2. 中国农业科学院北京畜牧兽医研究所, 北京 100193
2. Institute of Animal Science, Chinese Academy of Agricultural Sciences, Beijing 100193, China
自约9 000年前被驯化以来,猪在人类生存中发挥了重要作用[1]。猪作为一种农业动物,是人类主要的肉类蛋白来源[2]。事实上,截至2018年,猪肉已经成为世界上消费最多的肉类。然而,猪不仅被用作食用肉类,由于它们在解剖学、生理学和遗传学上与人类相似,并且脂肪沉积水平很高,因此猪是用于研究人类肥胖和能量代谢的合适模型[3-5]。此外,脂肪沉积是一项重要的经济性状,它与胴体质量、肉质和消费者的适口性相关,而背膘厚是脂肪沉积的一个重要指标[6]。因此,猪的背膘厚性状一直是研究的热点。Yang等[7]对1 200头约克夏和长白猪的345 570个核苷酸多态性(SNPs)进行了单标记回归分析,以确定与背膘厚相关的SNPs,结果发现SSC12:46226580、SSC6:149876737和SSC8:54567459等13个显著的SNPs与背膘厚有关。Gozalo-Marcilla等[8]通过对8个具有不同遗传背景的猪进行全基因组关联分析,结果发现MC4R、FGF23和PTN等27个基因组区域的264个SNPs与背膘厚有显著关联。这些研究说明了广大研究者正致力于寻找可靠的SNPs用来提高背膘厚的选择效率,提高经济效益,然而目前对于背膘厚的研究多集中于大白、长白等商品猪种,缺乏对地方优质品种的研究。
随着SNPs基因分型阵列、基因表达分析和其他高通量基因分型技术的出现,越来越多的与背膘厚度相关的候选基因被报道。趋化因子受体1(CCR1)属于G-蛋白偶联受体家族成员,可被稳态或炎症趋化因子选择性激活,在多种免疫细胞中表达,如:中性粒细胞、单核细胞/巨噬细胞、T细胞和NK细胞[9]。趋化因子通过与靶细胞膜上G-蛋白偶联受体(GPCR)超家族的特定趋化因子受体结合来发挥其生物学活性[10]。研究表明,CCR1及其配体在机体呼吸道系统[11]、免疫系统[12]、生殖系统[13]和生长发育中[14]均发挥着重要作用。Kogelman等[15]研究发现,CCR1基因在肥胖猪过度表达。DKK3是一种多功能蛋白,通过Wnt/β-catenin途径参与各种细胞过程,如细胞分化、增殖和凋亡[16],并导致多种疾病,包括癌症[17]和慢性心力衰竭[18]等。Wnt/β-catenin通路相关基因在脂肪分化和发育过程中表达[19]。因此,DKK3基因可能通过这一途径在脂肪中发挥不同的作用。本课题组在先前的研究中已经发现了CCR1和DKK3可作为背膘厚的候选基因[20]。
在养猪业中,通常测量背膘厚度以指示猪的生产和生长性能。因此,了解控制北京黑猪背膘厚的遗传决定因素对于改善肉品质、提高育种效率和获得遗传进展具有重要意义。本试验以413头北京黑猪为试验对象,将DKK3和CCR1基因作为影响背膘厚的候选基因,利用PCR直接测序技术筛选DKK3和CCR1基因上的SNPs,将筛选的SNPs与背膘厚状进行关联分析,筛选出对北京黑猪背膘厚有重要作用的SNPs位点,以期为北京黑猪肉用生产性能的改良和选育奠定基础,为分子标记辅助育种提供参考。
1 材料与方法 1.1 试验动物及样品采集本研究使用的北京黑猪均来自北京黑六牧业科技有限公司,为2020年8月至2020年11月屠宰测定的共413头北京黑猪,所有的黑猪健康状况良好、体重相近、饲养管理统一。采集猪的耳组织,置于2 mL冻存管中,并迅速置于液氮中速冻,后置于-80 ℃冰箱冷冻保存。
1.2 性状测定用电子游标卡尺准确测量右侧胴体六七肋处背膘厚和腰荐处背膘厚,并记录。
1.3 引物设计根据NCBI数据库中猪的DKK3基因序列(登录号:NM_001039749.1),利用Premier 5.0设计8对引物,覆盖DKK3基因的全部外显子区域。CCRl基因序列(登录号:NM_001001621.1),针对CCR1基因的全部外显子区域,共设计5对引物。引物由上海生工生物股份有限公司合成。设计引物见表 1。
|
|
表 1 DKK3和CCR1基因扩增引物 Table 1 Amplification primers of DKK3 and CCR1 genes |
取约10 mg背最长肌,置于2 mL研磨管中,后放入冷冻研磨机中进行研磨。使用天根生化有限公司生产的细胞/组织/血液DNA提取试剂盒提取总DNA,利用分光光度计(美国,Thermo Fisher)和1%琼脂糖检测合格后,以此为模板进行PCR扩增。扩增条件为:95 ℃预变性5 min;95 ℃变性30 s,退火30 s(退火温度见表 1),72 ℃延伸1.5 min,30个循环;72 ℃终延伸2 min。
1.5 SNPs多态性检验及统计分析取符合要求的PCR产物送上海生工生物股份有限公司进行测序。对测序结果用DNAMAN和Chromas软件进行核苷酸序列和波峰图对比分析,寻找突变位点。使用Excel计算等位基因频率和基因型频率。运用SAS 9.2统计软件中的Duncan’s多重检验分析不同位点各基因型对北京黑猪背膘厚的影响,结果用“平均值±标准差”表示,用P<0.05作为差异显著性判断标准。利用Hapioview4.1软件对筛选出的DKK3基因SNPs位点进行连锁不平衡分析。
