肌肉组织是动物机体的重要组成部分,主要包括骨骼肌、心肌和平滑肌,其中骨骼肌约占成年动物体重的40%~60%[1]。肌肉的生长发育是一个极其复杂和精密的过程,受到许多细胞因子和转录因子构成的调控网络共同作用,其中包括生肌调节因子(myogenic regulatory factors, MRFs)家族[2-4]、肌细胞增强因子2(myocyte enhancer factor 2, MEF2)家族和转录调控因子Pax(paired box)家族。近些年的研究表明,非编码RNA,包括长链非编码RNA(long noncoding RNA, lncRNA)、微小RNA(microRNA, miRNA)和环状RNA(circular RNA, circRNA)等,参与了肌肉生成的调控网络[5-7]。RNA结合蛋白(RNA binding proteins, RBPs)作为细胞生命活动中的重要成员,通过与编码和非编码RNA互作,也参与了肌肉生长发育的调控过程[8-9]。
RNA结合蛋白是细胞中一类重要的蛋白质,它们通过识别特殊的RNA结合域与RNA互作,广泛参与到RNA剪切、转运、序列编辑、胞内定位及翻译控制等多个转录后调控过程中,在基因调控过程中扮演着关键作用[10-12]。RNA结合蛋白的结构域包括K同源基序(K homology, KH)、RNA识别基序(RNA recognition motif, RRM)和锌指结构域(zinc finger domain, ZNF)等[13]。研究表明,RNA结合蛋白通过特异性结合靶mRNA分子3′非翻译区(UTRs)的顺式作用元件,调控靶基因的表达水平,进而参与组织发育、细胞周期及疾病发生等生物学过程[14-16]。作为RNA结合蛋白家族的重要成员,HuR(human antigen R)蛋白在机体内广泛表达,通过调控靶基因mRNA的稳定性或翻译效率影响靶基因的表达水平,从而参与调控细胞的生命活动。本文结合近年来RNA结合蛋白HuR的相关研究,主要综述了HuR的生物学特性、主要功能与作用方式及其在肌肉生长发育和相关疾病中的调控作用,同时对RNA结合蛋白HuR的今后研究方向进行了展望,以期让相关科研人员更全面地了解HuR蛋白的研究进展,为进一步研究其在肌肉生长发育调控中的作用提供有力支撑。
1 HuR的生物学特性RNA结合蛋白HuR是胚胎致死异常视觉(embryonic lethal abnormal vision, ELAV)基因家族成员之一,又被称为类胚胎致死性异常视觉基因1(ELAV1)。ELAV家族包括HuB、HuC、HuD和HuR 4个成员。HuB、HuC和HuD主要在神经组织中表达,而HuR则是在机体各组织中普遍表达[17-19]。HuR蛋白编码基因在人类上位于第19号染色体,小鼠上位于第8号染色体,山羊上位于第7号染色体,编码的蛋白均由326个氨基酸组成。HuR蛋白含有3个经典的RNA识别结构域[18],其中RRM1和RRM2结构域可以特异性的结合AU富含元件(AU-rich element, ARE)。RRM3对于稳定RNA-蛋白质复合物以及介导蛋白质-蛋白质相互作用非常重要。RRM2和RRM3之间还含有一个介导HuR进行核质穿梭的铰链区(HuR nuclear shuttle, HNS),HuR的HNS区域含有核定位信号(nuclear localization sequence, NLS)和核输出信号(nuclear export signal, NES),HuR主要存在于细胞核中,当细胞受到刺激时,HuR的铰链区会发生不同的蛋白修饰,从而促进HuR从细胞核穿梭到细胞质中[20-21]。
2 HuR的功能与作用方式大量研究表明,HuR作为基因转录后水平的重要调控因子,通过与靶mRNA分子3′末端非翻译区的ARE元件(AU rich element)结合来发挥其生物学功能,通常是通过提高靶mRNA的稳定性或翻译效率而上调靶基因在细胞中的表达水平[22]。此外有研究表明,HuR蛋白也可以与非编码RNA(如miRNA、lncRNA)或其它RNA结合蛋白相互作用,进而参与调控RNA剪接加工、多聚腺苷酸化形成、细胞凋亡等生命活动,以及肿瘤的发生发展等过程(图 1)[23-24]。
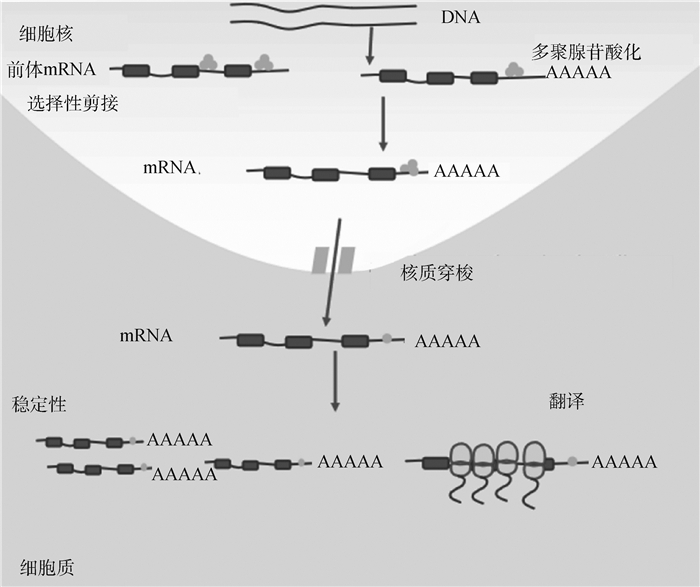
|
 代表RNA结合蛋白HuR 代表RNA结合蛋白HuR
 Represents RNA binding protein HuR
图 1 HuR的功能示意图[18]
Fig. 1
The schematic diagram of HuR function[18] Represents RNA binding protein HuR
图 1 HuR的功能示意图[18]
Fig. 1
The schematic diagram of HuR function[18]
|
研究发现,HuR通过调控剪接因子对mRNA前体内含子或外显子上剪接位点的识别,进而调节前体mRNA的剪接模式[25]。研究人员发现,TIA-1(T cell intracellular antigen 1)/TIAR(TIA1-related)蛋白与位于降钙素(calcitonin gene-related peptide, CGPR)外显子下游富含U的序列相互作用能够促进降钙素非特异性形成。HuR能够阻断拼接因子TIA-1或TIAR的活性,并与其竞争性地结合到内含子的U富集区域,促使CGRP mRNA前体非神经元外显子4发生跳跃,致使正常应该产生的肽激素被神经传递素所代替[26]。HuR通过改变SIRT1外显子8周围组蛋白修饰模式,使RNA聚合酶Ⅱ(RNA polymeraseⅡ, RNAPⅡ) 的延伸速率加快,促进SIRT1外显子8的切除,进而增加缺失SIRT1外显子8的SIRT1 mRNA的表达,而TIA-1/TIAL1通过改变外显子周围组蛋白修饰模式,使RNAPⅡ的延伸速率减慢,促进SIRT1外显子8的保留,使缺失SIRT1外显子8的SIRT1 mRNA的表达降低[27]。此外有研究表明,HuR蛋白还能够发挥剪接增强子作用,结合HuD外显子6下游的两个AU元件,促进HuD mRNA外显子6插入[28]。
2.2 HuR影响多聚腺苷酸化的形成研究证明,HuR能够选择性结合到靶mRNA的U富集区,影响靶mRNA的poly(A)形成,进而影响靶mRNA多聚腺苷酸化的修饰过程。多聚腺苷酸化需要两步反应,包括裂解和新生成的3′端上加poly(A)尾巴。这两步需要切割刺激因子(cleavage stimulation factor, CstF)和多聚腺苷酸特异性因子(cleavage and polyadenylation specificity factor, CPSF)的参与,而HuR蛋白能够与多聚腺苷酸化这两个特异性因子(CstF、CPSF)竞争性结合富含U的序列,从而阻断了mRNA的多聚腺苷酸化形成[29]。
