2. 通江县农业局, 巴中 636700;
3. 稻城县农牧农村和科技局, 甘孜 627750;
4. 乡城县农牧农村和科技局, 甘孜 627850;
5. 荣县农牧农村和科技局, 甘孜 627950;
6. 四川铁骑力士集团冯光德实验室, 绵阳 621006
2. Tongjiang Agriculture Bureau, Bazhong 636700, China;
3. Agriculture, Animal Husbandry, Rural and Science and Technology Bureau of Daocheng County, Ganzi 627750, China;
4. Agriculture, Animal Husbandry, Rural and Science and Technology Bureau of Xiangcheng County, Ganzi 627850, China;
5. Agriculture, Animal Husbandry, Rural and Science and Technology Bureau of Rong County, Ganzi 627950, China;
6. Fengguangde Lab, Techlex Group, Mianyang 621006, China
我国是猪种遗传多样性特别丰富的国家之一,2011年在《中国畜禽遗传资源志·猪志》中公布的地方猪遗传资源有76个[1],它们具有性成熟早、抗病力强、肉质优异以及耐粗饲等特点[2]。丰富的猪种遗传资源是培育新品种、配套系最重要的物质基础和基本育种素材[3],这不仅是中国也是世界动物遗传资源宝库的重要组成部分。地方猪遗传资源是关乎我国养猪业可持续发展的重大战略性资源,是实现种业安全的根本性保障。然而,由于大量高效外种猪的引进,使许多地方猪种数量急剧减少,有的已处于濒危状态或濒临灭绝[3]。四川悠久的养猪历史和复杂多样的地理环境造就了内江猪、成华猪、雅南猪、乌金猪(凉山类群)、青峪猪、丫杈猪和藏猪等优良地方猪种资源,均已列入《国家畜禽遗传资源品种名录(2021年版)》[4]。研究四川省地方猪种的遗传多样性,对于地方猪遗传资源的正确认识和评价、优势性状挖掘以及科学开发利用具有重要意义。
单核苷酸多态性(single nucleotide polymorphism,SNP)作为第三代分子标记,已被广泛用于生物群体遗传多样性分析、分子标记辅助选择的研究[5-18],全基因组SNP芯片技术则是群体遗传结构和多样性分析的一把利器[19-26]。“中芯一号”是江西农业大学黄路生院士团队研发的猪育种芯片,它包含繁殖、生长肥育、胴体、肉质、免疫等7大类422个性状的相关位点,涵盖猪从出生到屠宰共10个时间点,拥有51 368个SNPs位点,其中包含有81个亚洲特有地方品种位点,具有高效性和精准性优势,对我国特有猪品种的选育工作更具针对性[27-30]。为增加对四川省地方猪种遗传价值和经济价值的认识,充分发掘、评价和利用地方猪种优良种质特性,促进地方猪种资源保护,本研究采用“中芯一号”芯片分析了四川省地方猪种和山东省徒河黑猪重要经济性状关联位点的SNP基因型分布以及群体间遗传结构,为地方猪种资源的科学保护和利用提供分子水平的依据。
1 材料与方法 1.1 试验材料采集四川省6个地方猪种7个类群和徒河黑猪共562头猪的耳组织样本,包括23头成华猪、26头雅南猪、60头青峪猪、57头内江猪、151头丫杈猪、57头乌金猪(凉山类群)、51头平原藏猪、109头高原藏猪和28头徒河黑猪,四川省地方猪种来自四川省内各个猪种的保种场,徒河黑猪来自于四川省铁骑力士食品有限公司。采集的耳组织样置于装有75%酒精的2 mL冻存管中,-20 ℃保存。
1.2 试验方法1.2.1 DNA提取与分型 采用磁珠法提取DNA,首先用酒精消毒后的镊子从原始样本管夹出组织并用卫生纸吸干表面保护液,用消毒后的剪刀剪取大米粒大小的动物组织放入加入20 μL Proteinase K和300 μL Buffer WL的裂解管中,涡旋混匀振荡,将裂解管放入56 ℃恒温水浴锅裂解40 min,将裂解完成的裂解管放入离心机进行瞬时离心,转移上清液至96深孔板中,向96深孔板中加入相关试剂,然后用磁棒套进行磁珠吸附,最后将洗脱产物转移至1.5 mL收集管中低温保存。
提取好的DNA经紫外分光光度仪(NanoDrop2000)和凝胶电泳进行质量检测,检测合格后的DNA样本使用“中芯一号”芯片(北京康普森农业科技有限公司,北京)进行SNP基因分型。
1.2.2 芯片数据质控 利用Plink V1.90软件[31]对SNP基因型数据进行质控,只使用常染色体上的位点,再剔除检出率小于90%、最小等位基因频率(minor allele frequency,MAF)小于0.05的SNP位点,用于后续分析。
1.2.3 重要经济性状关联位点SNP基因型分析 利用Pink V1.90版本、R等软件对猪的7个影响肉质、生长和体型的基因及抗病性基因进行基因型分析统计,对经济性状有利的等位基因统一用Q表示,不利的等位基因统一用q表示。