2 结果 2.1 北京黑猪背膘厚表型统计本研究共测定了413头北京黑猪的背膘厚性状(表 2)。六七肋背膘厚的最大值为5.258 cm,最小值为2.735 cm,平均值为3.067 cm,标准差为0.619 cm;腰荐背膘厚的最大值为4.842 cm,最小值为2.437 cm,平均值为2.743 cm,标准差为0.695 cm。
|
|
表 2 北京黑猪背膘厚表型统计 Table 2 Phenotypic statistics of backfat thickness in Beijing black pigs |
DKK3基因外显子区域检测到10个SNPs位点,分别为g.47385307 G>T、g.47399612 A>G、g.47443779 T>C、g.47443783 C>T、g.47443858 C>T、g.47443903 A>G、g.47444258 C>T、g.47444275 C>T、g.47444912 G>T和g.47445304 T>C,其中g.47385307 G>T为可变剪切突变,g.47399612 A>G为错义突变,其余均为3′UTR突变。由表 3可知,北京黑猪DKK3基因g.47385307 G>T和g.47444912 G>T突变位点GG基因型为优势基因型(0.674和0.884),G为优势等位基因,g.47399612 A>G和g.47443903 A>G突变位点AA基因型为优势基因型(0.538和0.786),A为优势等位基因,g.47443779 T>C突变位点TT基因型为优势基因型(0.509),T为优势等位基因,g.47445304 T>C突变位点TC基因型为优势基因型(0.382),C为优势等位基因,其余的突变位点,CC基因型为优势基因型,C均为优势等位基因。
|
|
表 3 DKK3基因多态性位点基因型频率和等位基因频率 Table 3 The genotype and allele frequencies of polymorphism sites of DKK3 gene |
CCR1基因外显子区域检测到3个SNPs位点,分别为g.29229037 G>A、g.29229315 T>C和g.29229694 C>T,其中c g.29229037 G>A和g.29229694 C>T为同义突变,g.29229315 T>C为错义突变。由表 4可知,北京黑猪CCR1基因g.29229037 G>A突变位点GA基因型为优势基因型(0.363),G为优势等位基因,g.29229315 T>C突变位点TC基因型为优势基因型(0.393),T为优势等位基因,g.29229694 C>T突变位点CC基因型为优势基因型(0.506),C为优势等位基因。
|
|
表 4 CCR1基因多态性位点基因型频率和等位基因频率 Table 4 The genotype and allele frequencies of polymorphism sites of CCR1 gene |
利用SAS软件对北京黑猪CCR1和DKK3基因突变位点与六七肋背膘厚和腰荐部背膘厚进行差异显著性检验,由表 5可知,有4个3′UTR突变位点均与腰荐部背膘厚显著关联(P<0.05),分别为g.47443779 T>C、g.47444258 C>T、g.47444912 G>T和g.47445304 T>C,其余位点与六七肋背膘厚和腰荐部背膘厚均不显著关联(P>0.05),其中g.47443779 T>C位点TT基因型个体的腰荐背膘厚显著高于CC基因型个体(P<0.05),g.47444258 C>T位点CC和CT基因型个体的腰荐背膘厚显著高于TT基因型个体(P<0.05),g.47444912 G>T位点的TT基因型个体腰荐背膘厚显著高于GT基因型个体(P<0.05),g.47445304 T>C位点CC基因型个体的腰荐背膘厚显著高于TT基因型个体(P<0.05)。由表 6可知,CCR1基因只有1个突变位点基因型与六七肋背膘厚显著相关(P<0.05),g.29229037 G>A位点GG基因型个体的六七肋背膘厚显著高于AA基因型个体(P<0.05)。
|
|
表 5 北京黑猪DKK3基因10个SNPs与背膘厚的关联分析 Table 5 Association analysis of 10 SNPs in DKK3 gene and backfat thickness in Beijing black pigs |
|
|
表 6 北京黑猪CCR1基因3个SNPs与背膘厚的关联分析 Table 6 Association analysis of 3 SNPs in CCR1 gene and backfat thickness in Beijing black pigs |
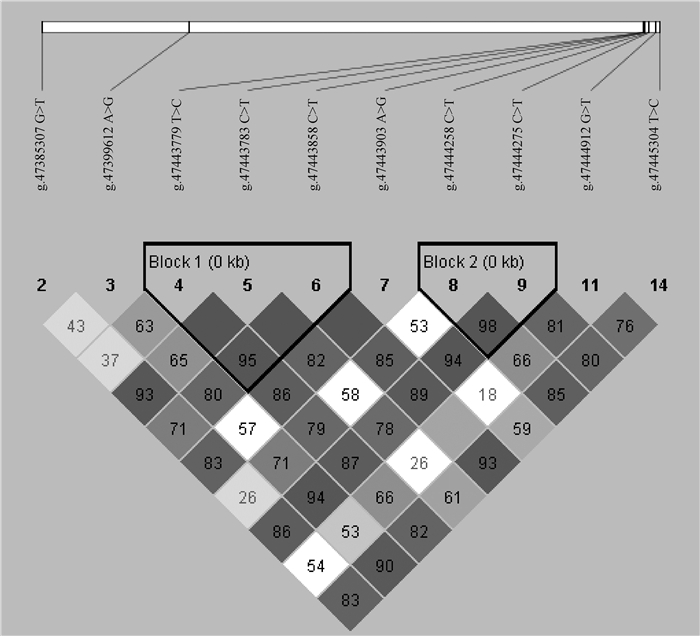
