猪在体型、生理和基因组特征上与人类相似,被认为是人类疾病研究的最佳动物模型[1]。作为主要的产肉家畜,商品猪的产仔数一直以来都是重要的经济指标,而产仔数的多少与胚胎是否能发育正常有着重要的联系。猪的胚胎发育是一个复杂的生理过程,需要经历一系列重要的生理阶段[2-3]以及自发胎儿丢失[4]、宫内生长受限[5]等严峻的考验后才能幸运地发育成新生儿,这导致了猪胚胎产前死亡率高达20%~45%[6]。虽然在胚胎发育过程中基因的严格表达与正确指导是胚胎能否正常发育的决定性条件,但是近年来的研究发现,表观遗传调控对胚胎的发育也起着必不可少的作用。
表观遗传学是指细胞核DNA序列没有改变的情况下,个体全基因组功能发生可遗传和可逆的改变。研究表明,表观遗传修饰并不局限于生物体的特定生命阶段,而是贯穿整个生命周期[7]。然而,它们更常见于胚胎发育和细胞增殖阶段[8]。在生殖细胞和早期胚胎中,表观遗传重编程发生在全基因组范围内[9]。通过这些表观遗传机制,细胞整合环境刺激来协调基因转录,基因表达等生理过程,对猪的胚胎发育至关重要。本文总结了目前研究得最成熟的表观遗传修饰——DNA甲基化,在猪胚胎发育过程中的研究进展,为今后进一步了解和研究DNA甲基化及其与其他表观遗传修饰的串扰在胚胎发育的作用提供借鉴和参考。
1 猪胚胎在妊娠期间的发育过程受精卵的形成标志着哺乳动物胚胎发育的开始。在最初的4 d里,猪的胚胎独立于母体环境进行卵裂分裂,储存在卵母细胞中的编码细胞周期和核酸合成调节因子的mRNA和蛋白质维持着这一早期胚胎发育。但大约在第3天(4细胞期)时,胚胎基因组开始激活,随后自主指导自身的发育,直到囊胚形成[10-11]。
大约在受精后第7天,胚胎从透明带中孵化出来,随后继续增殖发育,第10天时直径扩大到2~6 mm[11-12]。从妊娠第10天开始,胚胎的形态开始发生剧烈变化,从球状囊胚首先转变成管状胚胎,然后在妊娠第12天左右发育成丝状(100~150 mm),并进一步伸长,在妊娠第16天左右达到了1 000 mm[3, 13]。伸长后的胚胎能显著增加与子宫内膜的接触面积,从而为胚胎后续的附植提供了条件。
妊娠第11~13天是妊娠识别的关键时期,此阶段胚胎开始分泌能启动胚胎-母体识别的信号——雌二醇[14]来完成识别过程。成功进行妊娠识别后,胚胎开始进入附植窗口期,并在妊娠第18天左右完成附植过程。此时胚胎滋养层上皮与子宫腔上皮相互黏附,胎盘开始建立[3]。大约在妊娠28 d,胎盘完全形成,标志着猪的妊娠早期阶段结束。
在妊娠中期时,胎盘基质层的厚度显著增加,并且形成褶皱结构。褶皱的形成大大增加了胎盘与母体子宫的接触面积,从而使营养物质的运输效率显著提高,为胎儿的进一步发育提供了条件[15]。进入妊娠中期后,胎儿中已分化的器官继续发育完善,体重在妊娠后期迅速增加,并在分娩前体重达到最高水平。猪的胚胎发育过程中的其他重要事件总结如图 1所示。
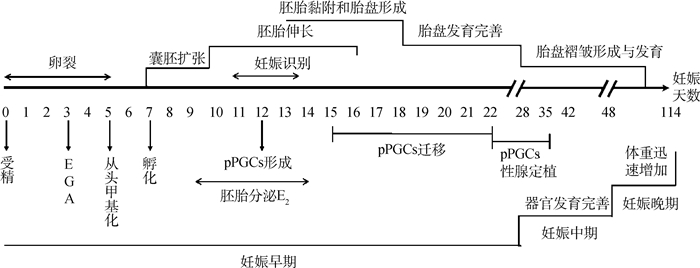
|
EGA. 胚胎基因组激活;pPGCs. 猪原始生殖细胞 EGA. Embryonic genome activation; pPGCs. Porcine primordial germ cells 图 1 猪胚胎发育过程中重要事件 Fig. 1 The crucial events during porcine embryonic development |
DNA甲基化是一种常见且重要的表观遗传修饰,虽然不改变DNA的一级序列,但也包含可遗传信息,并在基因的转录调控中起重要作用。3种不同的DNA甲基转移酶(DNA methyltransferase, DNMT)DNMT1、DNMT3a和DNMT3b催化DNA甲基化,其中DNMT1是主要的维持甲基化酶,DNMT3a和DNMT3b则负责从头甲基化[16-17]。而近年来发现DNMT1在卵母细胞中也可以介导从头甲基化的过程[18]。当甲基以时间和空间的精度被添加到除位于CpG岛上的CpG二核苷酸中的5′-胞嘧啶残基位置时,部分生物学过程会随之被改变[19],例如基因转录[20]、基因组印记[21]、组织分化[22]和表型变异[23]。此外,在非CpG位点也发现了DNA甲基化,这被称为非CpG甲基化,可由DNMT3a和DNMT3b介导[24-25]。目前已经在人类胚胎干细胞中发现了非CpG甲基化的存在[26-27]。
2.2 猪早期胚胎发育中DNA甲基化的动态过程DNA甲基化在分化的体细胞中相对稳定,但在哺乳动物的早期胚胎发生中是高度动态的。在受精过程中,高度甲基化的精子(内含父本基因组)经历了快速的主动去甲基化过程,这一过程主要由tet甲基胞嘧啶双加氧酶(tet methylcytosine dioxygenase, TET)来介导[28-29]。由于DNMT1不能进入细胞核,因此随着胚胎继续发生卵裂,其去甲基化继续以被动的形式发生[30],并在囊胚中达到整个胚胎发育过程的最低水平[31]。从头甲基化(de novo methylation)是早期胚胎发育的主要DNA甲基化模式,在DNMT3a和DNMT3b的介导下从猪的早期囊胚(受精后第5天)开始,并在附植期间建立[32-33]。有报道还观察到了在第7~8天的囊胚中出现系谱特异性的DNA甲基化,其中甲基化的主要部位是内细胞团,其次是滋养外胚层[34]。胚胎在附植前发生的第一次DNA甲基化重编程(DNA methylation reprogramming,DMR)消除了配子发生过程中所积累的与分化相关且不必要的表观遗传信息,从而使胚胎获得了能独立发育成一个新个体的多能性[35]。与小鼠和人一样,猪在pPGCs分化形成后不久也会发生第二次DMR,并且这一过程所持续的时间较长[36]。在从后肠迁移到性腺脊期间(妊娠约15~22天),高度甲基化的pPGCs开始去甲基化,并在第20天左右达到较低水平,随后开始重新升高,在28天时重新达到一个较高水平[37]。但在28天后,PGC中的甲基化程度重新降低,并在妊娠中期的第36天左右达到第二个低水平,随后继续升高,在第42天时恢复至第28天左右的水平[38](图 2)。随着上一代建立的DNA甲基化印记的消除,DNA甲基化模式在胚胎发生过程中通过DNMT3a和非催化旁系同源物DNMT3L在印记基因座和转座因子上重新建立[16, 39]。重编程的去甲基化过程涉及5-甲基胞嘧啶(5-methylcytosine,5 mC)的修饰和DNA修复。在这一过程中,5 mC修饰可以通过不同的机制(例如TET1介导的羟基化)来介导,从而启动主动去甲基化过程[40-41]。但与第一次全基因组DMR不同,pPGCs中的DMR只是局部基因组的去甲基化和擦除,并根据胎儿的性别形成了印记[42-43]。有研究发现,在妊娠第30和第45天猪胚胎的DNMT1(P < 0.01)、DNMT3a和DNMT3b(P < 0.05)表达均高于成年组织[44],这不仅与pPGCs中甲基化程度的动态变化相适应,并且与前人研究的DNMT1功能以及发现小鼠DNMT3a和DNMT3b在未分化的胚胎干细胞中高表达的事实相一致[45-46],表明猪胚胎内DNA甲基化的动态过程与其他哺乳动物之间是具有广谱性的。