1.2.4 群体遗传结构分析 利用Mega X[32](V10.0)进行聚类分析,使用邻接法(Neighbor-Joining, NJ),基于IBS遗传距离,对四川省地方猪种和徒河黑猪样本进行聚类。使用Plink V1.90软件对四川省6个地方猪种7个类群和徒河黑猪进行主成分分析(PCA),分析各猪种间遗传距离。同时基于全基因组SNP芯片结果,根据PCA分析结果,利用VCFtools(V0.1.17)软件计算地方猪种群间的Fst指数,分析群体间遗传关系远近。
2 结果 2.1 重要经济性状关联位点的SNP基因型对四川省地方猪和徒河黑猪群体中与重要经济性状关联位点的SNP基因型检测结果(表 1)显示,四川省地方猪群体中,采食量、酸肉和应激性状的杂合基因型Qq和隐性基因型qq总比例很低,均低于2.20%;料重比和抗仔猪腹泻性状的Qq、qq基因型总比例分别为11.80%和8.99%,其中qq基因型仅为0.38%和0.56%;多肋和公猪精液品质性状的Qq、qq基因型总比例较高,分别为95.32%和73.97%。徒河黑猪群体中,应激性状的等位基因q已完全剔除,采食量和酸肉性状的Qq、qq基因型总比例很低,均为3.57%;料重比、多肋、抗仔猪腹泻和公猪精液品质性状的Qq、qq基因型总比例较高,分别为82.14%、92.86%、50.00%、39.29%。
|
|
表 1 重要经济性状关联位点的SNP基因分型情况 Table 1 SNP genotyping of important economic traits |
四川省地方猪群体中(表 2),采食量性状仅乌金猪和高原藏猪分别有2个和1个非优势杂合基因型Qq,比例分别为3.51%和0.92%;应激性状仅丫杈猪、乌金猪和高原藏猪分别有7个、3个和1个非优势杂合基因型Qq,比例分别为4.64%、5.26%和0.92%;抗仔猪腹泻性状的非优势基因型主要集中在丫杈猪和乌金猪群体,比例分别为17.88%和28.07%;酸肉性状的非优势基因型只有高原藏猪群体中存在,Qq、qq基因型各3个,占比均为2.75%;料重比性状的非优势基因型主要集中在成华猪、青峪猪、丫杈猪和乌金猪群体,比例分别为17.39%、25.00%、22.52%和15.79%;各猪种多肋性状的非优势基因型总比例均高于80.00%,且多为隐性基因型qq,杂合基因型Qq比例丫杈猪最高(55.63%),乌金猪次之(28.07%),其余猪种比例均低于4.00%;各猪种公猪精液品质性状的非优势基因型比例较高,按比例高低顺序排列为内江猪(92.98%)、成华猪(91.30%)、平原藏猪(90.20%)、乌金猪(82.45%)、青峪猪(80.00%)、丫杈猪(73.51%)、雅南猪(73.08%)、高原藏猪(45.87%)。平原藏猪和高原藏猪各性状的基因型分布基本一致,主要不同之处在于高原藏猪的酸肉性状有6个非优势等位基因型,且公猪精液品质性状的qq基因型比例(6.42%)远低于平原藏猪(47.06%)。
|
|
表 2 四川省不同地方猪群体重要经济性状关联位点的SNP非优势基因型分布情况 Table 2 The distribution of non-advantage SNP genotypes of important economic traits in local pig breeds in Sichuan province |
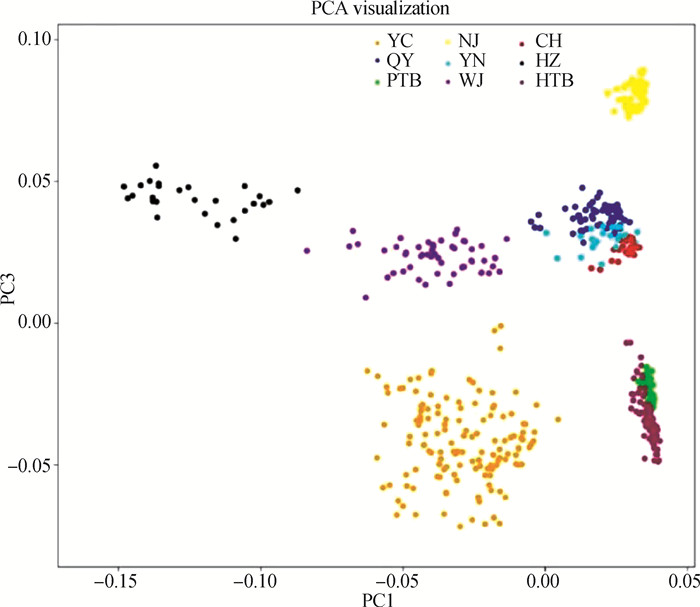
聚类分析结果显示(图 1),同一品种的所有个体均能够聚在一起,品种间没有交叉,平原藏猪和高原藏猪两个群体间有1头交叉。主成分分析可以用来解析群体间分层结构,结果显示(图 2),徒河黑猪与四川省地方猪种遗传距离较远,群体分层结构明显。四川省地方猪种中,丫杈猪、乌金猪、内江猪和藏猪(平原和高原)各自聚为一类,品种间群体分层结构明显;雅南猪、成华猪、青峪猪聚为一类,3个品种之间距离较近并有部分个体重叠。

|
YC.丫杈猪;QY.青峪猪;NJ.内江猪;YN.雅南猪;WJ.乌金猪;CH.成华猪;PTB.平原藏猪;HTB.高原藏猪;HZ.徒河黑猪。下同 YC.Yacha pig; QY. Qingyu pig; NJ. Neijiang pig; YN. Ya'nan pig; WJ. Wujin pig; CH. Chenghua pig; PTB. Plain Tibetan pig; HTB. Plateau Tibetan pig; HZ. Tuhe black pig. The same as below 图 1 聚类分析结果 Fig. 1 Results of cluster analysis |
|
图 2 PCA分析结果 Fig. 2 Results of principal component analysis (PCA) |
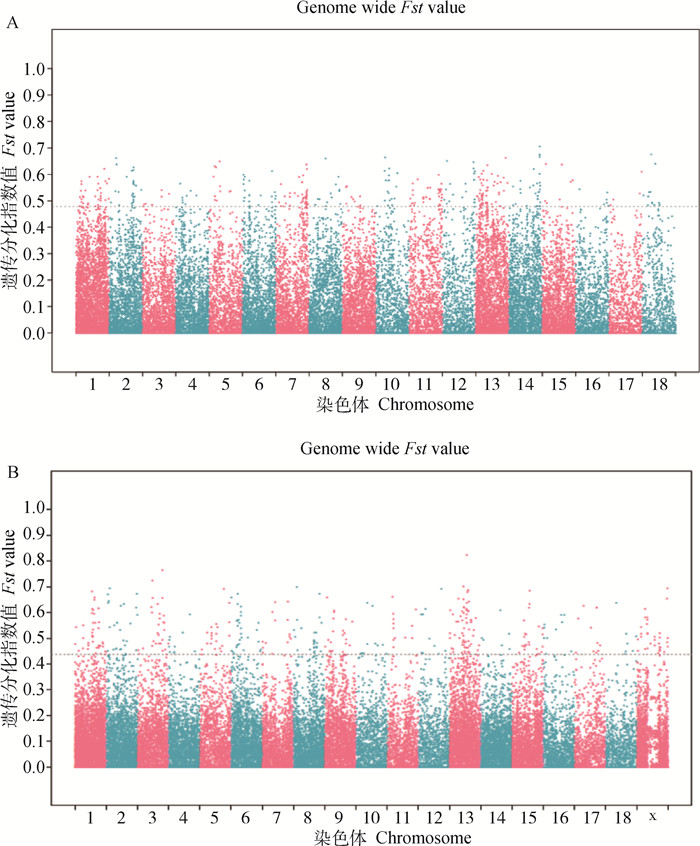
丫杈猪和青峪猪同属湖川山地猪,分别分布于四川北部山区和南部山区,在地域分布上存在很大差异,同时在性能表型上也存在显著差异[33],主成分分析结果显示青峪猪与丫杈猪形成了明显各自的簇(cluster),表明两个群体分层结构明显,且明显高于成华猪与雅南猪。为进一步解析丫杈猪和青峪猪的群体分化程度,在基因组层面分别计算了丫杈猪与青峪猪间、内江猪与乌金猪间的群体遗传分化指数(Fst)。结果显示,丫杈猪与青峪猪间的Fst指数达0.48(图 3A),高于内江猪与乌金猪间的0.44(图 3B),说明丫杈猪和青峪猪群体间存在高度遗传分化。