对DKK3的10个SNPs进行连锁不平衡分析,分析结果显示(图 1),共形成了2处连锁不平衡模块,分别是g.47443779 T>C、g.47443783 C>T和g.47443858 C>T形成了第一个模块,g.47444258 C>T和g.47444275 C>T形成了第二个模块。其中g.47443779 T>C和g.47443783 C>T处于完全连锁状态(D’=1), g.47443783 C>T和g.47443858 C>T处于完全连锁状态(D’=1),g.47444258 C>T和g.47444275 C>T呈现高度连锁状态(D’=0.98)。由表 7和表 8可知,block1形成4种单倍型,block2形成3种单倍型。
|
图 1 北京黑猪DKK3基因SNPs的连锁不平衡 Fig. 1 The linkage disequilibrium analysis of SNPs in DKK3 gene in Beijing black pigs |
|
|
表 7 DKK3基因block1单倍型及其频率 Table 7 The haplotypes and their frequencies of DKK3 gene block1 |
|
|
表 8 DKK3基因block2单倍型及其频率 Table 8 The haplotypes and their frequencies of DKK3 gene block2 |
作为家猪的重要经济性状,背膘厚在猪遗传学中得到了广泛的研究。高背膘厚不仅导致饲料效率低下,而且也不受消费者的青睐。在满足消费者需求的同时,生猪养殖的目标是在不影响猪肉口感的前提下,降低胴体脂肪,提高瘦肉率。目前的研究多集中于杜洛克[21-22]、大白猪[23]和约克夏猪[24]等商品猪背膘厚。北京黑猪作为中国培育的优良品种之一,毛黑色,中等体型和生长速度快,肉色鲜红,大理石纹评分高,被认为是高端猪肉之一[25]。然而对影响北京黑猪背膘厚相关SNPs的研究却鲜见报道。本研究以DKK3和CCR1基因为影响北京黑猪背膘厚的候选基因,探究其基因多态性对北京黑猪背膘厚性状的影响,以期能够筛选出可用于提高北京黑猪背膘厚性状的分子标记。
背膘厚度是家猪脂肪沉积的典型数量性状,显示出中等至高度的遗传性[26]。本研究利用SAS软件对检测到的突变位点的不同基因型分别与北京黑猪的背膘厚进行关联分析,希望为北京黑猪背膘厚研究提供更多的SNPs标记。在CCR1外显子检测到3个SNPs,其中2个为同义突变(g.29229037 G>A和g.29229694 C>T),1个为错义突变(g.29229315 T>C)。关联分析发现,g.29229037 G>A GG基因型个体的六七肋背膘厚显著高于AA基因型个体(P<0.05)。目前对于CCR1突变位点的研究发现其多与疾病有关。Toyoda等[27]发现, CCR1基因rs3181077位点与发作性睡病有关。此外,位于人类CCR1基因3′-侧翼区域的SNPs与Behçet综合征密切相关,Behçet综合征被认为是一种以眼/皮肤炎症、复发性口腔溃疡和生殖器溃疡为特征的自身免疫性疾病[28]。已有报道CCR1基因是人体脂肪组织中甘油三酯调控的关键候选基因[29]。本研究在北京黑猪群体中发现CCR1基因g.29229037 G>A突变与六七肋背膘厚显著相关。
在北京黑猪DKK3基因的外显子区共检测到10个SNPs,其中有4个突变位点(g.47443779 T>C、g.47444258 C>T、g.47444912 G>T和g.47445304 T>C)基因型与腰荐背膘厚显著相关(P<0.05)。DKK3与Wnt/B-catenin信号通路相关,Wnt/B-catenin信号通路相关基因在脂肪分化发育过程中表达[30]。该信号通路可阻断脂肪分化关键转录因子(C/EBPα和PPARγ)的表达,从而影响脂肪分化[31]。因此,DKK3基因可能通过这一途径在脂肪中发挥不同的作用。这4个位点突变均为3′UTR突变,说明3′UTR突变对于猪背膘厚性状具有重要影响。Zang等[32]在猪DGAT2基因中发现其3′UTR的多态位点与猪背膘厚和瘦肉率显著相关。研究表明,3′UTR包含miRNA的靶点[33]。miRNA是短的内源性RNA,通过与3′UTR中的位点配对来调节靶基因的蛋白质表达,从而发挥重要的基因调控作用[34-35]。miRNA已被证明在广泛的生物过程中发挥调节作用,例如发病机制、组织发育、脂肪细胞分化和脂肪生成等过程[36-39]。第一个报道的miRNA对脂质代谢的影响是在果蝇中,其中miR-14的缺失导致甘油三酯和甘油二酯的积累增加[40]。所以miRNA可能通过调控DKK3基因3′UTR进而影响脂质代谢,本课题组先前的研究发现miR-3477-5p可以通过调节靶基因DKK3进而影响脂肪生成[20]。下一步可以进行DKK3基因4个3′UTR突变的靶向miRNA研究。
相对于单个SNPs对表型的影响,一组具有连锁效应的SNPs所形成单倍型的作用更大[41]。因此本研究对DKK3的10个SNPs进行连锁不平衡分析,分析结果显示g.47443779 T>C-g.47443783 C>T处于完全连锁状态(D’=1),g.47443783 C>T-g.47443858 C>T处于完全连锁状态(D’=1),g.47444258 C>T和g.47444275 C>T呈现高度连锁状态(D’=0.98)。所以虽然g.47443783 C>T与g.47444275 C>T位点不同基因型与背膘厚关联时无显著差异,但是可以认为其是影响背膘厚的潜在SNPs。
