genetic screening for aneuploidy of embryos after in vitro fertilization in women aged at least 35 years: a prospective randomized trial[J]. Fertil Steril, 2010, 93(2): 364-373. DOI:10.1016/j.fertnstert.2008.10.072 |
| [50] |
HARDARSON T, HANSON C, LUNDIN K, et al. Preimplantation genetic screening in women of advanced maternal age caused a decrease in clinical pregnancy rate: a randomized controlled trial[J]. Hum Reprod, 2008, 23(12): 2806-2812. DOI:10.1093/humrep/den217 |
| [51] |
SULLIVAN-PYKE C, DOKRAS A. Preimplantation genetic screening and preimplantation genetic diagnosis[J]. Obstet Gynecol Clin North Am, 2018, 45(1): 113-125. DOI:10.1016/j.ogc.2017.10.009 |
| [52] |
CHUANG T H, HSIEH J Y, LEE M J, et al. Concordance between different trophectoderm biopsy sites and the inner cell mass of chromosomal composition measured with a next-generation sequencing platform[J]. Mol Hum Reprod, 2018, 24(12): 593-601. DOI:10.1093/molehr/gay043 |
| [53] |
KULIEV A, RECHITSKY S. Preimplantation genetic testing: current challenges and future prospects[J]. Expert Rev Mol Diagn, 2017, 17(12): 1071-1088. DOI:10.1080/14737159.2017.1394186 |
| [54] |
CIMADOMO D, CAPALBO A, UBALDI F M, et al. The impact of biopsy on human embryo developmental potential during preimplantation genetic diagnosis[J]. Biomed Res Int, 2016, 2016: 7193075. |
| [55] |
AOYAMA N, KATO K. Trophectoderm biopsy for preimplantation genetic test and technical tips: a review[J]. Reprod Med Biol, 2020, 19(3): 222-231. DOI:10.1002/rmb2.12318 |
| [56] |
MARA L, PILICHI S, SANNA A, et al. Sexing of in vitro produced ovine embryos by duplex PCR[J]. Mol Reprod Dev, 2004, 69(1): 35-42. DOI:10.1002/mrd.20147 |
| [57] |
ROSSANT J, TAM P P L. Early human embryonic development: blastocyst formation to gastrulation[J]. Dev Cell, 2022, 57(2): 152-165. DOI:10.1016/j.devcel.2021.12.022 |
| [58] |
LAWRENZ B, EL KHATIB I, LIÑÁN A, et al. The clinicians' dilemma with mosaicism-an insight from inner cell mass biopsies[J]. Hum Reprod, 2019, 34(6): 998-1010. DOI:10.1093/humrep/dez055 |
| [59] |
DEL REY J, VIDAL F, RAMÍREZ L, et al. Novel double factor PGT strategy analyzing blastocyst stage embryos in a single NGS procedure[J]. PLoS One, 2018, 13(10): e0205692. DOI:10.1371/journal.pone.0205692 |
| [60] |
LU N, QIAO Y, LU Z H, et al. Chimera: the spoiler in multiple displacement amplification[J]. Comput Struct Biotechnol J, 2023, 21: 1688-1696. DOI:10.1016/j.csbj.2023.02.034 |
| [61] |
VALENTE R S, MARSICO T V, SUDANO M J. Basic and applied features in the cryopreservation progress of bovine embryos[J]. Anim Reprod Sci, 2022, 239: 106970. DOI:10.1016/j.anireprosci.2022.106970 |
| [62] |
NAJAFZADEH V, SECHER J B M, PIHL M, et al. Vitrification yields higher cryo-survival rate than slow freezing in biopsied bovine in vitro produced blastocysts[J]. Theriogenology, 2021, 171: 44-54. DOI:10.1016/j.theriogenology.2021.04.020 |
| [63] |
MIKI T, EZOE K, KOURABA S, et al. Time from trophectoderm biopsy to vitrification affects the developmental competence of biopsied blastocysts[J]. Reprod Med Biol, 2022, 21(1): e12439. DOI:10.1002/rmb2.12439 |
| [64] |
WILSHER S, RIGALI F, COUTO G, et al. Vitrification of equine expanded blastocysts following puncture with or without aspiration of the blastocoele fluid[J]. Equine Vet J, 2019, 51(4): 500-505. DOI:10.1111/evj.13039 |
(编辑 郭云雁)



