奶牛的泌乳初期一般指产后15 d,又称为围产后期,产后16~100 d为泌乳盛期[1-2]。泌乳早期,特别是在奶牛产犊后的第1个月,高水平的产奶加上低采食量容易造成能量负平衡,更易造成代谢紊乱,影响奶牛健康和生育能力[3]。在泌乳30 d内,奶牛从泌乳初期过渡到泌乳盛期,在这两个状态的转变中,奶牛在生理和代谢上,都要经历很大的变化和一系列应激,包括泌乳启动、适应日粮、环境改变等。这些应激在很大程度上影响奶牛的健康水平及生产性能。因此,泌乳30 d对奶牛来说是一个挑战。
奶牛产犊后,牧场会提高精饲料比例,为瘤胃细菌提供更易获得的能量来源,瘤胃微生物组成也随之改变[4]。蒋涛[1]发现,泌乳初期和泌乳盛期奶牛瘤胃中细菌群落组成在属分类水平上存在明显差异,泌乳7 d瘤胃液中普雷沃菌属和罗氏菌属丰度显著高于泌乳50 d。同时,随着产奶量的增加,奶牛摄取的营养物质远远达不到产奶量的要求,这时就要动员体内储存的脂肪,以弥补泌乳早期的能量不足[5]。前期研究表明,牛奶脂肪酸中的C4~C14及一半的C16:0在乳腺中生成,其余长链脂肪酸(long chain fatty acid, LCFA)则来源于血液外源转化[6-7],而血液中的LCFA主要来自饲料、瘤胃微生物或体脂动员。在饲喂良好、体况正常的奶牛中,脂解和体脂动员的脂肪酸占牛奶脂肪酸的5%,但在泌乳早期,当奶牛处于能量负平衡时,比例可提高至20%[8]。樊永亮[2]发现,泌乳3和30 d乳中脂肪酸组成差异较大,随着泌乳时间的延长,C4~C16的含量逐渐上升,而C18:0、C18:1cis9、C18:2cis9, 12的含量逐渐下降。由此可见,生理阶段会影响奶牛瘤胃细菌的组成和乳腺中脂肪酸的代谢[9-11],但鲜有文献分析泌乳早期瘤胃微生物组成对牛乳中脂肪酸组成的影响。本研究旨在分析泌乳早期奶牛瘤胃微生物组成及牛乳脂肪酸组成的变化,探究瘤胃微生物组成与乳脂代谢之间的关联,为产后奶牛营养调控和提高原料乳品质提供理论基础。
1 材料与方法 1.1 试验动物与样品采集选取江苏省某奶牛场2~3胎次、健康无病且305 d产奶量、年龄、体况评分和预产期相近的荷斯坦牛50头,每日采用全混合日粮(TMR)饲喂2次,饲粮组成及营养水平见表 1。分别于每头牛的泌乳7 d(泌乳初期)及30 d(泌乳盛期)采集奶样和瘤胃液样品,记录采样当日干物质采食量及产奶量,每头奶牛采集100 mL混合奶样(早、中、晚的比例4∶3∶3),混匀后用于乳品质指标及乳脂肪酸的测定。饲喂2 h后,固定牛只,通过牛口腔导管采集瘤胃液20 mL,立即液氮保存,用于瘤胃微生物组的测定。
|
|
表 1 日粮组成及营养水平(干物质基础) Table 1 Composition and nutrient levels of the diet (dry matter basis) |
利用MilkoScan FT-1型多功能乳品分析仪(multifunctional dairy analyzer, Foss Electric, 丹麦)测定乳成分。
奶样离心后取上层脂肪,装入带盖玻璃瓶中,加入乙醚∶二氯甲烷∶己烷(1∶10∶89),加入C11:0,作为内标,加入乙酸甲酯,混匀后加入氢氧化钠甲醇溶液,旋涡震荡,室温静置10 min;离心后,转移上清至气相小瓶中,氮吹仪吹干至0.5 mL,过滤后取1 μL进行气相色谱检测,脂肪酸甲酯标准品为GLC-463, 674, 481B(NuChek Prep,美国),检测仪器为7890-B气相色谱仪,色谱柱为DB-HP毛细管气相色谱柱(100 m×0.25 mm×0.25 μm,Agilent,美国),具体检测步骤参照文献[12]进行,脂肪酸的含量为每种脂肪酸的峰面积占该样品中所有脂肪酸峰面积之和的百分比,使用单个脂肪酸含量计算饱和脂肪酸(saturated fatty acid, SFA)、单不饱和脂肪酸(monounsaturated fatty acid, MUFA)、多不饱和脂肪酸(polyunsaturated fatty acid, PUFA)、短链脂肪酸(short chain fatty acid, SCFA)、中链脂肪酸(medium chain fatty acid, MCFA)、LCFA、反式脂肪酸(trans fatty acid, TRANS)和奇数碳脂肪酸(odd-carbon fatty acid, ODFA)。
1.3 Illumina测序采用QIAamp PowerFecal DNA提取试剂盒(Qiagen,美国)提取瘤胃微生物DNA,具体方法参照说明书。使用NanoDrop1000(Thermo Fisher Scientific,美国)测定DNA浓度,使用0.8%(w/v)琼脂糖凝胶电泳分析DNA完整性。提取的DNA标准化至40 ng·μL-1进行测序。利用Illumina HiSeq V2平台对16S rRNA的V3-V4区进行测序(引物序列为:341F,5′-CCTACGGGNGGCWGCAG-3′;806R,5′-GGACTACHVGGGTATCTAAT-3′),测序服务委托广州基迪奥生物科技有限公司。
1.4 测序数据分析首先利用R语言中的DADA2工具包,对原始数据进行去重、校正、降噪、去嵌合体,获得扩增序列变体(amplicon sequence variant,ASV)的序列和丰度信息,比对SILVA数据库(132)获得物种分类注释信息。根据ASV丰度信息获得各层级的物种丰度信息。使用R语言ggplot2包(2.2.1)绘制物种丰度堆叠图;使用QIIME(1.9.1)分析α多样性指数,Chao1和Observed species代表了细菌群落的丰富度,Shannon和Simpson指数则反映了细菌群落的物种多样性;利用PICRUSt(2.1.4)进行细菌的KEGG代谢通路预测,分类并富集到KEGG的level 3水平;并利用R语言Vegan包(2.5.3)分析两组间物种丰度差异、α多样性指数的差异、主成分分析(PCA)及功能差异。