4 结论本试验检测北京黑猪群体中CCR1和DKK3基因的SNPs位点,并分析了这些SNPs位点与背膘厚性状的相关性。北京黑猪DDK3基因的g.47443779 T>C、g.47444258 C>T、g.47444912 G>T和g.47445304 T>C位点与北京黑猪的腰荐背膘厚显著相关;CCR1基因的g.29229037 G>A位点与北京黑猪的六七肋背膘厚显著相关。本研究为CCR1和DKK3基因作为北京黑猪背膘厚性状的候选基因应用于猪遗传育种提供了参考。
| [1] |
GIUFFRA E, KIJAS J M H, AMARGER V, et al. The origin of the domestic pig: Independent domestication and subsequent introgression[J]. Genetics, 2000, 154(4): 1785-1791. DOI:10.1093/genetics/154.4.1785 |
| [2] |
HE Y A, YANG X G, XIA J, et al. Consumption of meat and dairy products in China: A review[J]. Proc Natr Soc, 2016, 75(3): 385-391. DOI:10.1017/S0029665116000641 |
| [3] |
MEURENS F, SUMMERFIELD A, NAUWYNCK H, et al. The pig: A model for human infectious diseases[J]. Trends Microbiol, 2012, 20(1): 50-57. DOI:10.1016/j.tim.2011.11.002 |
| [4] |
BERTHO N, MEURENS F. The pig as a medical model for acquired respiratory diseases and dysfunctions: an immunological perspective[J]. Mol Immunol, 2021, 135: 254-267. DOI:10.1016/j.molimm.2021.03.014 |
| [5] |
BERGEN W G. Pigs (Sus Scrofa) in biomedical research[M]//WU G Y. Recent Advances in Animal Nutrition and Metabolism. Cham: Springer, 2022: 335-343.
|
| [6] |
SUZUKI K, KATOH K, ADOWAKI H K, et al. Genetic correlations among carcass cross-sectional fat area ratios, production traits, intramuscular fat, and serum leptin concentration in Duroc pigs[J]. J Anim Sci, 2009, 87(7): 2209-2215. DOI:10.2527/jas.2008-0866 |
| [7] |
YANG Q, WU P X, WANG K, et al. SNPs associated with body weight and backfat thickness in two pig breeds identified by a genome-wide association study[J]. Genomics, 2019, 111(6): 1583-1589. DOI:10.1016/j.ygeno.2018.11.002 |
| [8] |
GOZALO-MARCILLA M, BUNTJER J, JOHNSSON M, et al. Genetic architecture and major genes for backfat thickness in pig lines of diverse genetic backgrounds[J]. Genet Sel Evol, 2021, 53(1): 76. DOI:10.1186/s12711-021-00671-w |
| [9] |
WHITE G E, IQBAL A J, GREAVES D R. CC chemokine receptors and chronic inflammation—therapeutic opportunities and pharmacological challenges[J]. Pharmacol Rev, 2013, 65(1): 47-89. DOI:10.1124/pr.111.005074 |
| [10] |
CHENG J F, JACK R. CCR1 antagonists[J]. Mol Divers, 2008, 12(1): 17-23. DOI:10.1007/s11030-008-9076-x |
| [11] |