2.3 HuR调控mRNA的稳定性研究发现,在人的心肌细胞中,HuR蛋白能够识别并结合MEF2C基因的ARE序列,增加其mRNA稳定性,从而促进了心脏钠通道基因(cardiac sodium channel gene, SCN5A)的转录[30]。另外,HuR还可以通过与miRNA、lncRNA或circRNA相互作用调控靶mRNA的稳定性。环状RNA circE2F2(circRNA transcription factor 2, circE2F2)与HuR蛋白结合能够促进HuR结合在E2F2的3′UTR,进而增加E2F2的mRNA稳定性[31]。研究表明,circDLC1可以与MMP1竞争结合HuR,通过降低MMP1的稳定性,进而抑制MMP1的表达,最终抑制肝癌细胞的增殖和迁移[32]。食管鳞状细胞癌相关研究发现,HuR通过与miR-4319竞争性结合SEMA4D,增强SEMA4D的稳定性,从而调节食管癌细胞的增殖、凋亡和迁移[33]。当HOTAIR泛素化底物的水平较低时,HuR会优先与HOTAIR结合,并通过募集let-7-Ago2(argonaute RISC catalytic component 2)复合物来降低HOTAIR的稳定性[34]。此外,HuR自身的蛋白修饰也可以影响靶mRNA的稳定性。HuR蛋白的磷酸化能够影响其对应激蛋白SIRT1(stress-response protein)mRNA稳定性的调节,细胞周期检查点激酶(checkpoint kinase 2, Chk2)对HuR进行磷酸化的修饰,抑制了HuR与SIRT1 mRNA的结合[35]。在人类肝细胞癌的研究中发现,ADORA2A-AS1与肌动蛋白束蛋白(fascin actin-bundling protein 1, FSCN1)转录物竞争性结合HuR,通过影响FSCN1的稳定性来降低FSCN1的表达,从而阻断了AKT通路的激活,最终抑制肿瘤的发生[36]。此外有研究表明,LINC01119能够与HuR相互作用,形成LINC01119-HuR复合物,进而与脑源性神经营养因子(brain-derived neurotrophic factor, BDNF)mRNA结合,增强其稳定性,提高BDNF在转录和蛋白水平的表达,促进神经性疼痛(neuropathic pain, NP)的发生[37]。研究人员发现,LINC00668通过招募HuR增强蛋白激酶N2(protein kinase n2,PKN2)的稳定性,从而促进胃癌的转移[38]。
2.4 HuR调控mRNA的翻译有研究表明,HuR通过结合靶mRNA的3′UTR富含AU的ARE区,调节靶mRNA的翻译效率,进而参与细胞生命活动。在人类小肠上皮细胞中,HuR可以结合微囊蛋白1(caveolin-1, Cav-1)的3′UTR区调节Cav-1 mRNA的翻译,从而促进早期小肠上皮的恢复[39]。另外研究发现,HuR也可以与靶基因的5′UTR结合来调控翻译。HuR可特异性地结合到细胞周期素依赖性激酶抑制因子p27Kip1 mRNA的5′UTR序列中的内部核糖体进入位点(internal ribosome entry site, IRES),抑制p27Kip1的翻译[40]。相关研究发现,真核细胞翻译起始因子(eukaryotic translation initiation factor 3, EIF3)与HuR相互作用结合在Chk1 mRNA的3′UTR上,正向调节Chk1 mRNA的翻译,从而调控细胞周期进程[41]。此外有研究表明,HuR可以招募miRNA let-7(RNA-induced silencing complex, RISC)结合在c-Myc mRNA的3′UTR,进而抑制c-Myc mRNA下调[42]。
研究发现,HuR还可以与非编码RNA互作,调控靶mRNA的翻译。小鼠上研究发现,过表达的miR-195能够与HuR竞争性结合双皮质素样激酶1(double cortin-like kinase 1, DCLK1)的3′UTR并抑制DCLK1的翻译,进而破坏肠绒毛细胞的功能[43]。HuR也能与程序性细胞死亡因子4(programmed cell death 4, PDCD4)的3′UTR相互作用,阻止miR-21介导的PDCD4翻译抑制[44]。Abdelmohsen等[45]研究发现,环状多聚腺苷酸结合蛋白核1(circle poly(A) binding protein nuclear 1, PABPN1)能够与HuR蛋白结合,并抑制HuR蛋白与PABPN1基因mRNA的结合能力,从而抑制其翻译。在肺癌研究中发现,circBACH1通过促进HuR的核质易位,进而抑制p27的翻译,促进肝癌细胞增殖[46]。此外,在人类癌症细胞的研究中发现,circAGO2能够与HuR蛋白相互作用,促进其在促癌相关靶基因3′UTR的富集,从而减少HuR与AGO2的结合,抑制AGO2/miRNA介导的肿瘤进展相关的基因沉默[47]。在小鼠研究中发现,CircPABPN1竞争性结合HuR,阻断了其与ATG16L1的结合,抑制ATG16L翻译,从而调节肠上皮细胞自噬[48]。
3 HuR调控肌肉生长发育及相关疾病的研究 3.1 HuR通过影响mRNA的稳定性参与调控肌生成近些年来的研究表明,RNA结合蛋白HuR能够与编码或非编码RNA相互作用参与肌肉生长发育以及肌肉疾病的发生,其中研究较为广泛的作用机制是HuR通过调节靶基因mRNA稳定性参与肌肉生长发育的调控(表 1)。在分化的肌肉细胞中,RNA结合蛋白HuR能够识别结合肌源性调节因子(p21、MyoD、MyoG和MyHC)的3′UTR的ARE序列,增加其mRNA稳定水平,进而促进肌肉分化[49]。另外,研究人员还发现,HuR蛋白可以结合并稳定成肌细胞增殖的关键细胞周期控制因子CCND1[50],而Pitx2(paired like homeodomain 2)介导了HuR和CCND1之间的相互结合作用,在肌肉分化过程中,Pitx2被磷酸化,复合体Pitx2/HuR/CCND1解离,导致CCND1不稳定,引起细胞周期停滞,最终促进肌肉分化[51]。此外有研究表明,在未分化的肌肉细胞中,HuR位于细胞核中,当肌肉细胞分化开始后,细胞质中的HuR增加,当细胞分化结束后,HuR重新回到细胞核,HuR在细胞核质的表达量的变化与肌源性调节因子的核质表达量相一致[52],表明HuR的核质穿梭与肌生成的调控密切相关。在成肌细胞融合过程中,半胱天冬酶发挥作用,使10%~15%的HuR被裂解为HuR-CP1(HuR-cleavage product 1, 24 ku)和HuR-CP2(HuR-cleavage product 2, 8 ku)两个片段。在小鼠C2C12成肌细胞分化过程中,HuR-CP1表达增加,并与HuR导入因子转运蛋白-2(HuR-import factor transportin-2, TRN2)结合,阻止TRN2介导完整的HuR分子进入细胞核,从而增加细胞质中HuR表达量,通过调节成肌基因mRNA的稳定性来促进肌肉生成[53]。