|
A.青峪猪与丫杈猪间群体 Fst 指数;B. 内江猪与乌金猪间群体 Fst 指数 A. Fst index between Qingyu pigs and Yacha pigs; B. Fst index between Neijiang pigs and Wujin pigs 图 3 种群间Fst指数计算结果 Fig. 3 Calculation results of Fst index |
SNP分型技术已广泛应用于畜禽遗传学研究、品种选育、种质资源保护等领域,促进了畜禽产业的发展[34-45]。本研究中,四川省地方猪群体除高原藏猪外,酸肉性状已无不利的等位基因q,应激性状的不利等位基因q只存在于丫杈猪、乌金猪和高原藏猪的极个别个体中,这与地方猪肉质优异、抗应激性好的特性一致。采食量性状的不利等位基因q只存在于乌金猪和高原藏猪个别个体中。料重比和抗仔猪腹泻性状的非优势基因型比例较低,在不影响群体血缘的情况下,可通过针对性的选择,尽快淘汰qq基因型个体,同时加强杂合子Qq基因型个体后代的检测,提升Q等位基因比例。多肋和公猪精液品质性状的非优势基因型比例高,说明四川省地方猪种没有通过分子标记辅助选择技术在该位点上对这两个性状进行选育。
徒河黑猪群体中没有应激性状的非优势等位基因型,采食量和酸肉性状也分别只有1个Qq基因型和1个qq基因型个体,在不影响群体血缘的情况下可及时淘汰;料重比、多肋、抗仔猪腹泻和公猪精液品质性状的非优势基因型比例较高,说明之前没有通过分子标记辅助选择技术在该位点上对这3个性状进行选育,可根据血缘覆盖原则慢慢淘汰qq基因型个体,同时加强Qq基因型个体后代基因型检测,不断提升Q等位基因频率。
3.2 不同猪群间遗传距离分析研究品种间的遗传距离对于研究品种的形成以及猪品种的分类具有重要意义。本研究基于SNP芯片数据,对四川省7个地方猪类群和山东省徒河黑猪群体进行了主成分分析,并基于IBS遗传距离,使用邻接法(neighbor-joining, NJ)进行了聚类分析。山东省徒河黑猪与四川地方猪种间遗传距离较远,泾渭分明;雅南猪、成华猪、青峪猪聚为一类,品种间遗传距离较近,丫杈猪、乌金猪、内江猪和藏猪各自聚为一类。
1977年根据四川省地方猪种资源联合调查结果,四川盆地周围的丫杈猪、黔江猪、青峪猪及南江猪被统一归并命名为盆周山地猪,载入《四川省畜禽品种志》;1980年,四川的盆周山地猪、湖北的鄂西黑猪和与湖南西部接壤地区的猪种又被归并统一命名为湖川山地猪;2009年,经国家畜禽遗传资源委员会猪专业委员会组织专家讨论,认为丫杈猪体型较大,与盆周山地猪有一定差异,在湖川山地猪中单独作为一个类群[1]。丫杈猪和青峪猪在地域分布和肥育、胴体和肉质性能表型上都存在明显差异[33],本研究中,聚类和主成分分析均显示,青峪猪与丫杈猪形成了明显各自的簇类(cluster),表明两个猪群遗传距离较远,差异很大;群体遗传分化指数(Fst)计算结果也表明,两个群体间存在高度遗传分化,从基因组层面表明了丫杈猪与青峪猪不属于一个类群,且将其归类于湖川山地猪同一品种缺乏依据,有待商榷。
4 结论本研究结果显示,四川省地方猪群体的采食量、酸肉、应激、抗仔猪腹泻和料重比性状的非优势基因型比例较低,公猪精液品质和多肋性状的非优势基因型比例较高;徒河黑猪群体的应激性状已无q等位基因,采食量和酸肉性状的非优势基因型比例较低,公猪精液品质、抗仔猪腹泻、料重比和多肋性状的非优势基因型比例较高。同时,明晰了四川省地方猪种间遗传分化关系,发现丫杈猪与青峪猪都归类于湖川山地猪有待商榷,为地方猪种遗传资源保护和科学的种群分类提供了理论依据。
| [1] |
国家畜禽遗传资源委员会. 中国畜禽遗传资源志·猪志[M]. 北京: 中国农业出版社, 2011. China National Commission of Animal Genetic Resources. Animal genetic resources in China, pigs[M]. Beijing: China Agriculture Press, 2011. (in Chinese) |
| [2] |
魏菁, 张子群, 王琦, 等. 中国地方猪种种质资源的保护与利用探讨[J]. 畜禽业, 2021, 32(3): 43-44. WEI J, ZHANG Z Q, WANG Q, et al. Discussion on protection and utilization of local pig germplasm resources in China[J]. Livestock and Poultry Industry, 2021, 32(3): 43-44. (in Chinese) |
| [3] |