1.5 统计分析采用SPSS 16.0软件中的单因素方差分析模型(one-way ANOVA,LSD)分析不同泌乳天数奶牛生产性能(干物质采食量、日产奶量、乳脂率等)及乳脂肪酸含量的组间差异,并采用皮尔森相关系数(pearson correlation coefficient)评价瘤胃微生物丰度和乳脂肪酸含量的相关性(|r|≥0.25)。试验结果用“平均值±标准误(Mean±SE)”表示,P<0.01表示差异极显著,P<0.05表示差异显著,0.05<P<0.1表示有显著趋势。
2 结果 2.1 泌乳7和30 d生产性能的比较如表 2,泌乳7 d的干物质采食量为15.79 kg·d-1,日产奶量为26.81 kg,均极显著低于泌乳30 d的干物质采食量(18.87 kg·d-1)和日产奶量(37.47 kg)(P<0.01),而泌乳7 d的乳脂率(4.49%)、乳蛋白率(3.76%)和体细胞评分(3.19)极显著高于泌乳30 d的乳脂率(3.62%)、乳蛋白率(3.09%)和体细胞评分(1.74)(P<0.01)。体细胞数在两个泌乳日间存在降低的趋势(P<0.1),而乳中脂蛋白比和尿素氮含量在两个泌乳日间差异不显著(P>0.05)。
|
|
表 2 泌乳7和30 d生产性能的比较分析 Table 2 Comparative analysis of production performance between d7 and d30 |
在本研究中,用气相色谱法共检出49种乳脂肪酸,其中16种脂肪酸含量在泌乳初期变化显著(P<0.05),如表 3所示,泌乳7 d牛奶中的C6:0、C8:0、C10:0、C12:0、C14:0、C14:1trans9、C14:1cis9、C15:1trans10、MCFA和SFA的比例显著低于泌乳30 d(P<0.05),相反地,C17:0、C17:1cis10、C18:1trans6、C18:1trans11、C18:1cis9、C18:1cis11、C19:1trans10、C22:5cis4, 7, 10, 13, 16、LCFA、MUFA和TRANS的比例显著高于泌乳30 d(P<0.05)。
|
|
表 3 泌乳7和30 d差异乳脂肪酸的比较分析 Table 3 Comparative analysis of different milk fatty acids between d7 and d30 |
测序数据经质控处理后,每个瘤胃液样品平均获得52 506条reads,经DADA2软件处理后,每个样品有6 322个ASV,用于下游分析。瘤胃细菌群落的α多样性和β多样性分析显示两个泌乳日瘤胃细菌的α指数(Chao1、Observed、Shannon、Simpson)之间均不存在显著差异(P>0.05),表明泌乳7和30 d奶牛瘤胃细菌的丰富度与多样性无差异(图 1)。
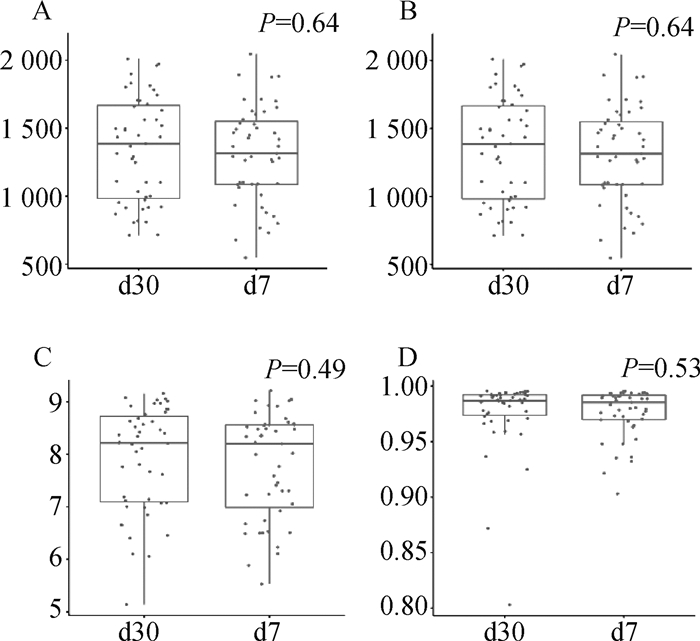
|
A. Chao1指数;B. Observed;C. Shannon;D. Simpson A. Chao1 index; B. Observed; C. Shannon; D. Simpson 图 1 瘤胃细菌菌群α多样性指数 Fig. 1 Alpha diversity index of rumen bacterial community |
PCA主坐标分析结果如图 2所示,第1主坐标(PC1)和第二主坐标(PC2)的贡献率分别为99.5%和0.3%,且两个泌乳日奶牛瘤胃细菌群落结构未明显区分开,说明奶牛泌乳7和30 d之间的瘤胃细菌组成结构差异不显著(P>0.05)。
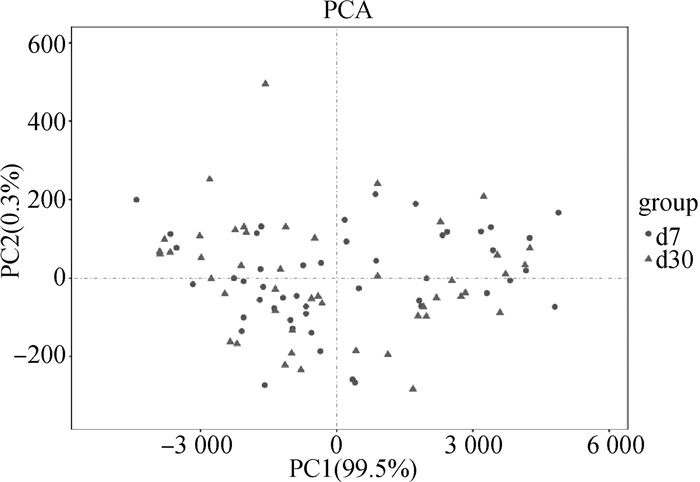
|
圆点表示泌乳7 d,三角表示泌乳30 d Circles represent d7, while triangles represent d30 图 2 奶牛瘤胃细菌菌群PCA图 Fig. 2 PCA graph of rumen bacterial community in dairy cows |
试验共检测到29个菌门,其中表达丰度最高的18个菌门如图 3所示。厚壁菌门(Firmicutes)和拟杆菌门(Bacteroidetes)占到总量的84%以上。在属水平上,94个瘤胃液样品共检测到178个属,普雷沃氏菌属(Prevotella_1)占比最高,可达到总菌属的13.93%。

|
图 3 奶牛瘤胃细菌物种门水平分布堆叠图 Fig. 3 Stack diagram of rumen bacterial species distribution at phylum level |
如图 4所示,在属水平上,相对丰度>1%的差异微生物有9种,与泌乳7 d相比,泌乳30 d奶牛瘤胃内的瘤胃梭菌属(Ruminiclostridium_1)、瘤胃球菌属(Ruminococcaceae_UCG_014和Ruminococcaceae_UCG_005)、梭菌属(Clostridium_sensu_stricto_1)和小杆菌属(Dialister)的相对丰度显著增加(P<0.05)。而球藻菌属(Sphaerochaeta)、弯曲菌属(Campylobacter)和真细菌属(Eubacterium_saphenum_group)的丰度显著降低(P<0.05)。
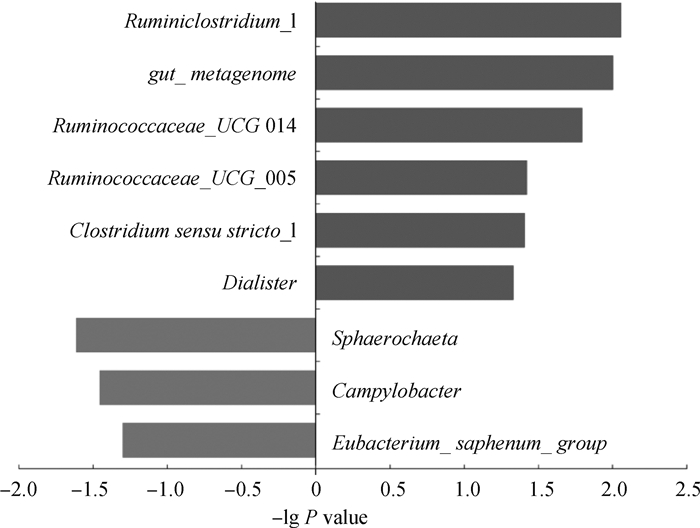
|
图 4 泌乳7和30 d瘤胃差异微生物 Fig. 4 Different rumen bacteria between d7 and d30 |
将两组间的差异微生物进行KEGG富集分析(表 4),发现泌乳天数不同,差异瘤胃微生物主要富集在脂质及免疫代谢通路中,如线粒体中脂肪酸的伸长、糖胺聚糖生物合成(P<0.1)等。
|
|
表 4 差异微生物的KEGG富集分析 Table 4 KEGG pathway enrichment of different rumen bacteria |