|
|
表 1 RNA结合蛋白调控肌肉生长发育的靶基因及作用方式 Table 1 The target genes and modes of action of RNA binding protein regulating muscle growth and development |
Lv等[54]在小鼠、猪和人中鉴定了一个新的促进肌肉生长的lncRNA——lncMGPF(lncRNA muscle growth promoting factor),该lncRNA主要通过两方面调控肌生成,一是作为miR-135a-5p分子海绵,增加肌细胞增强因子2C(MEF2C)表达; 二是通过调控HuR蛋白裂解促进HuR由细胞核到细胞质的迁移,增强HuR介导的肌源性调节基因MyoG、MyoD等mRNA的稳定性,进而提高肌细胞分化能力。有研究还发现,lncRNA-OIP5-AS1能够结合肌细胞增强因子MEF2C的3′UTR区域,增强MEF2C的稳定性,从而促进MEF2C的表达,其作用方式是,lncRNA-OIP5-AS1作为一个“分子支架”,招募RNA结合蛋白HuR结合到MEF2C的3′UTR,促进MEF2C的表达,从而促进成肌分化过程[55]。此外,有研究表明HuR还可通过与其它RNA结合蛋白相互作用参与调控肌肉生长发育。在肌肉发生早期,HuR和KSRP(KH-type splicing regulatory protein)以复合体的形式被募集到细胞周期启动子核磷蛋白(nucleophosmin, NPM)mRNA的3′UTR区域,而HuR/KSRP复合体可以招募两种核糖核酸酶,多聚腺苷酸特异性核糖核酸酶(poly(A) specific ribonuclease, PARN)和EXOSC5(exosome component 5),降低NPM mRNA的稳定性,进而促进肌肉纤维的形成[56]。Legnini等[57]研究发现,Linc-MD1作为miR-133b的宿主转录本,其在肌肉分化早期阶段表达。一方面,Linc-MD1能够与HuR蛋白结合并抑制Drosha酶作用,进而抑制miR-133合成来促进Linc-MD1的积累。另一方面,Linc-MD1又能作为miR-133的分子海绵,正向调控HuR蛋白的表达。此外,HuR蛋白可以与募集的miRNA协同来加强Linc-MD1的分子海绵活性,HuR和Linc-MD1之间形成的正向调控回路最终促进了肌肉的分化。
3.2 HuR通过解除miRNA介导的靶基因翻译抑制参与调控肌生成RNA结合蛋白HuR除了调节mRNA的稳定性,还可以通过解除miRNA介导的靶基因翻译抑制参与调控肌生成。研究发现,miR-1192可以结合到高迁移率族蛋白1(high mobility group box 1, HMGB1)mRNA的3′UTR,抑制HMGB1的翻译进而阻止肌生成过程,而HuR蛋白可以靶向结合HMGB1 3′UTR的miRNA结合位点邻近区域,阻止AGO2/miR-1192复合物的形成,从而解除miR-1192介导的翻译抑制,促进了肌生成[58]。研究表明,在癌症动物模型中,骨骼肌组织中STAT3(signal transducer and activator of transcription 3)的异常激活是导致肌肉萎缩的原因之一,而在炎症诱导的肌肉萎缩过程中,研究人员发现,HuR蛋白可以结合STAT3的3′UTR富含U的序列,阻止miR-330介导的STAT3翻译抑制,促进STAT3蛋白质的翻译,影响肌肉的发育[59]。
3.3 HuR调控肌肉发育相关疾病研究发现,RNA结合蛋白HuR还参与肌肉发育相关疾病的调节过程,包括先天性肌强直综合征和肌肉萎缩等。有研究表明,Q胶原蛋白(cooh-terminal collagen Q, COLQ)的突变会导致先天性肌强直综合征并伴随乙酰胆碱酯酶的缺失,而HuR在COLQ突变引起的先天性肌强直综合征中,起着稳定乙酰胆碱酯酶的作用,具体机制是当Q胶原蛋白缺失后,HuR能够识别并结合乙酰胆碱受体亚单位的3′UTR,增加其稳定性,促进其表达[64]。另外研究发现,HuR蛋白参与调控由癌症恶病质引起的肌肉萎缩,HuR特异性敲除可以促进小鼠Ⅰ型纤维的富集,进而使其免受癌症诱导的肌肉萎缩,过氧化物酶体增殖物激活受体γ共激活因子-1(peroxisome proliferator-activated receptor γ coactivator-1, PGC-1)是过氧化物酶体增殖物激活受体PPARγ(peroxisome proliferator-activated receptor γ, PPARγ)的转录共激活因子,HuR通过与mRNA衰减因子KSRP互作调节PGC-1 mRNA的稳定性,降低PGC-1的表达,进而抑制Ⅰ型肌纤维的形成,促进Ⅱ型肌纤维的形成,表明HuR可以通过促进糖酵解型肌纤维的形成,参与调节骨骼肌纤维类型[65]。多聚腺苷酸结合蛋白核1(polyadenylate binding protein nuclear 1, PABPN1)基因第一外显子突变能够导致肌肉特异性疾病——眼咽型肌营养不良(oculopharyngeal muscular dystrophy, OPMD),研究人员利用小鼠体内外模型研究发现,HuR蛋白能够识别并结合PABPN1 3′UTR的ARE序列,负向调控PABPN1的mRNA与蛋白水平,这一结果为以后研究OPMD治疗策略提供了参考[66]。
4 展望近年来,RNA结合蛋白与非编码RNA互作调控肌肉生长发育成为肌肉生物学研究新的焦点之一,作为机体内表达最广泛的RNA结合蛋白,HuR的生物学功能及在肌生成与肌肉疾病发生过程中的作用机制也逐渐被报道,其主要通过影响靶mRNA的稳定性和翻译参与调控肌肉生长发育及肌肉疾病的发生。但是,目前关于HuR的功能研究主要集中在模式动物上,畜禽动物上HuR的功能和作用机制尚未阐明。近些年,随着功能性非编码RNA不断被发掘,RNA结合蛋白和非编码RNA互作的调控机制研究也会越来越多。通过开展RNA结合蛋白及其与非编码RNA互作调控肌肉生长发育的相关研究,能够进一步完善肌肉生成分子调控网络,深化对肌肉生长发育潜在分子机制的认识,并可能为肌肉发育相关疾病的治疗提供新的思路和应对策略。