采编部, 王旭, 尹亚斌. 四川农业大学: 保护开发利用猪品种确保生猪稳产保供[J]. 中国畜牧业, 2021(1): 25. Editorial Department, WANG X, YIN Y B. Sichuan agricultural university: protect, develop and utilize pig breeds to ensure stable production and supply of live pigs[J]. China Animal Industry, 2021(1): 25. (in Chinese) |
| [4] |
葛桂华, 王小强, 李强, 等. 四川列入《国家畜禽遗传资源品种名录》情况介绍[J]. 四川畜牧兽医, 2021, 48(4): 21-22. GE G H, WANG X Q, LI Q, et al. Introduction of livestock and poultry genetic resources of Sichuan included in the "national list of livestock and poultry genetic resources"[J]. Sichuan Animal & Veterinary Sciences, 2021, 48(4): 21-22. (in Chinese) |
| [5] |
BOSSE M, MEGENS H J, MADSEN O, et al. Using genome-wide measures of coancestry to maintain diversity and fitness in endangered and domestic pig populations[J]. Genome Res, 2015, 25(7): 970-981. DOI:10.1101/gr.187039.114 |
| [6] |
QIAO R, LI X, HAN X, et al. Population structure and genetic diversity of four Henan pig populations[J]. Anim Genet, 2019, 50(3): 262-265. DOI:10.1111/age.12775 |
| [7] |
WANG K J, LIU Y F, XU Q, et al. A post-GWAS confirming GPAT3 gene associated with pig growth and a significant SNP influencing its promoter activity[J]. Anim Genet, 2017, 48(4): 478-482. DOI:10.1111/age.12567 |
| [8] |
KRUPA E, MORAVXČÍKOVÁ N, KRUPOVÁ Z, et al. Assessment of the genetic diversity of a local pig breed using pedigree and SNP data[J]. Genes (Basel), 2021, 12(12): 1972. DOI:10.3390/genes12121972 |
| [9] |
ZHANG Y F, ZHANG J J, GONG H F, et al. Genetic correlation of fatty acid composition with growth, carcass, fat deposition and meat quality traits based on GWAS data in six pig populations[J]. Meat Sci, 2019, 150: 47-55. DOI:10.1016/j.meatsci.2018.12.008 |
| [10] |
LEE J, PARK N, LEE D, et al. Evolutionary and functional analysis of Korean native pig using single nucleotide polymorphisms[J]. Mol Cells, 2020, 43(8): 728-738. |
| [11] |
LORENZINI R, FANELLI R, TANCREDI F, et al. Matching STR and SNP genotyping to discriminate between wild boar, domestic pigs and their recent hybrids for forensic purposes[J]. Sci Rep, 2020, 10(1): 3188. DOI:10.1038/s41598-020-59644-6 |
| [12] |
崔清明, 刘奇, 彭英林, 等. 基于RAD-seq简化基因组测序的大围子猪群体遗传学分析[J]. 中国畜牧杂志, 2021, 57(S1): 222-225. CUI Q M, LIU Q, PENG Y L, et al. Population genetics analysis of Daweizi pigs based on simplified RAD-seq genome sequencing[J]. Chinese Journal of Animal Science, 2021, 57(S1): 222-225. (in Chinese) |