将泌乳7和30 图6所示。瘤胃内Campylobacter丰度与乳中LCFA、MUFA和TRANS含量显著正相关(P<0.05),如C17:0、C17:1cis10、C18:1trans6、C18:1trans11、C18:1cis9、C18:1cis11和C19:1trans10,而与MCFA和SFA含量显著负相关(P<0.05),如C6:0、C8:0、C10:0、C12:0和C14:0。Dialister与C14:1cis9呈正相关(P<0.05),gut_metagenome与C18:1trans6和C22:5cis4, 7, 10, 13, 16含量呈负相关(P<0.05)。Ruminiclostridium_1、Ruminococcaceae_UCG_005均与C15:1trans10呈正相关(P<0.05),与C17:0、C18:1trans6、C18:1trans11、C19:1trans10和TRANS呈极显著负相关(P<0.01)。Ruminococcaceae_UCG_005和Ruminococcaceae_UCG_014均与C22:5cis4, 7, 10, 13, 16含量呈负相关(P<0.05)。图6中大部分微生物的丰度均与C17:0、C18:1trans6、C18:1trans11和C19:1trans10的含量相关。Campylobacter和Sphaerochaeta与这些脂肪酸含量显著正相关(P<0.05),而Ruminiclostridium_1和Ruminococcaceae_UCG_005与这些脂肪酸含量显著负相关(P<0.05)。

|
*. 差异显著(P<0.05);**. 差异极显著(P<0.01) *. Significant difference (P < 0.05); **. Extremely significant difference(P < 0.01) 图 5 乳脂肪酸与瘤胃微生物关联分析 Fig. 5 The correlation between rumen bacteria and milk fatty acids |
随着泌乳时间的延长,奶牛的采食量不断上升,其干物质采食量在泌乳8~10周可达到高峰。本研究发现,泌乳7 d采食量显著低于泌乳30 d,说明围产后期奶牛采食量显著低于泌乳盛期,围产后期采食量低可能是由于产犊后奶牛日粮成分的突然改变引发奶牛厌食,通常瘤胃要4~6周才能适应高精日粮,还可能与瘤胃pH的变化有关,表现为瘤胃运动减弱,食物郁积,排空弛缓,进而引起采食量下降[1]。此外,在泌乳早期,随着泌乳时间的延长,乳中的乳蛋白率和乳脂率逐渐下降[2]。陈永华等[13]发现,泌乳7 d的乳脂率为5.00%,高于常乳的乳脂率(3.46%),而泌乳7 d的乳蛋白率(3.11%)和常乳(3.01%)接近。雷晓薇等[14]发现,健康奶牛泌乳开始时,牛奶中的体细胞数会立刻上升,在泌乳35 d会迅速下降。泌乳初期乳中体细胞数高,并不一定表明奶牛出现了乳房炎的症状,这可能是一种泌乳初期的乳腺生理特征,此时,细胞可通过乳腺上皮细胞间紧密连接中的漏洞逃逸至牛奶中[15]。这些结论与本研究结果相一致,即泌乳7 d的乳脂率、乳蛋白率和体细胞评分显著高于泌乳30 d,此外,本研究还发现,泌乳7 d的日产奶量显著低于泌乳30 d。在之前的研究中也发现泌乳早期产奶量是逐渐升高的,这与采食量不断上升有关[16]。
3.2 泌乳早期奶牛乳脂肪酸组成的变化牛奶脂肪酸中的C4~C14及一半的C16:0在乳腺中生成,其余LCFA则来源于血液外源转化[7]。本研究发现泌乳7 d牛奶的中链饱和脂肪酸含量(C6~C14)显著低于泌乳30 d,而长链单不饱和脂肪酸(C18:1、C19:1trans10)则呈相反的趋势,泌乳7 d显著高于泌乳30 d。由此可见,与泌乳30 d相比,泌乳7 d时,乳腺合成的脂肪酸少,而LCFA多,可能与泌乳初期的体脂动员有关[5]。奶牛产犊后,泌乳的能量需要急剧增加,干物质采食量不足,机体被迫动用储备体脂获得能量。动员体脂进入血液后,形成非酯化脂肪酸,在泌乳早期,血液中40%以上的非酯化脂肪酸被用于合成乳脂[1]。此外,Bionaz和Loor[17]发现,泌乳初期的乳腺上皮细胞内,从血液中摄取LCFA相关的基因表达量较高。Kay等[11]比较了泌乳1周和8周时牛奶脂肪酸组成的差异,结果发现泌乳1周奶牛牛奶中的C6:0、C8:0、C10:0、C12:0、C14:0低于泌乳8周的,而泌乳1周的C17:0、C18:1cis9高于泌乳8周的,其变化趋势与本文的研究一致。C18:1cis9有降低血脂血压、抗血栓、抑敏抗炎、抑制癌症转移等生理功能,且乳品质越高,其LCFA、PUFA比例越高[18]。本研究发现,泌乳7 d牛奶中的C22:5cis4, 7, 10, 13, 16含量显著高于30 d,多项研究表明C22:5cis4, 7, 10, 13, 16对降低炎症和改善血脂有直接作用,其脂质衍生物可促进炎症因子的降解[19]。付力立[20]比较了泌乳首日与泌乳21 d牛奶的代谢组差异,结果表明,两个泌乳日乳中共存在97种差异显著的代谢物。泌乳首日乳中多种PUFA(C18:2cis9, 12、C20:4cis5, 8, 11, 14)的含量显著高于泌乳21 d,这些脂肪酸在炎症调控的生理机能中尤为重要,因此,泌乳初期中优质脂类的存在主要是为新生犊牛羸弱的身体提供被动免疫以及激活主动免疫,从而促进初生犊牛的存活。
3.3 泌乳早期瘤胃细菌组成的变化许多研究表明,瘤胃微生物区系在各泌乳阶段显著不同,Derakhshani等[21]比较了奶牛泌乳10、20及28 d瘤胃细菌的变化,发现产后随着干物质采食量的上升,Prevotellaceae、Streptococcus和Lactobacillus等与蛋白质降解、淀粉水解有关的菌群丰度显著升高,且这些微生物主要富集在能量代谢通路上,表明瘤胃微生物在一定程度上适应了高精料日粮。不同生理时期奶牛瘤胃微生物的结构和组成存在显著差异,可能因为不同泌乳时期奶牛瘤胃中微生物群落的适应性存在差异,或者是不同生理时期奶牛所面临的内部和外部应激不一样所致[22]。本研究发现,泌乳30 d奶牛瘤胃内Ruminiclostridium、Ruminococcaceae、Clostridium和Dialister的相对丰度显著高于泌乳7 d。