禽流感是由禽流感病毒(avian influenza virus, AIV)引起的一种从呼吸系统病变到严重全身败血症等多种症状的禽类传染病[1]。AIV属于正黏病毒科,流感病毒属,根据其对鸡的致病性,可分为低致病性AIV(low pathogenic avian influenza virus, LPAIV)和高致病性AIV(high pathogenic avian influenza virus, HPAIV)[2]。H3亚型AIV是LPAIV中分离率较高的一种亚型[3]。H3亚型流感病毒宿主范围较广,除感染禽类外,陆续发现该亚型流感病毒还能感染马、犬、猫等哺乳动物[4]。H3亚型流感病毒易发生基因重组[5],如猪H3N2病毒在猪这个“混合器”中通过与人流感病毒发生基因重排,可形成感染人的新型猪流感病毒[6]。近三年来,赵婉宸等[7]在中国华东地区(江苏、山东和安徽)共分离到6株鸭或鹅源的H3N2亚型AIV,但未见浙江省H3N2亚型AIV的报道。因此,对浙江地区开展家禽H3N2亚型AIV分子流行病学检测,密切关注其传播和进化情况,不仅对畜牧业生产十分重要,还具有重要的公共卫生学意义。
本试验以2021年浙江地区分离到的3株H3N2亚型AIV为研究对象,对其开展分子特征及遗传进化分析,以期为浙江乃至华东地区H3N2亚型AIV的流行病学监测提供数据参考。
1 材料与方法 1.1 实验材料和试剂反转录试剂盒购自赛默飞世尔科技公司;Trizol试剂、Phanta Max Super-fidelity DNA Polymerase等购自南京诺唯赞生物科技有限公司;9~12日龄SPF鸡胚购自宁波纯派农业科技有限公司;氯仿、异丙醇、乙醇购自国药集团化学试剂有限公司;引物由浙江尚亚生物技术有限公司合成。
1.2 样品检测与分离2021年,于浙江省四地(嘉兴、湖州、舟山、台州)采集禽类共923份咽肛拭子。采用RT-PCR技术对样品进行AIV的检测和亚型的鉴定。采用鸡胚分离法分离病毒[8]。采用鸡胚终点稀释法纯化病毒[9],采用血凝试验(HA)和PCR法对分离到的病毒进行鉴定[10]。经PCR流感病毒HA亚型检测,只检测到单一亚型的样品用Trizol法抽提病毒RNA,并送晶能生物技术(上海)有限公司进行二代测序(Illumina NovaSeq 6000测序平台),对测序结果利用Burrows-Wheeler Aligner(版本0.7.17)[11]和SAMtools(版本1.10)[12]进行序列的比对。
1.3 病毒全基因组序列分析将测序结果用BLAST工具进行同源序列搜索,采用DNAStar软件中MegAlign模块进行同源性分析;对NCBI和GISAID数据库中的相关序列进行筛选,筛选标准:1)删除两个数据库中的重复序列;2)须具有完整开放阅读框。从NCBI和GISAID数据库中最终获得H3N2 HA基因(717条)、H3N6 HA基因(21条)、H3N2 NA基因(605条)和H6N2 NA基因(331条)。利用软件RAxML(版本8.2.4)[13]绘制HA和NA基因进化树,并用iTOL v4[14]进行可视化分析。
2 结果 2.1 样品检测与病毒分离鉴定2021年,采自嘉兴、湖州、舟山和台州的小型养殖场的鸡源咽肛拭子分别为386、294、100和30份,采自嘉兴和台州的鸭源咽肛拭子分别为3和110份,共计923份样品。AIV总阳性率为7.69%(71/923),其中,采自嘉兴市、湖州市、舟山市和台州市样品的AIV阳性率分别为4.88%(19/389)、8.16%(24/294)、28%(28/100)和0(0/140)。鸡源样品AIV阳性率为8.40%(68/810),鸭源样品AIV阳性率为2.65%(3/113)。从嘉兴市样品中分离到1株鸭源H3N2亚型AIV(A/duck/Zhejiang/Y1/2021, ZJ/Y1),从湖州市样品中分离到2株鸡源H3N2亚型AIV(A/chicken/Zhejiang/57/2021, ZJ57和A/chicken/Zhejiang/64/2021, ZJ64)。
2.2 病毒全基因组同源分析BLAST比对结果显示,2株鸡源ZJ57和ZJ64的HA基因与A/duck/Hunan/161/2015(H3N6)相似性高达94.42%,鸭源ZJ/Y1的HA基因与A/duck/Hubei/ZYSYF18/2015(H3N6)相似性为97.25%,它们均与H3N6相似度较高;3株分离株的NA基因与H6N2相似性均在96.24%以上;PB2、M基因与H1Nx相似性为97.24%和97.66%以上;ZJ57和ZJ64的PB1基因、ZJ57的NS基因、ZJ/Y1的PA基因均与H10N7对应基因具有较高相似性(表 1)。3株H3N2亚型分离株之间的HA基因核苷酸相似性为93.4%~100.0%,NA基因之间的核苷酸相似性为94.0%~99.9%。
|
|
表 1 3株H3N2亚型AIV分离株的BLAST分析 Table 1 BLAST analysis of three H3N2-subtype AIV isolates |