| [1] |
KHANNA S, MERRIAM A P, GONG B D, et al. Comprehensive expression profiling by muscle tissue class and identification of the molecular niche of extraocular muscle[J]. FASEB J, 2003, 17(10): 1370-1372. DOI:10.1096/fj.02-1108fje |
| [2] |
BHAGAVATI S, SONG X S, SIDDIQUI M A Q. RNAi inhibition of Pax3/7 expression leads to markedly decreased expression of muscle determination genes[J]. Mol Cell Biochem, 2007, 302(1-2): 257-262. DOI:10.1007/s11010-007-9444-3 |
| [3] |
TAYLOR M V, HUGHES S M. Mef2 and the skeletal muscle differentiation program[J]. Semin Cell Dev Biol, 2017, 72: 33-44. DOI:10.1016/j.semcdb.2017.11.020 |
| [4] |
OTT M O, BOBER E, LYONS G, et al. Early expression of the myogenic regulatory gene, myf-5, in precursor cells of skeletal muscle in the mouse embryo[J]. Development, 1991, 111(4): 1097-1107. DOI:10.1242/dev.111.4.1097 |
| [5] |
ZHAN S Y, QIN C Y, LI D D, et al. A novel long noncoding RNA, lncR-125b, promotes the differentiation of goat skeletal muscle satellite cells by sponging miR-125b[J]. Front Genet, 2019, 10: 1171. DOI:10.3389/fgene.2019.01171 |
| [6] |
ZHAO W, YANG H L, LI J T, et al. MiR-183 promotes preadipocyte differentiation by suppressing Smad4 in goats[J]. Gene, 2018, 666: 158-164. DOI:10.1016/j.gene.2018.05.022 |
| [7] |
LI L, CHEN Y, NIE L, et al. MyoD-induced circular RNA CDR1as promotes myogenic differentiation of skeletal muscle satellite cells[J]. Biochim Biophys Acta Gene Regul Mech, 2019, 1862(8): 807-821. DOI:10.1016/j.bbagrm.2019.07.001 |
| [8] |
ZHOU G Q, YANG Y N, ZHANG X M, et al. Msx1 cooperates with Runx1 for inhibiting myoblast differentiation[J]. Protein Expr Purif, 2021, 179: 105797. DOI:10.1016/j.pep.2020.105797 |
| [9] |
SHI D L, GRIFONE R. RNA-binding proteins in the post-transcriptional control of skeletal muscle development, regeneration and disease[J]. Front Cell Dev Biol, 2021, 9: 738978. DOI:10.3389/fcell.2021.738978 |
| [10] |
APPONI L H, CORBETT A H, PAVLATH G K. RNA-binding proteins and gene regulation in myogenesis[J]. Trends Pharmacol Sci, 2011, 32(11): 652-658. DOI:10.1016/j.tips.2011.06.004 |
| [11] |
VAN NOSTRAND E L, FREESE P, PRATT G A, et al. A large-scale binding and functional map of human RNA-binding proteins[J]. Nature, 2020, 583(7818): 711-719. DOI:10.1038/s41586-020-2077-3 |
| [12] |
PEDROTTI S, GIUDICE J, DAGNINO-ACOSTA A, et al. The RNA-binding protein Rbfox1 regulates splicing required for skeletal muscle structure and function[J]. Hum Mol Genet, 2015, 24(8): 2360-2374. DOI:10.1093/hmg/ddv003 |
| [13] |
LUNDE B M, MOORE C, VARANI G. RNA-binding proteins: modular design for efficient function[J]. Nat Rev Mol Cell Biol, 2007, 8(6): 479-490. DOI:10.1038/nrm2178 |