Bach等[23]的发现与本试验结果相同,奶牛泌乳21 d时,瘤胃内的Ruminococcaceae UCG-014和Dialister的相对丰度高于泌乳7 d。此外,高产奶牛瘤胃内Ruminococcaceae和Dialister的丰度高于低产奶牛[24-25]。Ruminiclostridium和Ruminococcaceae是瘤胃内主要的纤维素降解菌,同时也是潜在的有益细菌,它们可参与消化道环境的调节,与炎症和细胞毒性因子的释放有关,并与免疫调节和健康的内稳态有关[26]。由此可见,泌乳30 d内,奶牛瘤胃的纤维素降解能力逐渐上升。纤维素降解菌可发酵饲粮,产生乙酸、丁酸、H2和CO2[27]。Sawanon等[28]发现,瘤胃内纤维素降解菌(瘤胃球菌)丰度与瘤胃内乙酸浓度显著正相关。在泌乳早期,瘤胃内发酵菌的数量和比例发生改变,瘤胃内的发酵参数也会发生改变[29]。Cersosimo等[30]发现,泌乳盛期(泌乳93 d)奶牛瘤胃内乙酸浓度显著高于泌乳初期(泌乳3 d)。由此可见,泌乳30 d内,瘤胃内的纤维素降解菌丰度上升,可导致瘤胃内乙酸浓度上升。瘤胃内Clostridium是一种蛋白质降解菌,其丰度与牛奶中的MUN含量正相关[21],这与本文报道不同,虽然Clostridium丰度在两个泌乳日差异显著,乳中尿素氮含量差异却不显著。
此外,本研究还发现,泌乳30 d瘤胃内Sphaerochaeta、Campylobacter和Eubacterium丰度显著低于泌乳7 d,Campylobacter是一种潜在的硝酸盐还原菌,当动物肠道机能紊乱时,硝酸盐还原菌会大量繁殖,此外,当饲料中添加硝酸盐时,瘤胃内Campylobacter的丰度会上升[31]。Eubacterium可释放丁酸盐或丙酸盐等代谢产物,与乙酸相比,这类代谢物可为奶牛提供更高的能量[32]。由此可见,泌乳30 d内,奶牛瘤胃机能逐渐恢复,潜在的致病菌丰度逐渐下降,这也与文献报道相一致[33]。
3.4 瘤胃细菌与乳脂肪酸的相关性许多研究表明,瘤胃菌群与奶牛的生产性能显著相关,Jami等[25]发现,产奶量与瘤胃微生物多样性密切相关,且Firmicutes/Bacteroidetes的比例与乳脂率呈极显著正相关。奶牛瘤胃内Clostridium、unclassified Lachnospiraceae和unclassified Erysipelotrichaceae的丰度与乳蛋白率显著正相关[21]。本研究发现,瘤胃Campylobacter与乳中MCFA(C6~C14)具有显著的相关性,而其余细菌与MCFA无显著相关性。这表明瘤胃细菌对乳中脂肪酸的从头合成影响不大,这与Buitenhuis等[34]报道一致,即瘤胃微生物对乳中MCFA含量影响较小,他们还发现瘤胃细菌对乳中ODFA和C18多不饱和脂肪酸的影响更为明显。虽然本研究并未发现瘤胃细菌对C18多不饱和脂肪酸的影响,但随着纤维素降解细菌丰度的增加,如Ruminiclostridium-1、Ruminococcaceae_UCG_005,乳中C17:0、C18:1trans6、C18:1trans11和C19:1trans10的含量会显著降低。而Campylobacter和Sphaerochaeta丰度则与乳中的这些脂肪酸的含量显著正相关。Vlaeminck等[35]研究表明,反刍动物乳中的ODFA一般来源于瘤胃细菌的细胞膜,当瘤胃微生物死亡进入小肠后,这些ODFA会被宿主消化吸收后沉积到奶中。瘤胃细菌可发酵饲粮产生挥发性脂肪酸,然后利用这些代谢产物合成细菌自身的脂类等物质。如瘤胃微生物在脂肪酸合成酶作用下,以丙酸和戊酸为前体合成ODFA。不同类型瘤胃微生物合成的脂肪酸组成不同。纤维素降解菌的升高可提高异构脂肪酸的比例,而反式异构脂肪酸和ODFA在淀粉分解菌中较丰富[27]。Liu等[36]发现,牦牛瘤胃内Ruminococcaceae_UCG-010和UCG-011与瘤胃内戊酸浓度负相关,而戊酸是微生物合成ODFA的底物,由此可见,瘤胃内的纤维素降解菌Ruminiclostridium-1、Ruminococcaceae_UCG_005与乳中C17:0含量负相关的原因可能与瘤胃内的戊酸浓度有关。Liu等[37]发现瘤胃内Ruminococcus albus、Ruminococcus flavefaciens和Eubacterium ruminantium丰度与乳中C13:0含量相关,提示乳中C13:0可能反映了瘤胃纤维素降解菌的数量。因此推测,C17:0可能反映了泌乳初期瘤胃中纤维素降解菌Ruminiclostridium-1、Ruminococcaceae_UCG_005的数量。
乳中的反式脂肪酸主要来源于奶牛瘤胃内饲粮PUFA的不完全生物氢化产物。Kennelly等[38]研究表明,牛奶中C18反式脂肪酸浓度的增加与饲喂高精料奶牛的乳脂降低有关,因为这类脂肪酸可抑制乳腺组织中脂肪酸的从头合成和甘油三酯的合成。研究发现,瘤胃微生物按照参与生物氢化的过程可分为A、B两类:A类微生物将C18:2n6及C18:3n3转换为C18:1trans11;B类微生物将C18:1trans11转化为C18:0。目前已发现Bifidobacterium、Clostridium、Eubacterium、Lactobacillus、Ruminococcus等微生物属于A类微生物[39]。Huws等[40]发现Ruminococcaceae和Lachnospiraceae在瘤胃生物氢化过程中起着重要的作用。本研究发现泌乳30 d乳中C18:1trans6和C18:1trans11的含量低于泌乳7 d,可能与瘤胃内Ruminococcaceae丰度变化相关。
4 结论泌乳7 d奶牛的干物质采食量和产奶量低于泌乳30 d,但其乳脂率、乳蛋白率和体细胞评分均高于泌乳30 d,且16种乳脂肪酸含量在两个泌乳日间变化显著。奶牛泌乳30 d瘤胃内纤维素降解菌丰度高于泌乳7 d,而硝酸盐还原菌Campylobacter和真细菌Eubacterium丰度低于泌乳7 d。相关性分析显示瘤胃微生物对乳中MCFA含量影响较小,且纤维素降解细菌(Ruminiclostridium 1、Ruminococcaceae UCG 005)与乳中C17:0、C18:1trans6、C18:1trans11和C19:1trans10的含量显著负相关。