| [13] |
刘刁, 芦春莲, 李尚, 等. 深县猪全基因组选择信号检测分析[J]. 中国畜牧杂志, 2021, 57(10): 135-139. LIU D, LU C L, LI S, et al. Detection and analysis of genomic selection signature of Shenxian pigs[J]. Chinese Journal of Animal Science, 2021, 57(10): 135-139. DOI:10.19556/j.0258-7033.20210329-07 (in Chinese) |
| [14] |
张云鹏, 高一, 张琪, 等. 松辽黑猪NR5A2基因内含子6多态性及其与繁殖性状的关联分析[J]. 中国畜牧杂志, 2021, 57(9): 119-122. ZHANG Y P, GAO Y, ZHANG Q, et al. Intron 6 polymorphism of NR5A2 gene in Songliao black pigs and its association with reproductive traits[J]. Chinese Journal of Animal Science, 2021, 57(9): 119-122. (in Chinese) |
| [15] |
吴俊静, 阮航, 乔木, 等. 猪PLIN2基因变异筛选及其与猪肌内脂肪含量性状的关联分析[J]. 中国畜牧杂志, 2021, 57(S1): 158-163. WU J J, RUAN H, QIAO M, et al. Variation screening of porcine PLIN2 gene and its association with porcine intramuscular fat content[J]. Chinese Journal of Animal Science, 2021, 57(S1): 158-163. (in Chinese) |
| [16] |
VAN BA N, NAM L Q, DO D N, et al. An assessment of genetic diversity and population structures of fifteen Vietnamese indigenous pig breeds for supporting the decision making on conservation strategies[J]. Trop Anim Health Prod, 2020, 52(3): 1033-1041. DOI:10.1007/s11250-019-02090-y |
| [17] |
XU P, WANG X P, NI L G, et al. Genome-wide genotyping uncovers genetic diversity, phylogeny, signatures of selection, and population structure of Chinese Jiangquhai pigs in a global perspective[J]. J Anim Sci, 2019, 97(4): 1491-1500. DOI:10.1093/jas/skz028 |
| [18] |
JOAQUIM L B, CHUD T C S, MARCHESI J A P, et al. Genomic structure of a crossbred Landrace pig population[J]. PLoS One, 2019, 14(2): e0212266. DOI:10.1371/journal.pone.0212266 |
| [19] |
AI H S, HUANG L S, REN J. Genetic diversity, linkage disequilibrium and selection signatures in Chinese and western pigs revealed by genome-wide SNP markers[J]. PLoS One, 2013, 8(2): e56001. DOI:10.1371/journal.pone.0056001 |
| [20] |
EDEA Z, HONG J K, JUNG J H, et al. Detecting selection signatures between Duroc and Duroc synthetic pig populations using high-density SNP chip[J]. Anim Genet, 2017, 48(4): 473-477. DOI:10.1111/age.12559 |