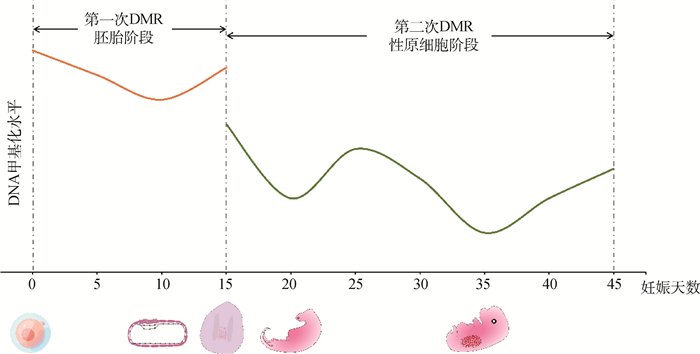
|
图 2 猪胚胎发育过程中DNA甲基化的动态变化 Fig. 2 Dynamic changes in DNA methylation during porcine embryonic development |
N6-甲基脱氧腺嘌呤(N6-methyldeoxyadenosine, 6 mA)是一个在原核生物中研究得较多的DNA甲基化标记,主要被原核生物用来保护自身基因组免受外来DNA的入侵[47]。由于其丰度极低和技术限制的原因,过去人们难以检测到真核生物DNA中的6 mA,对真核生物中的6 mA的关注度一直以来都十分有限[48]。随着检测技术的提高,6 mA修饰不仅在果蝇[49]、斑马鱼[50-51]的胚胎发生过程中被检测到,而且在哺乳动物如小鼠[51]和猪[50]的胚胎发育过程中呈现出动态变化,并被发现在哺乳动物胚胎干细胞发育和分化过程中发挥关键作用[52-53]。在猪早期胚胎发生过程,特别是处于全基因组DNA去甲基化时期的4-细胞期和桑葚胚阶段也积累到了相对较高的丰度水平(6 mA/A的比例达到了0.1%~0.2%)[50],此阶段对应猪胚胎发育的EGA事件。随后6 mA的丰度迅速降低,这与He等[49]发现的6 mA在果蝇EGA期间丰度的变化趋势基本一致,因此认为,6 mA的正常修饰可能在猪早期EGA事件中的表观遗传重编程过程中起着重要作用,其调控机制或许与果蝇基本相同。有人提出DNA中的6 mA修饰具有调节DNA结构和转录以及跨代传递信息等表观遗传调控功能,并且有可能在植入前胚胎和pPGCs中起到补充5 mC的抑制作用[54-55]。但在猪胚胎发育过程中6 mA是否也具有这些功能,需要更多的研究来验证。
2.3 DNA甲基化对猪SCNT的胚胎发育影响近年来,随着全球首例猪心移植和猪肾移植的成功,猪作为人体器官移植潜在的主要提供者受到越来越多的关注。异种器官移植的前提是需要构建适当的基因修饰猪,而体细胞核移植(somatic cell nuclear transfer, SCNT)一直以来都是构建基因修饰猪的重要方法,能将分化细胞快速重编程为全能胚胎,在养猪业、人类生物医学和生命科学中具有重要的应用价值[56-57]。然而,只有约1%的成功率(出生克隆仔猪数量/移植克隆胚胎数量)使得SCNT的进一步应用受阻,这与早期猪SCNT胚胎中全基因组DNA甲基化的动态模式异常有着密不可分的联系[58-59]。
提高SCNT的成功率的办法之一是把供体细胞DNA甲基化水平降低到全能胚胎细胞的水平。因此,抑制供体细胞中DNA甲基化酶的活性是提高SCNT胚胎发育成功率的一个重要方法。研究发现,供体细胞中的Dnmt1 s(Dnmt1的一种体细胞形式亚型)是SCNT介导的DNA甲基化重编程的障碍,损害了克隆胚胎的发育[60]。当Dnmt1 s被敲除后,其自身和克隆胚胎基因组的甲基化被重建,与多能性有关的基因的表达也得到了显著提高[60]。提高DNA去甲基化酶的活性也可以达到降低DNA甲基化水平的效果。例如克隆胚胎中过表达甲基-CpG结合结构域蛋白(methyl-CpG-binding domain proteins, MBD)中的MBD3(一种能触发DNA去甲基化的酶)能显著改善克隆胚胎的囊胚率和每个囊胚的细胞数以及降低与多能性有关的基因启动子的DNA甲基化水平[61]。此外,供体细胞的类型和表观遗传修饰模式也会显著影响SCNT胚胎的发育能力。例如猪骨髓间充质干细胞由于只有部分分化,因此相比于猪胚胎成纤维细胞(porcine embryonic fibroblast, pEF)更容易重新编程为多能状态[62]。Zhai等[63]发现,来自猪骨髓间充质干细胞的SCNT胚胎比来自pEF的SCNT胚胎拥有更多的活跃表观遗传标记和较少的抑制性表观遗传标记,其NANOG和POU5F1(与胚胎多能性有关的两个基因)的启动子区域DNA甲基化水平均低于pEF的SCNT胚胎,且染色质状态更活跃,因此拥有更高的卵裂率和囊胚率。
SCNT技术的成功率难以提高也与印记基因的表达异常有关。猪SCNT胚胎发育异常和印记基因H19和IGF2的甲基化异常有着密不可分的联系,因此,一些能挽救H19和IGF2甲基化异常状态的物质(例如Trichostatin A(TSA)[64]、RG108和scriptaid[65])或维持H19和IGF2甲基化正常的基因(例如tripartite motif containing 28(TRIM28)[66]) 都能间接影响SCNT的胚胎发育状态。此外,Yu等[67]对猪的诱导多能干细胞(induced pluripotent stem cells, iPSCs)和相关的克隆胚胎进行了甲基化组和转录组分析,发现印记基因的异常沉默,特别是反转录转座子衍生的印记基因retrotransposon Gag like 1(RTL1)的沉默,是导致妊娠失败的主要原因之一。若恢复RTL1在iPSCs中的表达可以挽救胎儿的丢失。因此, RTL1在动物克隆中或许是反映供体细胞印记状态的一个很好的标记物。但在猪中,目前只有少数印记基因的调控机制被研究过,而胚胎发育过程可能受一个复杂而协调的印记基因网络的调节。因此,深入研究印记基因的具体分子机制,揭示这一调控网络对提高SCNT技术的成功率来说具有重要意义。
其他表观遗传修饰类型也能影响供体细胞的DNA甲基化水平,例如miR-148A被证明可以通过靶向抑制pEF的DNMT1表达,从而降低SCNT胚胎的DNA甲基化整体水平,增强SCNT胚胎的发育潜力[68]。一些组蛋白甲基化抑制剂也被发现不但可以改变组蛋白的甲基化程度,而且可以抑制供体细胞的DNA甲基化水平,从而提高SCNT的成活率和囊胚率,这也提示可以联合调节不同表观遗传修饰类型(例如同时降低DNA甲基化和组蛋白甲基化水平)来进一步提高SCNT胚胎的克隆成活率。近年来发现的能够改善SCNT胚胎成活率的物质如表 1所示。
|
|
表 1 影响 SCNT 胚胎 DNA 甲基化水平的物质 Table 1 Substances that can affect the level of DNA methylation in SCNT embryos |
孕期母体营养可通过改变后代的表观遗传修饰影响后代的基因表达。在妊娠早期营养摄入不足或过量都有可能会影响子宫内膜或胎儿的DNA甲基化程度,从而影响胎儿的正常发育[74-75]。例如,Altmann等[76]发现,母猪摄入蛋白过高或过低会导致妊娠第95天的胎儿肝中DNMT1和DNMT3b的基因表达变低,并且导致全基因组甲基化水平降低。而肌肉组织则不太相同。相比于对照组来说,高蛋白日粮组的胎儿的DNMT1和DNMT3a的表达显著增加,低蛋白日粮的胎儿中DNMT3a表达显著降低[76]。此外,如果在母猪的整个妊娠期间饲喂添加了与DNA甲基化有关的微量元素叶酸、维生素B6和B12、蛋氨酸、胆碱和锌的日粮会使雄性胎儿在妊娠后期体重增加,但出生后体重下降,并伴随着由肌源性关键驱动因子(Pax7、MyoD1、mygenin)组成的代偿性转录反应,但在雌性胎儿中却没有这一现象[77]。这说明母体营养的改变在胎儿不同的组织和性别内所造成的DNA甲基化的改变程度是不尽相同的,而其具体的发生机制或许也有所差异。