| [1] |
蒋涛. 瘤胃微生物重塑对围产后期奶牛采食量和采食行为的影响[D]. 北京: 中国农业大学, 2018. JIANG T. Influence of rumen microbial reshaping on feed intake and feeding behavior of Holstein Cows during the late perinatal period[D]. Beijing: China Agricultural University, 2018. (in Chinese) |
| [2] |
樊永亮. 奶牛泌乳早期乳脂肪酸变化及乳腺组织转录调控分析[D]. 扬州: 扬州大学, 2017. FAN Y L. Fatty acids changes of milk and transcriptional regulation analysis of mammary gland tissue in the early milk of dairy cows[D]. Yangzhou: Yangzhou University, 2017. (in Chinese) |
| [3] |
JOUANY J P. Optimizing rumen functions in the close-up transition period and early lactation to drive dry matter intake and energy balance in cows[J]. Anim Reprod Sci, 2006, 96(3-4): 250-264. DOI:10.1016/j.anireprosci.2006.08.005 |
| [4] |
ELOLIMY A A, ARROYO J M, BATISTEL F, et al. Association of residual feed intake with abundance of ruminal bacteria and biopolymer hydrolyzing enzyme activities during the peripartal period and early lactation in Holstein dairy cows[J]. J Anim Sci Biotechnol, 2018, 9: 43. DOI:10.1186/s40104-018-0258-9 |
| [5] |
XU W, VERVOORT J, SACCENTI E, et al. Relationship between energy balance and metabolic profiles in plasma and milk of dairy cows in early lactation[J]. J Dairy Sci, 2020, 103(5): 4795-4805. DOI:10.3168/jds.2019-17777 |
| [6] |
POULSEN N A, HEIN L, KARGO M, et al. Realization of breeding values for milk fatty acids in relation to seasonal variation in organic milk[J]. J Dairy Sci, 2020, 103(3): 2434-2441. DOI:10.3168/jds.2019-17065 |
| [7] |
陈美庆, 张养东, 郑楠, 等. 牛奶中脂肪酸的合成机理及影响因素研究进展[J]. 动物营养学报, 2021, 33(8): 4244-4254. CHEN M Q, ZHANG Y D, ZHENG N, et al. Advance in synthesis mechanism and influencing factors of milk fatty acids[J]. Chinese Journal of Animal Nutrition, 2021, 33(8): 4244-4254. DOI:10.3969/j.issn.1006-267x.2021.08.005 (in Chinese) |
| [8] |
MANGWE M, BRYANT R, GREGORINI P. Rumen fermentation and fatty acid composition of milk of mid lactating dairy cows grazing chicory and ryegrass[J]. Animals (Basel), 2020, 10(1): 169. |
| [9] |
SOULAT J, KNAPP E, MOULA N, et al. Effect of dry-period diet on the performance and metabolism of dairy cows in early lactation[J]. Animals (Basel), 2020, 10(5): 803. |
| [10] |
ZANFERARI F, VENDRAMINI T H A, RENTAS M F, et al. Effects of chitosan and whole raw soybeans on ruminal fermentation and bacterial populations, and milk fatty acid profile in dairy cows[J]. J Dairy Sci, 2018, 101(12): 10939-10952. DOI:10.3168/jds.2018-14675 |
| [11] |