ZHAO K S, DONG R, YU Y F, et al. Cigarette smoke-induced lung inflammation in COPD mediated via CCR1/JAK/STAT /NF-κB pathway[J]. Aging, 2020, 12(10): 9125-9138. DOI:10.18632/aging.103180 |
| [12] |
KHOLODNYUK I, RIVKINA A, HIPPE L, et al. Chemokine receptors CCR1 and CCR2 on peripheral blood mononuclear cells of newly diagnosed patients with the CD38-positive chronic lymphocytic leukemia[J]. J Clin Med, 2020, 9(7): 2312. DOI:10.3390/jcm9072312 |
| [13] |
YANG Y F, ZHANG X M, ZHOU C Y, et al. Elevated immunoreactivity of RANTES and CCR1 correlate with the severity of stages and dysmenorrhea in women with deep infiltrating endometriosis[J]. Acta Histochem, 2013, 115(5): 434-439. DOI:10.1016/j.acthis.2012.10.006 |
| [14] |
柴继田. 基于猪CCR1和CCR5受体高通量细胞筛选模型的构建及其条件优化[D]. 扬州: 扬州大学, 2015. CHAI J T. The construction and optimization of porcine CCR1 and CCR5 receptor of high-throughput cell screening model[D]. Yangzhou: Yangzhou University, 2015. (in Chinese) |
| [15] |
KOGELMAN L J A, CIRERA S, ZHERNAKOVA D V, et al. Identification of co-expression gene networks, regulatory genes and pathways for obesity based on adipose tissue RNA Sequencing in a porcine model[J]. BMC Med Genomics, 2014, 7: 57. DOI:10.1186/1755-8794-7-57 |
| [16] |
VEECK J, DAHL E. Targeting the Wnt pathway in cancer: the emerging role of Dickkopf-3[J]. Biochim Biophys Acta, 2012, 1825(1): 18-28. |
| [17] |
LIU Q W, LI J Y, ZHANG X C, et al. Human amniotic mesenchymal stem cells inhibit hepatocellular carcinoma in tumour-bearing mice[J]. J Cell Mol Med, 2020, 24(18): 10525-10541. DOI:10.1111/jcmm.15668 |
| [18] |
CAO Q, ZHANG J X, GAO L, et al. Dickkopf-3 upregulation mediates the cardioprotective effects of curcumin on chronic heart failure[J]. Mol Med Rep, 2018, 17(5): 7249-7257. |
| [19] |
LI H X, LUO X, LIU R X, et al. Roles of Wnt/β-catenin signaling in adipogenic differentiation potential of adipose-derived mesenchymal stem cells[J]. Mol Cell Endocrinol, 2008, 291(1-2): 116-124. DOI:10.1016/j.mce.2008.05.005 |
| [20] |
LIU X, GONG J F, WANG L G, et al. Genome-wide profiling of the microrna transcriptome regulatory network to identify putative candidate genes associated with backfat deposition in pigs[J]. Animals (Basel), 2019, 9(6): 313. |
| [21] |