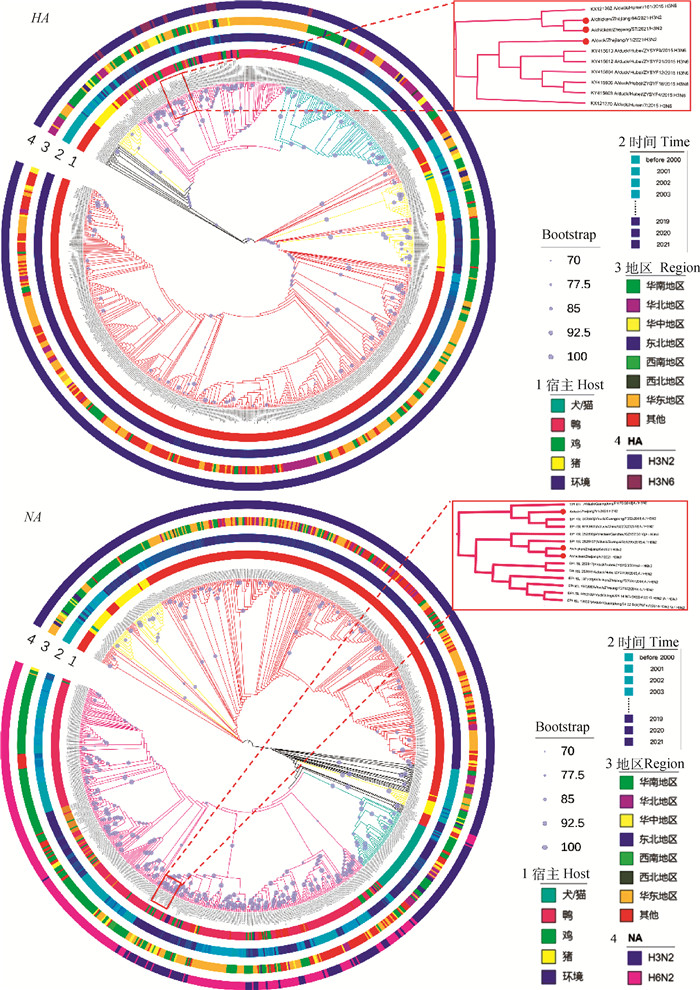
遗传进化分析表明:从宿主范围来看,H3N2亚型流感病毒的宿主范围可分为不同分支,禽源分支在遗传进化上与猪源、人源和犬/猫源均不在同一分支;H3N2亚型AIV的主要宿主为鸭(图 1);从地区分布来看,H3N2亚型流感病毒在全国均有分布,其中,H3N2亚型AIV主要流行于华东地区;从时间分布来看,AIV主要流行于2010年以后。3株H3N2亚型分离株的HA和NA基因均属于禽源分支,鸡源ZJ57和ZJ64的HA基因与A/duck/Hunan/161/2015(H3N6)遗传距离最近,而鸭源ZJ/Y1与A/duck/Hubei/ZYSYF9/2015(H3N6)遗传距离最近,鸡源ZJ57和ZJ64的HA基因与NA基因与A/duck/Guangxi/S31299/2015(H6N2)遗传距离最近,而鸭源ZJ/Y1与A/duck/Guangdong/F1172/2018 (H3N6)遗传距离最近(图 1)。
|
图 1 3株H3N2亚型流感病毒HA和NA基因遗传进化树 Fig. 1 Phylogenetic trees of HA and NA genes of three H3N2-subtype influenza viruses |
2.4.1 HA和NA基因分子特征分析 3株H3N2亚型AIV的HA基因编码区均由1 701个核苷酸组成,编码566个氨基酸。HA1区编码344个氨基酸,HA2区编码221个氨基酸。分离株的HA蛋白裂解位点序列均为PEKQTR↓GLF,具有典型的低致病性禽流感病毒特征。3株分离株的HA中影响流感病毒与受体特异性结合的226Q、228G、190E和225G未发生突变,位于结合位点周围的4个高度保守的残基(Y98、W153、H183、Y195)也未发生突变,提示分离株跨物种传播潜力较低。分离株HA蛋白的潜在N型糖基化位点均为6个,与浙江省禽源和人源分离株相比,存在糖基化位点增添和缺失的情况(表 2)。3株H3N2亚型AIV的NA基因编码区均由1 410个核苷酸组成,编码469个氨基酸,颈部区无氨基酸缺失。NA蛋白共有5个预测的N型糖基化位点,分别为第61位(NITE)、69位(NNTI)、86位(NWSK)、146位(NGTI)和234位(NGTC)。
|
|
表 2 3株H3N2亚型禽流感病毒HA蛋白N型糖基化位点预测 Table 2 Prediction of N-type glycosylation sites in HA protein of three H3N2-subtype AIVs |
2.4.2 内部片段基因分子特征分析 3株H3N2亚型禽流感分离株PB2蛋白中与哺乳动物适应性相关的第627位关键位点均为Glu,符合禽流感特征,而人源A/Homo sapien/China/LS340/2019(H3N2)株为Lys。PB1蛋白中与哺乳动物致病性相关的第66位氨基酸位为Ser(人源H3N2为Leu),而PB2中在跨物种传播起关键作用的701位氨基酸位点、PB1蛋白中与小鼠致病性相关的第198和317位氨基酸位点、PA蛋白中能使毒力增强的第97位氨基酸位点、M1蛋白中能增强病毒对小鼠致病性的第30和215位氨基酸位点均未发生突变。
3 讨论AIV为单股负链RNA病毒[15],由于其在RNA复制过程中缺乏校对机制,导致点突变频率很高。除通过快速突变产生遗传多样性外,同源重组是另外一种重要的进化机制,它驱动病毒基因变异的形成,使AIV能够克服选择压力而适应新的环境和宿主[16]。本研究结果显示,已报道的H3N2亚型AIV主要流行于2010年后,集中分布在华东地区,鸭是其主要宿主。分离株各基因片段来源复杂,说明它们可能是经过长时间进化而发生自然重排后形成的重组病毒:分离株的HA基因与湖南和湖北两地H3N6亚型毒株距离较近,ZJ57和ZJ64的NA基因与广西的H6N2毒株遗传关系较近,ZJ/Y1与广东的H3N6毒株遗传距离最近,进一步说明3株H3N2亚型AIVs片段来源较复杂,可能发生了基因重组。
流感病毒受体结合特异性会影响其跨物种传播。人H3流感病毒HA蛋白226L和228S特征使其优先与人细胞表面的α2-6半乳糖苷唾液酸(SAα2-6Gal)受体结合,而禽源优先与α2-3半乳糖苷唾液酸(SAα2-3Gal)受体结合[17],PB2中的627E和701K氨基酸位点也在流感病毒跨种间传播中起关键性作用[18]。本研究中3株H3N2亚型禽流感病毒分离株HA蛋白的226Q和228G特征表明其更倾向于结合禽源的SAα2-3Gal受体,提示分离株的跨物种传播潜力较低。PB2蛋白序列中与哺乳动物适应性相关的第627位关键位点均为Glu,符合禽流感病毒特征。PB1中的66S、198K和701M氨基酸位点,M1中的30D和215A能增强流感病毒致病性[19-22]。但PB1蛋白中第66位氨基酸位点均由Leu突变为Ser,但是否会增强对哺乳动物的致病性还需进一步验证。