KAY J K, WEBER W J, MOORE C E, et al. Effects of week of lactation and genetic selection for milk yield on milk fatty acid composition in Holstein cows[J]. J Dairy Sci, 2005, 88(11): 3886-3893. DOI:10.3168/jds.S0022-0302(05)73074-5 |
| [12] |
樊永亮, 张成龙, 张少卿, 等. 中国荷斯坦牛一个完整泌乳期中乳脂肪酸变化规律研究[J]. 黑龙江畜牧兽医, 2016(8): 95-97, 101. FAN Y L, ZHANG C L, ZHANG S Q, et al. Study on the change regularity of milk fatty acids in southern Holstein cattle in a complete lactation period[J]. Heilongjiang Animal Science and Veterinary Medicine, 2016(15): 95-97, 101. DOI:10.13881/j.cnki.hljxmsy.2016.1374 (in Chinese) |
| [13] |
陈永华, 毛永江, 常玲玲. 中国荷斯坦牛初乳、常乳与乳房炎乳乳成分及理化性质的比较研究[J]. 饲料广角, 2011(14): 35-37. CHEN Y H, MAO Y J, CHANG L L. The Comparasion of milk composition and physical characteristics among colostrum, normal milk and mastitis of Chinese Holstein Cattle[J]. Feed China, 2011(14): 35-37. DOI:10.3969/j.issn.1002-8358.2011.14.023 (in Chinese) |
| [14] |
雷晓薇, 王根林, 韩兆玉. 应用体细胞计数监测奶牛隐性乳房炎[J]. 畜牧与兽医, 2003, 35(12): 35-37. LEI X W, WANG G L, HAN Z Y. Prediction of subclinical mastitis of dairy cows using milk somatic cell count[J]. Animal Husbandry & Veterinary Medicine, 2003, 35(12): 35-37. DOI:10.3969/j.issn.0529-5130.2003.12.024 (in Chinese) |
| [15] |
WANG J X, HE Y T, PANG K, et al. Changes in milk yield and composition of colostrum and regular milk from four buffalo breeds in China during lactation[J]. J Sci Food Agric, 2019, 99(13): 5799-5807. DOI:10.1002/jsfa.9849 |
| [16] |
张慧敏, 王梦琦, 朱小瑞, 等. 中国荷斯坦牛乳中尿素氮变化规律的研究[J]. 家畜生态学报, 2016, 37(11): 36-41. ZHANG H M, WANG M Q, ZHU X R, et al. Study on variability of milk urea nitrogen of Chinese Holstein[J]. Journal of Domestic Animal Ecology, 2016, 37(11): 36-41. DOI:10.3969/j.issn.1673-1182.2016.11.007 (in Chinese) |
| [17] |
BIONAZ M, LOOR J. ACSL1, AGPAT6, FABP3, LPIN1, and SLC27A6 are the most abundant isoforms in bovine mammary tissue and their expression is affected by stage of lactation[J]. J Nutr, 2008, 138(6): 1019-1024. DOI:10.1093/jn/138.6.1019 |
| [18] |
高学军, 徐诗瑶, 王小艳, 等. 泌乳中期不同乳品质奶牛乳中脂肪酸含量差异分析[J]. 东北农业大学学报, 2013, 44(9): 7-11. GAO X J, XU S Y, WANG X Y, et al. Difference analysis of fatty acid contents on milk from different qualities of cows in mid-lactation period[J]. Journal of Northeast Agricultural University, 2013, 44(9): 7-11. DOI:10.3969/j.issn.1005-9369.2013.09.002 (in Chinese) |
| [19] |
BELIDE S, SHRESTHA P, KENNEDY Y, et al. Engineering docosapentaenoic acid (DPA) and docosahexaenoic acid (DHA) in Brassica juncea[J]. Plant Biotechnol J, 2022, 20(1): 19-21. DOI:10.1111/pbi.13739 |
| [20] |