| [14] |
IZQUIERDO J M. Hu antigen R (HuR) functions as an alternative pre-mRNA splicing regulator of Fas apoptosis-promoting receptor on exon definition[J]. J Biol Chem, 2008, 283(27): 19077-19084. DOI:10.1074/jbc.M800017200 |
| [15] |
YU X H, LI Y J, DING Y M, et al. HuR promotes ovarian cancer cell proliferation by regulating TIMM44 mRNA stability[J]. Cell Biochem Biophys, 2020, 78(4): 447-453. DOI:10.1007/s12013-020-00939-w |
| [16] |
ZANG Y Z, LI J, WAN B L, et al. circRNA circ-CCND1 promotes the proliferation of laryngeal squamous cell carcinoma through elevating CCND1 expression via interacting with HuR and miR-646[J]. J Cell Mol Med, 2020, 24(4): 2423-2433. DOI:10.1111/jcmm.14925 |
| [17] |
SIMONE L E, KEENE J D. Mechanisms coordinating ELAV/Hu mRNA regulons[J]. Curr Opin Genet Dev, 2013, 23(1): 35-43. DOI:10.1016/j.gde.2012.12.006 |
| [18] |
HINMAN M N, LOU H. Diverse molecular functions of Hu proteins[J]. Cell Mol Life Sci, 2008, 65(20): 3168-3181. DOI:10.1007/s00018-008-8252-6 |
| [19] |
SAMSON M L. Rapid functional diversification in the structurally conserved ELAV family of neuronal RNA binding proteins[J]. BMC Genomics, 2008, 9(1): 392. DOI:10.1186/1471-2164-9-392 |
| [20] |
FAN X C, STEITZ J A. HNS, a nuclear-cytoplasmic shuttling sequence in HuR[J]. Proc Natl Acad Sci U S A, 1998, 95(26): 15293-15298. DOI:10.1073/pnas.95.26.15293 |
| [21] |
TOBA G, WHITE K. The third RNA recognition motif of Drosophila ELAV protein has a role in multimerization[J]. Nucleic Acids Res, 2008, 36(4): 1390-1399. DOI:10.1093/nar/gkm1168 |
| [22] |
张岩, 梅柱中. RNA结合蛋白HuR的研究进展[J]. 广东医学, 2012, 33(24): 3820-3822. ZHANG Y, MEI Z Z. Research progress of RNA binding protein HuR[J]. Guangdong Medical Journal, 2012, 33(24): 3820-3822. (in Chinese) |
| [23] |
SUN Q Y, TRIPATHI V, YOON J H, et al. MIR100 host gene-encoded lncRNAs regulate cell cycle by modulating the interaction between HuR and its target mRNAs[J]. Nucleic Acids Res, 2018, 46(19): 10405-10416. DOI:10.1093/nar/gky696 |
| [24] |
HU Y P, JIN Y P, WU X S, et al. LncRNA-HGBC stabilized by HuR promotes gallbladder cancer progression by regulating miR-502-3p/SET/AKT axis[J]. Mol Cancer, 2019, 18(1): 167. DOI:10.1186/s12943-019-1097-9 |
| [25] |
CIEPLY B, PARK J W, NAKAUKA-DDAMBA A, et al. Multiphasic and dynamic changes in alternative splicing during induction of pluripotency are coordinated by numerous RNA-binding proteins[J]. Cell Rep, 2016, 15(2): 247-255. DOI:10.1016/j.celrep.2016.03.025 |
| [26] |
ZHU H, HASMAN R A, BARRON V A, et al. A nuclear function of Hu proteins as neuron-specific alternative RNA processing regulators[J]. Mol Biol Cell, 2006, 17(12): 5105-5114. DOI:10.1091/mbc.e06-02-0099 |
| [27] |
ZHAO W H, ZHAO J F, HOU M M, et al. HuR and TIA1/TIAL1 are involved in regulation of alternative splicing of SIRT1 pre-mRNA[J]. Int J Mol Sci, 2014, 15(2): 2946-2958. DOI:10.3390/ijms15022946 |