EUSEBI P G, GONZÁLEZ-PRENDES R, QUINTANILLA R, et al. A genome-wide association analysis for carcass traits in a commercial Duroc pig population[J]. Anim Genet, 2017, 48(4): 466-469. DOI:10.1111/age.12545 |
| [22] |
DING R R, ZHUANG Z W, QIU Y B, et al. Identify known and novel candidate genes associated with backfat thickness in Duroc pigs by large-scale genome-wide association analysis[J]. J Anim Sci, 2022, 100(2): skac012. DOI:10.1093/jas/skac012 |
| [23] |
FABBRI M C, ZAPPATERRA M, DAVOLI R, et al. Genome-wide association study identifies markers associated with carcass and meat quality traits in Italian Large White pigs[J]. Anim Genet, 2020, 51(6): 950-952. DOI:10.1111/age.13013 |
| [24] |
CHEN D J, WU P X, YANG Q, et al. Genome-wide association study for backfat thickness at 100 kg and loin muscle thickness in domestic pigs based on genotyping by sequencing[J]. Physiol Genomics, 2019, 51(7): 261-266. DOI:10.1152/physiolgenomics.00008.2019 |
| [25] |
武群清. 北京黑猪肉质性状的研究及其背最长肌转录组差异分析[D]. 长沙: 湖南农业大学, 2017. WU Q Q. The study on pork quality and transcriptional profiling of longissimus dorsi muscle on Beijing black pig[D]. Changsha: Hunan Agricultural University, 2017. (in Chinese) |
| [26] |
GUO Y M, QIU H Q, XIAO S J, et al. A genome-wide association study identifies genomic loci associated with backfat thickness, carcass weight, and body weight in two commercial pig populations[J]. J Appl Genet, 2017, 58(4): 499-508. DOI:10.1007/s13353-017-0405-6 |
| [27] |
TOYODA H, MIYAGAWA T, KOIKE A, et al. A polymorphism in CCR1/CCR3 is associated with narcolepsy[J]. Brain Behav Immun, 2015, 49: 148-155. DOI:10.1016/j.bbi.2015.05.003 |
| [28] |
KIRINO Y, BERTSIAS G, ISHIGATSUBO Y, et al. Genome-wide association analysis identifies new susceptibility loci for Behçet's disease and epistasis between HLA-B * 51 and ERAP1[J]. Nat Genet, 2013, 45(2): 202-207. DOI:10.1038/ng.2520 |
| [29] |
HAAS B E, HORVATH S, PIETILÄINEN K H, et al. Adipose co-expression networks across Finns and Mexicans identify novel triglyceride-associated genes[J]. BMC Med Genomics, 2012, 5: 61. DOI:10.1186/1755-8794-5-61 |
| [30] |
ALEXANDER T, NOLTE C, KRUMLAUF R. Hox genes and segmentation of the hindbrain and axial skeleton[J]. Annu Rev Cell Dev Biol, 2009, 25: 431-456. DOI:10.1146/annurev.cellbio.042308.113423 |