一份基因表达微阵列测序结果表明,在短而独特的围孕期(妊娠1~9 d)对母猪进行限饲(比正常饲喂量减少30%)会改变围附植期胚胎中大量基因的表达(496个基因的表达上调,291个基因的表达下调),其中就包括了DNMT1表达的显著降低[78]。进一步研究表明,这一期间的限饲可以使胚胎在围附植期中DNMT1的甲基化水平以及TRIM28和ZFP57的mRNAs和蛋白总丰度升高[75, 79]。这3个基因都是DNA甲基化维持的重要基因,说明母猪围孕期限饲会对围附植期猪胚胎的DNA甲基化水平产生较大的影响。相类似地,围孕期限饲也能影响母猪子宫内膜中与DNA甲基化有关的基因的表达。目前已发现限饲组中的子宫内膜DNMT1、DNMT3a、TRIM28和ZFP57的表达水平均显著降低[80]。这说明围孕期的营养不良有可能影响了子宫内膜的DNA从头甲基化和甲基化维持过程,并有可能对胎儿的生长发育产生影响。这些研究提示,应做好母猪妊娠期间的营养保育工作。
此外,有研究发现,对受体母猪进行营养管理也是提高克隆猪胚胎发育率的有效途径。若在妊娠14~75天日粮添加L-精氨酸,能提高移植克隆猪胚胎受体母猪血浆中精氨酸和精氨酸代谢产物(包括一氧化氮、亚精胺和腐胺)的浓度,从而提高受体母猪的妊娠率,并显著提高了克隆仔猪出生总数占移植克隆猪胚胎总数的比例[81]。这也提示了提高克隆猪的囊胚率和生存率可以通过改善受体母猪的营养管理方式这一途径。
3 总结与展望在猪的胚胎发育中,DNA甲基化的动态修饰起着必不可少的调节作用。若DNA甲基化发生异常,可能会造成胚胎发育受阻甚至是畸形或死亡,而这也是SCNT和体外生殖胚胎成活率低的主要原因之一。此外,对于孕期母体的营养状况调控也会通过DNA甲基化对胚胎造成影响。因此,对DNA甲基化进行深入研究,了解早期猪胚胎中DNA甲基化的异常全基因组动态模式,能为提高猪胚胎的成活率提供借鉴。此外,不同类型的表观遗传调控,如DNA甲基化、组蛋白修饰等已有较好的独立研究,然而不同表观遗传修饰之间更表现出相互作用,从而构成调控网络的可能。目前,DNA甲基化和组蛋白修饰途径已被发现可以相互发生串扰[82]。因此,在未来深入了解猪胚胎在发育过程中DNA甲基化与其他表观遗传修饰途径的相互作用机制对于提高猪的产仔数和在人类生物医学以及生命科学中的应用价值仍是非常重要的。
| [1] |
PRATHER R S, LORSON M, ROSS J W, et al. Genetically engineered pig models for human diseases[J]. Annu Rev Anim Biosci, 2013, 1: 203-219. DOI:10.1146/annurev-animal-031412-103715 |
| [2] |
BICK J T, FLÖTER V L, ROBINSON M D, et al. Small RNA-seq analysis of single porcine blastocysts revealed that maternal estradiol-17beta exposure does not affect miRNA isoform (isomiR) expression[J]. BMC Genomics, 2018, 19(1): 590. DOI:10.1186/s12864-018-4954-9 |
| [3] |
BAZER F W, JOHNSON G A. Pig blastocyst-uterine interactions[J]. Differentiation, 2014, 87(1-2): 52-65. DOI:10.1016/j.diff.2013.11.005 |
| [4] |
BIDARIMATH M, TAYADE C. Pregnancy and spontaneous fetal loss: A pig perspective[J]. Mol Reprod Dev, 2017, 84(9): 856-869. DOI:10.1002/mrd.22847 |
| [5] |
WU G, BAZER F W, WALLACE J M, et al. BOARD-INVITED REVIEW: Intrauterine growth retardation: implications for the animal sciences[J]. J Anim Sci, 2006, 84(9): 2316-2337. DOI:10.2527/jas.2006-156 |
| [6] |
POPE W F. Uterine asynchrony: A cause of embryonic loss[J]. Biol Reprod, 1988, 39(5): 999-1003. DOI:10.1095/biolreprod39.5.999 |
| [7] |
EGGER G, LIANG G N, APARICIO A, et al. Epigenetics in human disease and prospects for epigenetic therapy[J]. Nature, 2004, 429(6990): 457-463. DOI:10.1038/nature02625 |
| [8] |
JAENISCH R, BIRD A. Epigenetic regulation of gene expression: How the genome integrates intrinsic and environmental signals[J]. Nat Genet, 2003, 33 Suppl: 245-254. |
| [9] |
FENG S H, JACOBSEN S E, REIK W. Epigenetic reprogramming in plant and animal development[J]. Science, 2010, 330(6004): 622-627. DOI:10.1126/science.1190614 |
| [10] |
ALBERIO R. Regulation of cell fate decisions in early mammalian embryos[J]. Annu Rev Anim Biosci, 2020, 8: 377-393. DOI:10.1146/annurev-animal-021419-083841 |
| [11] |