| [28] |
WANG H W, MOLFENTER J, ZHU H, et al. Promotion of exon 6 inclusion in HuD pre-mRNA by Hu protein family members[J]. Nucleic Acids Res, 2010, 38(11): 3760-3770. DOI:10.1093/nar/gkq028 |
| [29] |
ZHU H, ZHOU H L, HASMAN R A, et al. Hu proteins regulate polyadenylation by blocking sites containing U-rich sequences[J]. J Biol Chem, 2007, 282(4): 2203-2210. DOI:10.1074/jbc.M609349200 |
| [30] |
ZHOU A Y, SHI G B, KANG G J, et al. RNA binding protein, HuR, regulates SCN5A expression through stabilizing MEF2C transcription factor mRNA[J]. J Am Heart Assoc, 2018, 7(9): e007802. DOI:10.1161/JAHA.117.007802 |
| [31] |
ZHANG M Y, XU Y, ZHANG Y J, et al. Circular RNA circE2F2 promotes malignant progression of ovarian cancer cells by upregulating the expression of E2F2 protein via binding to HuR protein[J]. Cell Signal, 2021, 84: 110014. DOI:10.1016/j.cellsig.2021.110014 |
| [32] |
LIU H L, LAN T, LI H, et al. Circular RNA circDLC1 inhibits MMP1-mediated liver cancer progression via interaction with HuR[J]. Theranostics, 2021, 11(3): 1396-1411. DOI:10.7150/thno.53227 |
| [33] |
WANG Y, ZHAO H L, ZHI W W. SEMA4D under the posttranscriptional regulation of HuR and miR-4319 boosts cancer progression in esophageal squamous cell carcinoma[J]. Cancer Biol Ther, 2020, 21(2): 122-129. DOI:10.1080/15384047.2019.1669996 |
| [34] |
YOON J H, ABDELMOHSEN K, KIM J, et al. Scaffold function of long non-coding RNA HOTAIR in protein ubiquitination[J]. Nat Commun, 2013, 4(1): 2939. DOI:10.1038/ncomms3939 |
| [35] |
ABDELMOHSEN K, PULLMANN R JR, LAL A, et al. Phosphorylation of HuR by Chk2 regulates SIRT1 expression[J]. Mol Cell, 2007, 25(4): 543-557. DOI:10.1016/j.molcel.2007.01.011 |
| [36] |
PU J, ZHANG Y, WANG A M, et al. ADORA2A-AS1 restricts hepatocellular carcinoma progression via binding HuR and repressing FSCN1/AKT axis[J]. Front Oncol, 2021, 11: 754835. DOI:10.3389/fonc.2021.754835 |
| [37] |
ZHANG L, FENG H, JIN Y W, et al. Long non-coding RNA LINC01119 promotes neuropathic pain by stabilizing BDNF transcript[J]. Front Mol Neurosci, 2021, 14: 673669. DOI:10.3389/fnmol.2021.673669 |
| [38] |
LI J T, DONG W, JIANG Q X, et al. LINC00668 cooperated with HuR dependent upregulation of PKN2 to facilitate gastric cancer metastasis[J]. Cancer Biol Ther, 2021, 22(4): 311-323. DOI:10.1080/15384047.2021.1905138 |
| [39] |
CAO S, XIAO L, WANG J Y, et al. The RNA-binding protein HuR regulates intestinal epithelial restitution by modulating Caveolin-1 gene expression[J]. Biochem J, 2021, 478(1): 247-260. DOI:10.1042/BCJ20200372 |
| [40] |
KULLMANN M, GÖPFERT U, SIEWE B, et al. ELAV/Hu proteins inhibit p27 translation via an IRES element in the p27 5'UTR[J]. Genes Dev, 2002, 16(23): 3087-3099. DOI:10.1101/gad.248902 |