付力立. 基于代谢组学研究奶牛初乳与常乳组分差异及补饲酵母硒的影响[D]. 雅安: 四川农业大学, 2019. FU L L. Differential metabolome between colostrum and normal milk in dairy cows and effects of supplementary yeast selenium-based on metabonomics[D]. Ya'an: Sichuan Agricultural University, 2019. (in Chinese) |
| [21] |
DERAKHSHANI H, TUN H M, CARDOSO F C, et al. Linking peripartal dynamics of ruminal microbiota to dietary changes and production parameters[J]. Front Microbiol, 2017, 7: 2143. |
| [22] |
高凤. 奶牛肠道微生物群落结构与多样性研究[D]. 邯郸: 河北工程大学, 2017. GAO F. The study on intestinal microbial community structure and diversity of dairy cows[D]. Handan: Hebei University of Engineering, 2017. (in Chinese) |
| [23] |
BACH A, LÓPEZ-GARCÍA A, GONZÁLEZ-RECIO O, et al. Changes in the rumen and colon microbiota and effects of live yeast dietary supplementation during the transition from the dry period to lactation of dairy cows[J]. J Dairy Sci, 2019, 102(7): 6180-6198. DOI:10.3168/jds.2018-16105 |
| [24] |
TONG J J, ZHANG H, YANG D L, et al. Illumina sequencing analysis of the ruminal microbiota in high-yield and low-yield lactating dairy cows[J]. PLoS One, 2018, 13(11): e0198225. DOI:10.1371/journal.pone.0198225 |
| [25] |
JAMI E, WHITE B A, MIZRAHI I. Potential role of the bovine rumen microbiome in modulating milk composition and feed efficiency[J]. PLoS One, 2014, 9(1): e85423. DOI:10.1371/journal.pone.0085423 |
| [26] |
WANG Y P, NAN X M, ZHAO Y G, et al. Ruminal degradation of rumen-protected glucose influences the ruminal microbiota and metabolites in early-lactation dairy cows[J]. Appl Environ Microbiol, 2021, 87(2): e01908-20. |
| [27] |
刘可园. 奇数和支链脂肪酸与奶牛瘤胃微生物及其发酵指标的关系[D]. 哈尔滨: 东北农业大学, 2016. LIU K Y. Odd-and Branched-Chain Fatty Acids profilesin relation to rumen microbial populations and fermentation patterns in dairy cows[D]. Harbin: Northeast Agricultural University, 2016. (in Chinese) |
| [28] |
SAWANON S, KOIKE S, KOBAYASHI Y. Evidence for the possible involvement of Selenomonas ruminantium in rumen fiber digestion[J]. FEMS Microbiol Lett, 2011, 325(2): 170-179. DOI:10.1111/j.1574-6968.2011.02427.x |
| [29] |
王晓旭, 赵晨旭, 陈灰, 等. 围产期奶牛瘤胃微生物区系的变化及其对瘤胃挥发性脂酸含量的影响[C]//中国畜牧兽医学会——第三届中国兽医临床大会论文集. 兰州: 中国畜牧兽医学会, 2012: 106-110. WANG X X, ZHAO C X, CHEN H, et al. Correlation between composition of the bacterial community and concentration of volatile fatty acids in the rumen during the transition period and ketosis in dairy cows[C]//Proceedings of the 3rd Chinese Veterinary Clinical Congress of Chinese Association of Animal Husbandry and Veterinary Medicine. Lanzhou: Chinese Association of Animal Science and Veterinary Medicine, 2012: 106-110. (in Chinese) |
| [30] |