禽流感病毒HA和NA蛋白都修饰有N-连接寡糖(N型糖基化位点)[23],流感病毒HA蛋白头部的糖基化位点的结构和数量不相同,是导致病毒抗原多样性的一个重要原因,它能够掩盖或者修饰抗原位点,减少先天免疫系统给病毒的生存压力[24]。本研究发现浙江人源A/Homo sapien/China/LS340/ 2019(H3N2)株HA蛋白和NA蛋白的糖基化位点较丰富,而禽源A/duck/Zhejiang/D13/2013(H3N2)[25]糖基化位点相对较少,3株分离株的糖基化位点均存在不同情况的缺失和增加,其是否会对病毒抗原多样性造成影响还需进一步探究。
4 结论2021年,浙江地区禽流感阳性率为7.70%,分离到3株H3N2亚型AIVs,其基因片段来源复杂,推测可能由不同亚型毒株经过长时间进化而发生自然重排后形成。因未发生与哺乳动物适应性位点相关的突变,表明其跨种传播潜力较低,但PB1中66位氨基酸突变为S,其是否会增加哺乳动物的致病性仍需进一步探究。
| [1] |
黄艳艳, 李悦, 张琳, 等. 6株H9N2亚型禽流感病毒的分子演化和抗原变异分析[J]. 畜牧兽医学报, 2018, 49(10): 2205-2214. HUANG Y Y, LI Y, ZHANG L, et al. Molecular and antigenic analyses of six H9N2-subtype avian influenza viruses isolated from Broiler chicken farms of Shandong province in 2017[J]. Acta Veterinaria et Zootechnica Sinica, 2018, 49(10): 2205-2214. DOI:10.11843/j.issn.0366-6964.2018.10.016 (in Chinese) |
| [2] |
赵国, 赵坤坤, 鹿欣伦, 等. 华东地区家禽低致病性禽流感的病原学检测与带毒情况的分析[J]. 畜牧兽医学报, 2010, 41(9): 1133-1137. ZHAO G, ZHAO K K, LU X L, et al. Infection of low pathogenic avian influenza in poultry in eastern China[J]. Acta Veterinaria et Zootechnica Sinica, 2010, 41(9): 1133-1137. (in Chinese) |
| [3] |
PENG Y, XIE Z X, LIU J B, et al. Visual detection of H3 subtype avian influenza viruses by reverse transcription loop-mediated isothermal amplification assay[J]. Virol J, 2011, 8: 337. DOI:10.1186/1743-422X-8-337 |
| [4] |
LIU M, HE S Q, WALKER D, et al. The influenza virus gene pool in a poultry market in South Central China[J]. Virology, 2003, 305(2): 267-275. DOI:10.1006/viro.2002.1762 |
| [5] |
SONG M S, OH T K, MOON H J, et al. Ecology of H3 avian influenza viruses in Korea and assessment of their pathogenic potentials[J]. J Gen Virol, 2008, 89(4): 949-957. DOI:10.1099/vir.0.83462-0 |
| [6] |
谭小艳, 顾大勇, 何建安, 等. 2016-2019年深圳口岸入境人群感染甲型流感的流行病学特征分析[J]. 中国人兽共患病学报, 2020, 36(11): 922-927. TAN X Y, GU D Y, HE J A, et al. Epidemiological characteristics of influenza A infection among the inbound population at Shenzhen ports, 2016-2019[J]. Chinese Journal of Zoonoses, 2020, 36(11): 922-927. (in Chinese) |
| [7] |