ARRELL V L, DAY B N, PRATHER R S. The transition from maternal to zygotic control of development occurs during the 4-cell stage in the domestic pig, Sus scrofa: quantitative and qualitative aspects of protein synthesis[J]. Biol Reprod, 1991, 44(1): 62-68. DOI:10.1095/biolreprod44.1.62 |
| [12] |
BAZER F W, SPENCER T E, JOHNSON G A, et al. Uterine receptivity to implantation of blastocysts in mammals[J]. Front Biosci (Schol Ed), 2011, 3(2): 745-767. |
| [13] |
TAYADE C, BLACK G P, FANG Y, et al. Differential gene expression in endometrium, endometrial lymphocytes, and trophoblasts during successful and abortive embryo implantation[J]. J Immunol, 2006, 176(1): 148-156. DOI:10.4049/jimmunol.176.1.148 |
| [14] |
KACZMAREK M M, NAJMULA J, GUZEWSKA M M, et al. miRNAs in the peri-implantation period: Contribution to embryo-maternal communication in pigs[J]. Int J Mol Sci, 2020, 21(6): 2229. DOI:10.3390/ijms21062229 |
| [15] |
胡群, 叶南, 史泽宇, 等. 猪妊娠过程中胎盘发育及其调控基因研究进展[J]. 中国畜牧兽医, 2018, 45(6): 1633-1638. DOI:10.16431/j.cnki.1671-7236.2018.06.026 |
| [16] |
LAW J A, JACOBSEN S E. Establishing, maintaining and modifying DNA methylation patterns in plants and animals[J]. Nat Rev Genet, 2010, 11(3): 204-220. DOI:10.1038/nrg2719 |
| [17] |
BESTOR T H. The DNA methyltransferases of mammals[J]. Hum Mol Genet, 2000, 9(16): 2395-2402. DOI:10.1093/hmg/9.16.2395 |
| [18] |
LI Y, ZHANG Z, CHEN J, et al. Stella safeguards the oocyte methylome by preventing de novo methylation mediated by DNMT1[J]. Nature, 2018, 564(7734): 136-140. DOI:10.1038/s41586-018-0751-5 |
| [19] |
SCHNEIDER E, PLIUSHCH G, EL HAJJ N, et al. Spatial, temporal and interindividual epigenetic variation of functionally important DNA methylation patterns[J]. Nucleic Acids Res, 2010, 38(12): 3880-3890. DOI:10.1093/nar/gkq126 |
| [20] |
MEDVEDEVA Y A, KHAMIS A M, KULAKOVSKIY I V, et al. Effects of cytosine methylation on transcription factor binding sites[J]. BMC Genomics, 2014, 15: 119. DOI:10.1186/1471-2164-15-119 |
| [21] |
DURCOVA-HILLS G, HAJKOVA P, SULLIVAN S, et al. Influence of sex chromosome constitution on the genomic imprinting of germ cells[J]. Proc Natl Acad Sci U S A, 2006, 103(30): 11184-11188. DOI:10.1073/pnas.0602621103 |
| [22] |
VARLEY K E, GERTZ J, BOWLING K M, et al. Dynamic DNA methylation across diverse human cell lines and tissues[J]. Genome Res, 2013, 23(3): 555-567. DOI:10.1101/gr.147942.112 |
| [23] |
LI M Z, WU H L, LUO Z G, et al. An atlas of DNA methylomes in porcine adipose and muscle tissues[J]. Nat Commun, 2012, 3: 850. DOI:10.1038/ncomms1854 |
| [24] |
GOWHER H, JELTSCH A. Enzymatic properties of recombinant Dnmt3a DNA methyltransferase from mouse: the enzyme modifies DNA in a non-processive manner and also methylates non-CpA sites[J]. J Mol Biol, 2001, 309(5): 1201-1208. DOI:10.1006/jmbi.2001.4710 |
| [25] |
RAMSAHOYE B H, BINISZKIEWICZ D, LYKO F, et al. Non-CpG methylation is prevalent in embryonic stem cells and may be mediated by DNA methyltransferase 3a[J]. Proc Natl Acad Sci U S A, 2000, 97(10): 5237-5242. DOI:10.1073/pnas.97.10.5237 |
| [26] |