| [21] |
WANG Y, ZHANG T R, WANG C D. Detection and analysis of genome-wide copy number variation in the pig genome using an 80 K SNP Beadchip[J]. J Anim Breed Genet, 2020, 137(2): 166-176. DOI:10.1111/jbg.12435 |
| [22] |
MUÑOZ M, BOZZI R, GARCÍA-CASCO J, et al. Genomic diversity, linkage disequilibrium and selection signatures in European local pig breeds assessed with a high density SNP chip[J]. Sci Rep, 2019, 9(1): 13546. DOI:10.1038/s41598-019-49830-6 |
| [23] |
冯雪燕, 刁淑琪, 刘玉强, 等. 基于SNP芯片的海南猪全基因组选择信号分析[J]. 畜牧兽医学报, 2022, 53(2): 349-359. FENG X Y, DIAO S Q, LIU Y Q, et al. Analysis of selection signatures for Hainan pigs across the whole genome based on SNP BeadChips[J]. Acta Veterinaria et Zootechnica Sinica, 2022, 53(2): 349-359. (in Chinese) |
| [24] |
路玉洁, 莫家远, 綦文晶, 等. 几个中国地方猪群体遗传结构和产仔数性状选择信号分析[J]. 畜牧兽医学报, 2022, 53(2): 360-369. LU Y J, MO J Y, QI W J, et al. Population genetic structure and selection signatures associated with litter size trait in several Chinese indigenous pig breeds[J]. Acta Veterinaria et Zootechnica Sinica, 2022, 53(2): 360-369. (in Chinese) |
| [25] |
刘彬, 沈林園, 陈映, 等. 基于SNP芯片分析青峪猪保种群体的遗传结构[J]. 畜牧兽医学报, 2020, 51(2): 260-269. LIU B, SHEN L Y, CHEN Y, et al. Analysis of genetic structure of conservation population in Qingyu pig based on SNP chip[J]. Acta Veterinaria et Zootechnica Sinica, 2020, 51(2): 260-269. (in Chinese) |
| [26] |
WANG X, WANG C, HUANG M, et al. Genetic diversity, population structure and phylogenetic relationships of three indigenous pig breeds from Jiangxi Province, China, in a worldwide panel of pigs[J]. Anim Genet, 2018, 49(4): 275-283. |
| [27] |
路玉洁, 莫家远, 朱思燃, 等. 利用中芯一号SNP芯片检测隆林猪全基因组拷贝数变异[J]. 中国畜牧兽医, 2022, 49(1): 23-31. LU Y J, MO J Y, ZHU S R, et al. Detection of genome-wide copy number variation using porcine 50K SNP beadchips in Longlin pigs[J]. China Animal Husbandry & Veterinary Medicine, 2022, 49(1): 23-31. (in Chinese) |
| [28] |
杨广礼, 章焕, 田慧月, 等. 野猪、松辽黑母猪及其杂交一代多性状功能基因分子遗传标记研究[J]. 中国畜牧兽医, 2021, 48(3): 946-953. YANG G L, ZHANG H, TIAN H Y, et al. Study on molecular genetic marker of multi-traits functional genes in wild boars, Songliao black sows and their hybrid first generation[J]. China Animal Husbandry & Veterinary Medicine, 2021, 48(3): 946-953. (in Chinese) |