CERSOSIMO L M, BAINBRIDGE M L, KRAFT J, et al. Influence of periparturient and postpartum diets on rumen methanogen communities in three breeds of primiparous dairy cows[J]. BMC Microbiol, 2016, 16: 78. DOI:10.1186/s12866-016-0694-7 |
| [31] |
ZHAO L P, MENG Q X, REN L P, et al. Effects of nitrate addition on rumen fermentation, bacterial biodiversity and abundance[J]. Asian-Australas J Anim Sci, 2015, 28(10): 1433-1441. DOI:10.5713/ajas.15.0091 |
| [32] |
AUFFRET M D, STEWART R D, DEWHURST R J, et al. Identification of microbial genetic capacities and potential mechanisms within the rumen microbiome explaining differences in beef cattle feed efficiency[J]. Front Microbiol, 2020, 11: 1229. DOI:10.3389/fmicb.2020.01229 |
| [33] |
HUANG S, JI S K, WANG F R, et al. Dynamic changes of the fecal bacterial community in dairy cows during early lactation[J]. AMB Expr, 2020, 10(1): 167. DOI:10.1186/s13568-020-01106-3 |
| [34] |
BUITENHUIS B, LASSEN J, NOEL S J, et al. Impact of the rumen microbiome on milk fatty acid composition of Holstein cattle[J]. Genet Sel Evol, 2019, 51(1): 23. DOI:10.1186/s12711-019-0464-8 |
| [35] |
VLAEMINCK B, FIEVEZ V, CABRITA A R J, et al. Factors affecting odd- and branched-chain fatty acids in milk: a review[J]. Anim Feed Sci Technol, 2006, 131(3-4): 389-417. DOI:10.1016/j.anifeedsci.2006.06.017 |
| [36] |
LIU C, WU H, LIU S J, et al. Dynamic alterations in yak rumen bacteria community and metabolome characteristics in response to feed type[J]. Front Microbiol, 2019, 10: 1116. DOI:10.3389/fmicb.2019.01116 |
| [37] |
LIU K Y, LI Y, LUO G B, et al. Relations of ruminal fermentation parameters and microbial matters to odd- and branched-chain fatty acids in rumen fluid of dairy cows at different milk stages[J]. Animals (Basel), 2019, 9(12): 1019. |
| [38] |
KENNELLY J J, ROBINSON B, KHORASANI G R. Influence of carbohydrate source and buffer on rumen fermentation characteristics, milk yield, and milk composition in early-lactation Holstein cows[J]. J Dairy Sci, 1999, 82(11): 2486-2496. DOI:10.3168/jds.S0022-0302(99)75500-1 |
| [39] |
DEWANCKELE L, TORAL P G, VLAEMINCK B, et al. Invited review: role of rumen biohydrogenation intermediates and rumen microbes in diet-induced milk fat depression: an update[J]. J Dairy Sci, 2020, 103(9): 7655-7681. DOI:10.3168/jds.2019-17662 |
| [40] |
HUWS S A, KIM E J, LEE M R F, et al. As yet uncultured bacteria phylogenetically classified as Prevotella, Lachnospiraceae incertae sedis and unclassified Bacteroidales, Clostridiales and Ruminococcaceae may play a predominant role in ruminal biohydrogenation[J]. Environ Microbiol, 2011, 13(6): 1500-1512. DOI:10.1111/j.1462-2920.2011.02452.x |
(编辑 范子娟)