LAURENT L, WONG E, LI G L, et al. Dynamic changes in the human methylome during differentiation[J]. Genome Res, 2010, 20(3): 320-331. DOI:10.1101/gr.101907.109 |
| [27] |
LISTER R, PELIZZOLA M, DOWEN R H, et al. Human DNA methylomes at base resolution show widespread epigenomic differences[J]. Nature, 2009, 462(7271): 315-322. DOI:10.1038/nature08514 |
| [28] |
SANTOS F, HENDRICH B, REIK W, et al. Dynamic reprogramming of DNA methylation in the early mouse embryo[J]. Dev Biol, 2002, 241(1): 172-182. DOI:10.1006/dbio.2001.0501 |
| [29] |
LEE H J, HORE T A, REIK W. Reprogramming the methylome: Erasing memory and creating diversity[J]. Cell Stem Cell, 2014, 14(6): 710-719. DOI:10.1016/j.stem.2014.05.008 |
| [30] |
LI E. Chromatin modification and epigenetic reprogramming in mammalian development[J]. Nat Rev Genet, 2002, 3(9): 662-673. DOI:10.1038/nrg887 |
| [31] |
SMITH Z D, CHAN M M, MIKKELSEN T S, et al. A unique regulatory phase of DNA methylation in the early mammalian embryo[J]. Nature, 2012, 484(7394): 339-344. DOI:10.1038/nature10960 |
| [32] |
KAFRI T, ARIEL M, BRANDEIS M, et al. Developmental pattern of gene-specific DNA methylation in the mouse embryo and germ line[J]. Genes Dev, 1992, 6(5): 705-714. DOI:10.1101/gad.6.5.705 |
| [33] |
MONK M, BOUBELIK M, LEHNERT S. Temporal and regional changes in DNA methylation in the embryonic, extraembryonic and germ cell lineages during mouse embryo development[J]. Development, 1987, 99(3): 371-382. DOI:10.1242/dev.99.3.371 |
| [34] |
FULKA J, FULKA H, SLAVIK T, et al. DNA methylation pattern in pig in vivo produced embryos[J]. Histochem Cell Biol, 2006, 126(2): 213-217. DOI:10.1007/s00418-006-0153-x |
| [35] |
JEONG Y S, YEO S, PARK J S, et al. DNA methylation state is preserved in the sperm-derived pronucleus of the pig zygote[J]. Int J Dev Biol, 2007, 51(8): 707-714. DOI:10.1387/ijdb.072450yj |
| [36] |
ZHU Q F, SANG F, WITHEY S, et al. Specification and epigenomic resetting of the pig germline exhibit conservation with the human lineage[J]. Cell Rep, 2021, 34(6): 108735. DOI:10.1016/j.celrep.2021.108735 |
| [37] |
HYLDIG S M W, OSTRUP O, VEJLSTED M, et al. Changes of DNA methylation level and spatial arrangement of primordial germ cells in embryonic day 15 to embryonic day 28 pig embryos[J]. Biol Reprod, 2011, 84(6): 1087-1093. DOI:10.1095/biolreprod.110.086082 |
| [38] |
GÓMEZ-REDONDO I, PLANELLS B, CÁNOVAS S, et al. Genome-wide DNA methylation dynamics during epigenetic reprogramming in the porcine germline[J]. Clin Epigenet, 2021, 13(1): 27. DOI:10.1186/s13148-021-01003-x |
| [39] |
REIK W. Stability and flexibility of epigenetic gene regulation in mammalian development[J]. Nature, 2007, 447(7143): 425-432. DOI:10.1038/nature05918 |
| [40] |
HAJKOVA P, JEFFRIES S J, LEE C, et al. Genome-wide reprogramming in the mouse germ line entails the base excision repair pathway[J]. Science, 2010, 329(5987): 78-82. DOI:10.1126/science.1187945 |
| [41] |