赵婉宸, 李扬, 才天宇, 等. 2019-2021年华东地区6株禽源H3亚型流感病毒的遗传进化分析[J/OL]. 中国动物传染病学报, 2022. [2022-03-05]. https://doi.org/10.19958/j.cnki.cn31-2031/s.20220302.005. ZHAO W C, LI Y, CAI T Y, et al. Genetic evolution analysis of six avian H3 subtype influenza viruses in Eastern China from 2019 to 2021[J/OL]. Chinese Journal of Animal Infectious Diseases, 2022. [2022-03-05]. https://doi.org/10.19958/j.cnki.cn31-2031/s.20220302.005. (in Chinese) |
| [8] |
刘婷婷, 谢芝勋, 宋德贵, 等. 11株H3N2亚型禽流感病毒广西分离株全基因序列测定与分析[J]. 中国兽医学报, 2017, 37(5): 859-865. LIU T T, XIE Z X, SONG D G, et al. Whole genome sequencing and analysis of eleven strains of avian influenza virus subtype H3N2 isolated in Guangxi province[J]. Chinese Journal of Veterinary Science, 2017, 37(5): 859-865. (in Chinese) |
| [9] |
仇保丰, 刘武杰, 胡顺林, 等. 混合感染的多种亚型禽流感病毒的纯化与鉴定[J]. 微生物学报, 2010, 50(1): 107-112. CHOU B F, LIU W J, HU S L, et al. Purification and identification of avian influenza viruses causing mixed multi-infection[J]. Acta Microbiologica Sinica, 2010, 50(1): 107-112. (in Chinese) |
| [10] |
张锐. 禽流感病毒的流行株序列分析及PA单克隆抗体的制备[D]. 杭州: 浙江大学, 2020. ZHANG R. Sequence analysis of avian influenza virus epidemic strains and preparation of a monoclonal antibody against PA protein[D]. Hangzhou: Zhejiang University, 2020. (in Chinese) |
| [11] |
LI H, DURBIN R. Fast and accurate short read alignment with Burrows-Wheeler transform[J]. Bioinformatics, 2009, 25(14): 1754-1760. DOI:10.1093/bioinformatics/btp324 |
| [12] |
LI H, HANDSAKER B, WYSOKER A, et al. The sequence alignment/map format and SAMtools[J]. Bioinformatics, 2009, 25(16): 2078-2079. DOI:10.1093/bioinformatics/btp352 |
| [13] |
STAMATAKIS A. RAxML version 8:a tool for phylogenetic analysis and post-analysis of large phylogenies[J]. Bioinformatics, 2014, 30(9): 1312-1313. DOI:10.1093/bioinformatics/btu033 |
| [14] |
LETUNIC I, BORK P. Interactive tree of life (iTOL) v3:an online tool for the display and annotation of phylogenetic and other trees[J]. Nucleic Acids Res, 2016, 44(W1): W242-W245. DOI:10.1093/nar/gkw290 |
| [15] |
何世成, 彭志, 王卫国, 等. 2011-2015年环洞庭湖区H3亚型禽流感病毒的分离鉴定与遗传演化[J]. 畜牧兽医学报, 2019, 50(2): 382-389. HE S C, PENG Z, WANG W G, et al. Isolation, identification and genetic evolution of H3 subtype avian influenza virus in Dongting lake region from 2011 to 2015[J]. Acta Veterinaria et Zootechnica Sinica, 2019, 50(2): 382-389. (in Chinese) |