| [29] |
王文君, 王小康, 徐国强, 等. 影响猪精子耐冻性的候选基因分析[J]. 华中农业大学学报, 2021, 40(3): 203-211. WANG W J, WANG X K, XU G Q, et al. Analysis of candidate genes affecting semen freezability of boar sperm[J]. Journal of Huazhong Agricultural University, 2021, 40(3): 203-211. (in Chinese) |
| [30] |
吴清清, 刘雯悦, 王媛, 等. 猪繁殖与呼吸综合征抗病指示性状的分子标记筛查及检测[J]. 华中农业大学学报, 2022, 41(1): 186-193. WU Q Q, LIU W Y, WANG Y, et al. Screening and detection of molecular markers for porcine reproductive and respiratory syndrome resistance traits[J]. Journal of Huazhong Agricultural University, 2022, 41(1): 186-193. (in Chinese) |
| [31] |
CHANG C C, CHOW C C, TELLIER L C, et al. Second-generation PLINK: rising to the challenge of larger and richer datasets[J]. GigaScience, 2015, 4: 7. |
| [32] |
KUMAR S, STECHER G, LI M, et al. MEGA X: molecular evolutionary genetics analysis across computing platforms[J]. Mol Biol Evol, 2018, 35(6): 1547-1549. |
| [33] |
王言, 钟志君, 何志平, 等. 四川6个地方猪种的育肥和胴体性能以及肉质和风味物质比较研究[J]. 畜牧与兽医, 2021, 53(1): 7-12. WANG Y, ZHONG Z J, HE Z P, et al. Comparative study on the fattening, carcass performance, meat quality and flavor substance of six local pig breeds in the Sichuan area[J]. Animal Husbandry & Veterinary Medicine, 2021, 53(1): 7-12. (in Chinese) |
| [34] |
宋玉朴, 孙永峰, 冯自强, 等. SNP分型检测技术及其在畜禽遗传和育种中的应用研究进展[J]. 中国畜牧杂志, 2021, 57(7): 37-42. SONG Y P, SUN Y F, FENG Z Q, et al. Advances in SNP typing techniques and their application in genetic and breeding of livestock and poultry[J]. Chinese Journal of Animal Science, 2021, 57(7): 37-42. (in Chinese) |
| [35] |
赵玉强, 陈鑫, 李清春, 等. 大白猪ENO3基因多态性及其与生长性状的关联分析[J]. 中国畜牧兽医, 2022, 49(1): 188-196. ZHAO Y Q, CHEN X, LI Q C, et al. Polymorphism of ENO3 gene and its correlation with growth traits in Yorkshire pigs[J]. China Animal Husbandry & Veterinary Medicine, 2022, 49(1): 188-196. (in Chinese) |
| [36] |
CENDRON F, MASTRANGELO S, TOLONE M, et al. Genome-wide analysis reveals the patterns of genetic diversity and population structure of 8 Italian local chicken breeds[J]. Poult Sci, 2021, 100(2): 441-451. |
| [37] |