MORGAN H D, DEAN W, COKER H A, et al. Activation-induced cytidine deaminase deaminates 5-methylcytosine in DNA and is expressed in pluripotent tissues: implications for epigenetic reprogramming[J]. J Biol Chem, 2004, 279(50): 52353-52360. DOI:10.1074/jbc.M407695200 |
| [42] |
POPP C, DEAN W, FENG S H, et al. Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency[J]. Nature, 2010, 463(7284): 1101-1105. DOI:10.1038/nature08829 |
| [43] |
GUIBERT S, FORNÉ T, WEBER M. Global profiling of DNA methylation erasure in mouse primordial germ cells[J]. Genome Res, 2012, 22(4): 633-641. DOI:10.1101/gr.130997.111 |
| [44] |
LUO Z G, ZHANG K, CHEN L, et al. Molecular characterization and tissue expression profile of the Dnmts gene family in pig[J]. J Integr Agric, 2017, 16(6): 1367-1374. DOI:10.1016/S2095-3119(16)61512-5 |
| [45] |
SHARIF J, MUTO M, TAKEBAYASHI S I, et al. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA[J]. Nature, 2007, 450(7171): 908-912. DOI:10.1038/nature06397 |
| [46] |
OKANO M, XIE S P, LI E. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases[J]. Nat Genet, 1998, 19(3): 219-220. DOI:10.1038/890 |
| [47] |
WION D, CASADESÚS J. N6-methyl-adenine: An epigenetic signal for DNA-protein interactions[J]. Nat Rev Microbiol, 2006, 4(3): 183-192. DOI:10.1038/nrmicro1350 |
| [48] |
RATEL D, RAVANAT J L, BERGER F, et al. N6-methyladenine: the other methylated base of DNA[J]. Bioessays, 2006, 28(3): 309-315. DOI:10.1002/bies.20342 |
| [49] |
HE S M, ZHANG G Q, WANG J J, et al. 6 mA-DNA-binding factor Jumu controls maternal-to-zygotic transition upstream of Zelda[J]. Nat Commun, 2019, 10(1): 2219. DOI:10.1038/s41467-019-10202-3 |
| [50] |
LIU J Z, ZHU Y X, LUO G Z, et al. Abundant DNA 6 mA methylation during early embryogenesis of zebrafish and pig[J]. Nat Commun, 2016, 7: 13052. DOI:10.1038/ncomms13052 |
| [51] |
FERNANDES S B, GROVA N, ROTH S, et al. N6-methyladenine in eukaryotic DNA: Tissue distribution, early embryo development, and neuronal toxicity[J]. Front Genet, 2021, 12: 657171. DOI:10.3389/fgene.2021.657171 |
| [52] |
LI Z, ZHAO S, NELAKANTI R V, et al. N6-methyladenine in DNA antagonizes SATB1 in early development[J]. Nature, 2020, 583(7817): 625-630. DOI:10.1038/s41586-020-2500-9 |
| [53] |
WU T P, WANG T, SEETIN M G, et al. DNA methylation on N6-adenine in mammalian embryonic stem cells[J]. Nature, 2016, 532(7599): 329-333. DOI:10.1038/nature17640 |
| [54] |
BOULIAS K, GREER E L. Means, mechanisms and consequences of adenine methylation in DNA[J]. Nat Rev Genet, 2022, 23(7): 411-428. DOI:10.1038/s41576-022-00456-x |
| [55] |
ZHU Q F, STÖGER R, ALBERIO R. A Lexicon of DNA modifications: Their roles in embryo development and the germline[J]. Front Cell Dev Biol, 2018, 6: 24. DOI:10.3389/fcell.2018.00024 |
| [56] |
ZHAO J G, WHYTE J, PRATHER R S. Effect of epigenetic regulation during swine embryogenesis and on cloning by nuclear transfer[J]. Cell Tissue Res, 2010, 341(1): 13-21. DOI:10.1007/s00441-010-1000-x |
| [57] |
PRATHER R S, SHEN M D, DAI Y F. Genetically modified pigs for medicine and agriculture[J]. Biotechnol Genet Eng Rev, 2008, 25: 245-265. |