| [31] |
ROSS S E, HEMATI N, LONGO K A, et al. Inhibition of adipogenesis by wnt signaling[J]. Science, 2000, 289(5481): 950-953. DOI:10.1126/science.289.5481.950 |
| [32] |
ZANG L, WANG Y D, SUN B X, et al. Identification of a 13 bp indel polymorphism in the 3'-UTR of DGAT2 gene associated with backfat thickness and lean percentage in pigs[J]. Gene, 2016, 576(2): 729-733. DOI:10.1016/j.gene.2015.09.047 |
| [33] |
BARTEL D P. MicroRNAs: target recognition and regulatory functions[J]. Cell, 2009, 136(2): 215-233. DOI:10.1016/j.cell.2009.01.002 |
| [34] |
DJURANOVIC S, NAHVI A, GREEN R. miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay[J]. Science, 2012, 336(6078): 237-240. DOI:10.1126/science.1215691 |
| [35] |
BARTEL D P. MicroRNAs[J]. Cell, 2004, 116(2): 281-297. DOI:10.1016/S0092-8674(04)00045-5 |
| [36] |
WANG G, ZOU H B, LAI C Y, et al. Repression of MicroRNA-124-3p alleviates high-fat diet-induced hepatosteatosis by targeting pref-1[J]. Front Endocrinol (Lausanne), 2020, 11: 589994. DOI:10.3389/fendo.2020.589994 |
| [37] |
WANG J K, WANG Z, LI G D. MicroRNA-125 in immunity and cancer[J]. Cancer Lett, 2019, 454: 134-145. DOI:10.1016/j.canlet.2019.04.015 |
| [38] |
何春, 张琦悦, 孙浩伟, 等. miRNA和lncRNA在动物脂肪沉积中的研究进展[J]. 生物工程学报, 2020, 36(8): 1504-1514. HE C, ZHANG Q Y, SUN H W, et al. Role of miRNA and lncRNA in animal fat deposition-a review[J]. Chinese Journal of Biotechnology, 2020, 36(8): 1504-1514. (in Chinese) |
| [39] |
LIU F T, LIU Y L, DU Y Q, et al. MiRNA-130a promotes inflammation to accelerate atherosclerosis via the regulation of proliferator-activated receptor γ (PPARγ) expression[J]. Anatol J Cardiol, 2021, 25(9): 630-637. DOI:10.5152/AnatolJCardiol.2021.56721 |
| [40] |
XU P Z, VERNOOY S Y, GUO M, et al. The Drosophila microRNA Mir-14 suppresses cell death and is required for normal fat metabolism[J]. Curr Biol, 2003, 13(9): 790-795. DOI:10.1016/S0960-9822(03)00250-1 |
| [41] |
李常红, 张明国, 胡玮宜, 等. 不同猪种IGF1R基因扩增及SNPs的筛选[J]. 吉林农业大学学报, 2018, 40(6): 734-739. LI C H, ZHANG M G, HU W Y, et al. Study on amplification of IGF1R gene and SNPs screening in different pig breeds[J]. Journal of Jilin Agricultural University, 2018, 40(6): 734-739. (in Chinese) |
(编辑 郭云雁)