| [41] |
DONG Z Z, LIU J G, ZHANG J T. Translational regulation of Chk1 expression by eIF3a via interaction with the RNA-binding protein HuR[J]. Biochem J, 2020, 477(10): 1939-1950. DOI:10.1042/BCJ20200025 |
| [42] |
KIM H H, KUWANO Y, SRIKANTAN S, et al. HuR recruits let-7/RISC to repress c-Myc expression[J]. Genes Dev, 2009, 23(15): 1743-1748. DOI:10.1101/gad.1812509 |
| [43] |
KWON M S, CHUNG H K, XIAO L, et al. MicroRNA-195 regulates Tuft cell function in the intestinal epithelium by altering translation of DCLK1[J]. Am J Physiol Cell Physiol, 2021, 320(6): C1042-C1054. DOI:10.1152/ajpcell.00597.2020 |
| [44] |
PORIA D K, GUHA A, NANDI I, et al. RNA-binding protein HuR sequesters microRNA-21 to prevent translation repression of proinflammatory tumor suppressor gene programmed cell death 4[J]. Oncogene, 2016, 35(13): 1703-1715. DOI:10.1038/onc.2015.235 |
| [45] |
ABDELMOHSEN K, PANDA A C, MUNK R, et al. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1[J]. RNA Biol, 2017, 14(3): 361-369. DOI:10.1080/15476286.2017.1279788 |
| [46] |
LIU B Q, YANG G S, WANG X, et al. CircBACH1 (hsa_circ_0061395) promotes hepatocellular carcinoma growth by regulating p27 repression via HuR[J]. J Cell Physiol, 2020, 235(10): 6929-6941. DOI:10.1002/jcp.29589 |
| [47] |
CHEN Y J, YANG F, FANG E H, et al. Circular RNA circAGO2 drives cancer progression through facilitating HuR-repressed functions of AGO2-miRNA complexes[J]. Cell Death Differ, 2019, 26(7): 1346-1364. DOI:10.1038/s41418-018-0220-6 |
| [48] |
LI X X, XIAO L, CHUNG H K, et al. Interaction between HuR and circPABPN1 Modulates Autophagy in the Intestinal Epithelium by Altering ATG16L1 Translation[J]. Mol Cell Biol, 2020, 40(6): e00492-12. |
| [49] |
FIGUEROA A, CUADRADO A, FAN J S, et al. Role of HuR in skeletal myogenesis through coordinate regulation of muscle differentiation genes[J]. Mol Cell Biol, 2003, 23(14): 4991-5004. DOI:10.1128/MCB.23.14.4991-5004.2003 |
| [50] |
LAL A, MAZAN-MAMCZARZ K, KAWAI T, et al. Concurrent versus individual binding of HuR and AUF1 to common labile target mRNAs[J]. EMBO J, 2004, 23(15): 3092-3102. DOI:10.1038/sj.emboj.7600305 |
| [51] |
GHERZI R, TRABUCCHI M, PONASSI M, et al. Akt2-mediated phosphorylation of Pitx2 controls Ccnd1 mRNA decay during muscle cell differentiation[J]. Cell Death Differ, 2010, 17(6): 975-983. DOI:10.1038/cdd.2009.194 |
| [52] |
VAN DER GIESSEN K, GALLOUZI I E. Involvement of transportin 2-mediated HuR import in muscle cell differentiation[J]. Mol Biol Cell, 2007, 18(7): 2619-2629. DOI:10.1091/mbc.e07-02-0167 |
| [53] |
BEAUCHAMP P, NASSIF C, HILLOCK S, et al. The cleavage of HuR interferes with its transportin-2-mediated nuclear import and promotes muscle fiber formation[J]. Cell Death Differ, 2010, 17(10): 1588-1599. DOI:10.1038/cdd.2010.34 |
| [54] |