CHEN A Y A, HUANG C W, LIU S H, et al. Single nucleotide polymorphisms of immunity-related genes and their effects on immunophenotypes in different pig breeds[J]. Genes (Basel), 2021, 12(9): 1377. |
| [38] |
SATO S, UEMOTO Y, KIKUCHI T, et al. SNP- and haplotype-based genome-wide association studies for growth, carcass, and meat quality traits in a Duroc multigenerational population[J]. BMC Genet, 2016, 17: 60. |
| [39] |
MEYERMANS R, GORSSEN W, WIJNROCX K, et al. Unraveling the genetic diversity of Belgian milk sheep using medium-density SNP genotypes[J]. Anim Genet, 2020, 51(2): 258-265. |
| [40] |
BERIHULAY H, LI Y, LIU X, et al. Genetic diversity and population structure in multiple Chinese goat populations using a SNP panel[J]. Anim Genet, 2019, 50(3): 242-249. |
| [41] |
MOOSANEZHAD KHABISI M, ASADI FOOZI M, LV F H, et al. Genome-wide DNA arrays profiling unravels the genetic structure of Iranian sheep and pattern of admixture with worldwide coarse-wool sheep breeds[J]. Genomics, 2021, 113(6): 3501-3511. |
| [42] |
赵旭庭, 徐盼, 马政, 等. 枫泾猪、梅山猪和沙乌头猪全基因组ROH检测及候选基因鉴定[J]. 江苏农业学报, 2021, 37(5): 1234-1243. ZHAO X T, XU P, MA Z, et al. Genome-wide scan for run of homozygosity and identification of corre-sponding candidate genes in Fengjing pigs, Meishan pigs and Shawutou pigs[J]. Jiangsu Journal of Agricultural Sciences, 2021, 37(5): 1234-1243. (in Chinese) |
| [43] |
陈佐权, 饶琳, 谢磊, 等. 杜洛克与二花脸杂交猪群体SNP偏分离分析[J]. 畜牧兽医学报, 2021, 52(9): 2384-2393. CHEN Z Q, RAO L, XIE L, et al. Transmission ratio distortion analysis of SNP in a Duroc×Erhualian intercross pigs population[J]. Acta Veterinaria et Zootechnica Sinica, 2021, 52(9): 2384-2393. (in Chinese) |
| [44] |
吴行雕, 石道宏, 黄世会, 等. 皱皮香猪HAS2基因SNPs的鉴定及生物信息学分析[J]. 畜牧兽医学报, 2021, 52(12): 3390-3402. WU X D, SHI D H, HUANG S H, et al. SNPs identification and bioinformatics analysis of the hyaluronan synthase 2 gene in Xiang Pig with wrinkled skin[J]. Acta Veterinaria et Zootechnica Sinica, 2021, 52(12): 3390-3402. (in Chinese) |
| [45] |
蔡春波, 张雪莲, 张万峰, 等. 运用SNP芯片评估马身猪保种群体的遗传结构[J]. 畜牧兽医学报, 2021, 52(4): 920-931. CAI C B, ZHANG X L, ZHANG W F, et al. Evaluation of genetic structure in Mashen pigs conserved population based on SNP chip[J]. Acta Veterinaria et Zootechnica Sinica, 2021, 52(4): 920-931. (in Chinese) |
(编辑 郭云雁)