| [16] |
CHEN X Y, GUO F C, PAN J B, et al. Rare homologous recombination in H3N2 avian influenza A viruses[J]. J Infect, 2020, 80(3): 350-371. |
| [17] |
LIN Y P, XIONG X L, WHARTON S A, et al. Evolution of the receptor binding properties of the influenza A(H3N2) hemagglutinin[J]. Proc Natl Acad Sci U S A, 2012, 109(52): 21474-21479. |
| [18] |
SHINYA K, HAMM S, HATTA M, et al. PB2 amino acid at position 627 affects replicative efficiency, but not cell tropism, of Hong Kong H5 N1 influenza A viruses in mice[J]. Virology, 2004, 320(2): 258-266. |
| [19] |
CONENELLO G M, ZAMARIN D, PERRONE L A, et al. A single mutation in the PB1-F2 of H5N1 (HK/97) and 1918 influenza A viruses contributes to increased virulence[J]. PLoS Pathog, 2007, 3(10): e141. |
| [20] |
KATZ J M, LU X H, TUMPEY T M, et al. Molecular correlates of influenza A H5N1 virus pathogenesis in mice[J]. J Virol, 2000, 74(22): 10807-10810. |
| [21] |
SONG M S, PASCUA P N Q, LEE J H, et al. The polymerase acidic protein gene of influenza a virus contributes to pathogenicity in a mouse model[J]. J Virol, 2009, 83(23): 12325-12335. |
| [22] |
FAN S F, DENG G H, SONG J S, et al. Two amino acid residues in the matrix protein M1 contribute to the virulence difference of H5N1 avian influenza viruses in mice[J]. Virology, 2009, 384(1): 28-32. |
| [23] |
TATE M D, JOB E R, DENG Y M, et al. Playing hide and seek: how glycosylation of the influenza virus hemagglutinin can modulate the immune response to infection[J]. Viruses, 2014, 6(3): 1294-1316. |
| [24] |
BROWN L E, FFRENCH R A, GAWLER J M, et al. Distinct epitopes recognized by I-Ad-restricted T-cell clones within antigenic site E on influenza virus hemagglutinin[J]. J Virol, 1988, 62(1): 305-312. |
| [25] |
WU H B, WU N P, PENG X R, et al. Molecular characterization and phylogenetic analysis of H3 subtype avian influenza viruses isolated from domestic ducks in Zhejiang Province in China[J]. Virus Genes, 2014, 49(1): 80-88. |
(编辑 孟培)