| [58] |
LIU Y, LI J, LØVENDAHL P, et al. In vitro manipulation techniques of porcine embryos: A meta-analysis related to transfers, pregnancies and piglets[J]. Reprod Fertil Dev, 2015, 27(3): 429-439. DOI:10.1071/RD13329 |
| [59] |
DESHMUKH R S, ØSTRUP O, ØSTRUP E, et al. DNA methylation in porcine preimplantation embryos developed in vivo and produced by in vitro fertilization, parthenogenetic activation and somatic cell nuclear transfer[J]. Epigenetics, 2011, 6(2): 177-187. DOI:10.4161/epi.6.2.13519 |
| [60] |
SONG X X, LIU Z H, HE H B, et al. Dnmt1s in donor cells is a barrier to SCNT-mediated DNA methylation reprogramming in pigs[J]. Oncotarget, 2017, 8(21): 34980-34991. DOI:10.18632/oncotarget.16507 |
| [61] |
WANG X W, SHI J S, CAI G Y, et al. Overexpression of MBD3 improves reprogramming of cloned pig embryos[J]. Cell Reprogram, 2019, 21(5): 221-228. DOI:10.1089/cell.2019.0008 |
| [62] |
LI Z C, HE X Y, CHEN L W, et al. Bone marrow mesenchymal stem cells are an attractive donor cell type for production of cloned pigs as well as genetically modified cloned pigs by somatic cell nuclear transfer[J]. Cell Reprogram, 2013, 15(5): 459-470. DOI:10.1089/cell.2013.0010 |
| [63] |
ZHAI Y H, LI W, ZHANG Z R, et al. Epigenetic states of donor cells significantly affect the development of somatic cell nuclear transfer (SCNT) embryos in pigs[J]. Mol Reprod Dev, 2018, 85(1): 26-37. DOI:10.1002/mrd.22935 |
| [64] |
HUAN Y J, ZHU J, HUANG B, et al. Trichostatin A rescues the disrupted imprinting induced by somatic cell nuclear transfer in pigs[J]. PLoS One, 2015, 10(5): e0126607. DOI:10.1371/journal.pone.0126607 |
| [65] |
XU W H, LI Z C, YU B, et al. Effects of DNMT1 and HDAC inhibitors on gene-specific methylation reprogramming during porcine somatic cell nuclear transfer[J]. PLoS One, 2013, 8(5): e64705. DOI:10.1371/journal.pone.0064705 |
| [66] |
ZHAI Y H, ZHANG M, AN X L, et al. TRIM28 maintains genome imprints and regulates development of porcine SCNT embryos[J]. Reproduction, 2021, 161(4): 411-424. DOI:10.1530/REP-20-0602 |
| [67] |
YU D W, WANG J, ZOU H Y, et al. Silencing of retrotransposon-derived imprinted gene RTL1 is the main cause for postimplantational failures in mammalian cloning[J]. Proc Natl Acad Sci U S A, 2018, 115(47): E11071-E11080. |
| [68] |
WANG P, LI X P, CAO L H, et al. MicroRNA-148a overexpression improves the early development of porcine somatic cell nuclear transfer embryos[J]. PLoS One, 2017, 12(6): e0180535. DOI:10.1371/journal.pone.0180535 |
| [69] |
QU J D, WANG X Y, JIANG Y J, et al. Optimizing 5-aza-2'-deoxycytidine treatment to enhance the development of porcine cloned embryos by inhibiting apoptosis and improving DNA methylation reprogramming[J]. Res Vet Sci, 2020, 132: 229-236. DOI:10.1016/j.rvsc.2020.06.020 |
| [70] |
JEONG P S, YANG H J, PARK S H, et al. Combined chaetocin/trichostatin A treatment improves the epigenetic modification and developmental competence of porcine somatic cell nuclear transfer embryos[J]. Front Cell Dev Biol, 2021, 9: 709574. DOI:10.3389/fcell.2021.709574 |