LV W, JIN J J, XU Z Y, et al. lncMGPF is a novel positive regulator of muscle growth and regeneration[J]. J Cachexia Sarcopenia Muscle, 2020, 11(6): 1723-1746. DOI:10.1002/jcsm.12623 |
| [55] |
YANG J H, CHANG M W, PANDEY P R, et al. Interaction of OIP5-AS1 with MEF2C mRNA promotes myogenic gene expression[J]. Nucleic Acids Res, 2020, 48(22): 12943-12956. DOI:10.1093/nar/gkaa1151 |
| [56] |
CAMMAS A, SANCHEZ B J, LIAN X J, et al. Destabilization of nucleophosmin mRNA by the HuR/KSRP complex is required for muscle fibre formation[J]. Nat Commun, 2014, 5(1): 4190. DOI:10.1038/ncomms5190 |
| [57] |
LEGNINI I, MORLANDO M, MANGIAVACCHI A, et al. A feedforward regulatory loop between HuR and the long noncoding RNA linc-MD1 controls early phases of myogenesis[J]. Mol Cell, 2014, 53(3): 506-514. DOI:10.1016/j.molcel.2013.12.012 |
| [58] |
DORMOY-RACLET V, CAMMAS A, CELONA B, et al. HuR and miR-1192 regulate myogenesis by modulating the translation of HMGB1 mRNA[J]. Nat Commun, 2013, 4(1): 2388. DOI:10.1038/ncomms3388 |
| [59] |
MUBAID S, MA J F, OMER A, et al. HuR counteracts miR-330 to promote STAT3 translation during inflammation-induced muscle wasting[J]. Proc Natl Acad Sci U S A, 2019, 116(35): 17261-17270. DOI:10.1073/pnas.1905172116 |
| [60] |
SALISBURY E, SAKAI K, SCHOSER B, et al. Ectopic expression of cyclin D3 corrects differentiation of DM1 myoblasts through activation of RNA CUG-binding protein, CUGBP1[J]. Exp Cell Res, 2008, 314(11-12): 2266-2278. DOI:10.1016/j.yexcr.2008.04.018 |
| [61] |
HAUSBURG M A, DOLES J D, CLEMENT S L, et al. Post-transcriptional regulation of satellite cell quiescence by TTP-mediated mRNA decay[J]. eLife, 2015, 4: e03390. DOI:10.7554/eLife.03390 |
| [62] |
PANDA A C, ABDELMOHSEN K, YOON J H, et al. RNA-binding protein AUF1 promotes myogenesis by regulating MEF2C expression levels[J]. Mol Cell Biol, 2014, 34(16): 3106-3119. DOI:10.1128/MCB.00423-14 |
| [63] |
JIN D H, HIDAKA K, SHIRAI M, et al. RNA-binding motif protein 24 regulates myogenin expression and promotes myogenic differentiation[J]. Genes Cells, 2010, 15(11): 1158-1167. DOI:10.1111/j.1365-2443.2010.01446.x |
| [64] |
WANG H, ZHAO X, YUN W, et al. Effect of inhibiting p38 on HuR involving in β-AChR post-transcriptional mechanisms in denervated skeletal muscle[J]. Cell Mol Neurobiol, 2019, 39(7): 1029-1037. DOI:10.1007/s10571-019-00698-0 |
| [65] |
JANICE SÁNCHEZ B, TREMBLAY A M K, LEDUC-GAUDET J P, et al. Depletion of HuR in murine skeletal muscle enhances exercise endurance and prevents cancer-induced muscle atrophy[J]. Nat Commun, 2019, 10(1): 4171. DOI:10.1038/s41467-019-12186-6 |
| [66] |
PHILLIPS B L, BANERJEE A, SANCHEZ B J, et al. Post-transcriptional regulation of Pabpn1 by the RNA binding protein HuR[J]. Nucleic Acids Res, 2018, 46(15): 7643-7661. DOI:10.1093/nar/gky535 |
(编辑 郭云雁)