| [71] |
ZHAI Y H, ZHANG Z R, YU H, et al. Dynamic methylation changes of DNA and H3K4 by RG108 improve epigenetic reprogramming of somatic cell nuclear transfer embryos in pigs[J]. Cell Physiol Biochem, 2018, 50(4): 1376-1397. DOI:10.1159/000494598 |
| [72] |
JIN J X, LEE S, TAWEECHAIPAISANKUL A, et al. The HDAC inhibitor LAQ824 enhances epigenetic reprogramming and in vitro development of porcine SCNT embryos[J]. Cell Physiol Biochem, 2017, 41(3): 1255-1266. DOI:10.1159/000464389 |
| [73] |
JEONG P S, SIM B W, PARK S H, et al. Chaetocin improves pig cloning efficiency by enhancing epigenetic reprogramming and autophagic activity[J]. Int J Mol Sci, 2020, 21(14): 4836. DOI:10.3390/ijms21144836 |
| [74] |
WATERLAND R A. Assessing the effects of high methionine intake on DNA methylation[J]. J Nutr, 2006, 136(6 Suppl)): 1706S-1710S. |
| [75] |
ZGLEJC K, FRANCZAK A. Peri-conceptional under-nutrition alters the expression of TRIM28 and ZFP57 in the endometrium and embryos during peri-implantation period in domestic pigs[J]. Reprod Domest Anim, 2017, 52(4): 542-550. DOI:10.1111/rda.12943 |
| [76] |
ALTMANN S, MURANI E, SCHWERIN M, et al. Maternal dietary protein restriction and excess affects offspring gene expression and methylation of non-SMC subunits of condensin I in liver and skeletal muscle[J]. Epigenetics, 2012, 7(3): 239-252. DOI:10.4161/epi.7.3.19183 |
| [77] |
OSTER M, TRAKOOLJUL N, REYER H, et al. Sex-specific muscular maturation responses following prenatal exposure to methylation-related micronutrients in pigs[J]. Nutrients, 2017, 9(1): 74. DOI:10.3390/nu9010074 |
| [78] |
FRANCZAK A, ZGLEJC-WASZAK K, MARTYNIAK M, et al. Peri-conceptional nutritional restriction alters transcriptomic profile in the peri-implantation pig embryos[J]. Anim Reprod Sci, 2018, 197: 305-316. DOI:10.1016/j.anireprosci.2018.08.045 |
| [79] |
ZGLEJC-WASZAK K, WASZKIEWICZ E M, FRANCZAK A. Periconceptional undernutrition affects the levels of DNA methylation in the peri-implantation pig endometrium and in embryos[J]. Theriogenology, 2019, 123: 185-193. DOI:10.1016/j.theriogenology.2018.10.002 |
| [80] |
FRANCZAK A, ZGLEJC K, WASZKIEWICZ E, et al. Periconceptional undernutrition affects in utero methyltransferase expression and steroid hormone concentrations in uterine flushings and blood plasma during the peri-implantation period in domestic pigs[J]. Reprod Fertil Dev, 2017, 29(8): 1499-1508. DOI:10.1071/RD16124 |
| [81] |
LI Z C, YUE Z M, AO Z, et al. Maternal dietary supplementation of arginine increases the ratio of total cloned piglets born to total transferred cloned embryos by improving the pregnancy rate of recipient sows[J]. Anim Reprod Sci, 2018, 196: 211-218. DOI:10.1016/j.anireprosci.2018.08.013 |
| [82] |
CEDAR H, BERGMAN Y. Linking DNA methylation and histone modification: Patterns and paradigms[J]. Nat Rev Genet, 2009, 10(5): 295-304. DOI:10.1038/nrg2540 |
(编辑 孟培)



