2. 国家肉牛遗传评估中心, 北京 100193
2. National Center of Beef Cattle Genetic Evaluation, Beijing 100193, China
扩大群体规模可以提高遗传参数和估计育种值(estimated breeding value,EBV)预测的准确性,加快育种进展,提高育种效率[1]。国际上有很多通过将联系紧密的育种场联合起来进行联合育种的成功经验,例如丹麦[2-3]土地面积小,猪场少,通过在全国范围内形成联合育种体系,采用人工授精的方式使优秀种公猪的遗传物质在全国各场之间交流,形成了广泛的遗传联系,加快了遗传进展[4]。中国肉用西门塔尔牛适应性好,抗病力强,耐粗饲,分布范围广,在我国多种生态条件下均能表现出良好的生产性能,目前已成立育种联合会持续开展育种工作。我国西门塔尔牛肉用群体规模大,已占我国肉牛存栏量的60%~70%[5],但由于分布广泛[6],育种进展受到限制[7],为加快我国肉牛遗传改良进度,提高中国肉用西门塔尔牛育种效率,可将育种目标相近、场间关联程度高的场联合起来[8],进行全国范围内的联合育种。Kennedy和Trus[9]及Mathur等[10-11]提出利用关联率(connectedness rate)反映场间关联程度,依据关联率划分出关联组,把关联组内的场视为一个群体进行遗传参数估计和EBV的联合估计。联合育种扩大了群体规模,提高了遗传参数和EBV估计的准确性[12-13],有利于加快遗传进展。巴西采用了基于对比决定系数的牛群群体标准方法,分析了1999—2003年及2004—2008年期间遗传关联率随时间变化的趋势,结果表明,内洛尔牛场间关联率较高,大多数群体间的遗传关联率高于0.4[14]。内洛尔牛群体间的平均关联率在2004—2008年从0.77增加到0.80。陈军等[15]分析了北京、天津和上海荷斯坦牛群体的关联程度,结果显示,京津、京沪、津沪荷斯坦牛群体间的遗传关联率分别为23.95%、17.10%和14.28%。本研究选择的38家育种场和种公牛站分布于内蒙古、吉林、河南、湖北等15个省、自治区,育种场规模为300~1 000头,种公牛站规模为100~200头,大部分牛场采用舍饲方式饲养,内蒙古个别牛场采用放养+舍饲方式饲养。中国肉用西门塔尔牛的初生重性状对肉牛繁育场和散户的经济效益有重要影响,是育种过程中的重要目标性状。
本研究通过对中国肉用西门塔尔牛场间关联率进行分析,划分关联组,对初生重性状进行遗传力和EBV估计,为实现高效的联合育种体系提供参考。
1 材料与方法 1.1 试验材料本研究选取了全国38家育种场和种公牛站中出生于2000—2019年的3 991头中国肉用西门塔尔牛作为研究对象。研究性状为初生重,数据由国家肉牛遗传评估中心提供。使用R语言剔除初生重性状的异常值,得到初生重表型数据和系谱数据有效记录3 840条。初生重性状的描述性统计见表 1。
|
|
表 1 中国肉用西门塔尔牛初生重性状描述性统计 Table 1 Descriptive statistics of Chinese Simmental beef cattle |
Mathur等[9-10]认为,使用场间关联率表示场间关联程度的方法简单准确,场间关联率公式如下:
| $ C{R_{ij}} = \frac{{{\mathop{ cov}} \left( {\widehat {{h_{ij}}}} \right)}}{{\sqrt {{\mathop{ var}} \left( {\widehat {{h_i}}} \right){\mathop{ var}} \left( {\widehat {{h_j}}} \right)} }} $ |
其中,var(
育种值和遗传参数的估计模型为:y=Xb+Za+e,其中,y为观察值向量;b为固定效应向量;a为随机效应向量,a~N(0, Aσa2),A为个体间亲缘关系矩阵,σa2是加性遗传方差;e为残差效应向量,e~ N(0, Iσe2),I为单位矩阵,σe2是残差方差。固定效应只包括场效应;X、Z为a、b的设计矩阵。
使用DMU软件的DMU4模块计算场间关联率[16],使用R(V3.4)语言pheatmap2包对场间关联率结果绘制热图进行聚类分析,依据关联程度划分关联组, 应用非求导约束最大似然法[17-18]进行遗传参数估计。
2 结果 2.1 中国肉用西门塔尔牛关联率情况及关联组划分本研究计算了38个中国肉用西门塔尔牛场的场间关联率,图 1展示了各场间关联率的聚类热图,横纵坐标均为场号,图中的两个浅色区域代表形成的两个关联组。关联组1包括6个场的1 917头中国肉用西门塔尔牛个体,关联组2包括8个场的385头中国肉用西门塔尔牛个体(表 2)。全国各场间平均关联率仅为1.91%,两关联组平均关联率分别达到11.23%和12.54%,属于较高关联水平[19-20](表 3),可以用于跨场联合遗传估计。
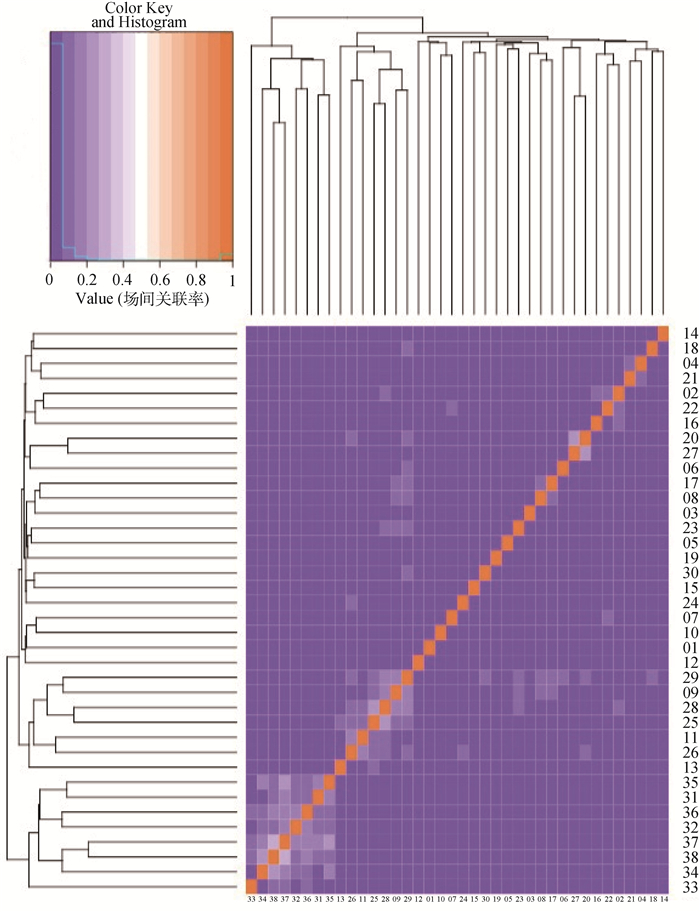
|
图 1 中国肉用西门塔尔牛场间关联率聚类分析热图 Fig. 1 Clustering heatmap of connectedness rate between farms in Chinese Simmental beef cattle |
|
|
表 2 中国肉用西门塔尔牛关联组划分情况 Table 2 Genetic evaluation groups for Chinese Simmental beef cattle |
|
|
表 3 中国肉用西门塔尔牛关联率情况 Table 3 Connectedness rate of Chinese Simmental beef cattle |
分别对单场和两个关联组的初生重性状进行遗传力估计,结果见表 4,单场估计遗传力范围为0.32~0.44,两个关联组遗传力估计值分别为0.47和0.43,关联组遗传力较单场高。单场遗传力标准误范围为0.033~0.052,两关联组遗传力标准误分别为0.021和0.019,关联组遗传力估计标准误相比单场明显降低。
|
|
表 4 单场和联合评估中国肉用西门塔尔牛初生重性状遗传力 Table 4 The estimation of heritability for birth weight trait of Chinese Simmental beef cattle from single herd and joint genetic evaluation |
分别对关联组1和关联组2进行EBV估计。表 5对比了联合估计和单场EBV排名顺序,可见两种方式得到EBV排序前10的个体有所差别。表 6比较了单场和联合估计EBV的准确性。关联组1中,联合估EBV的准确性为0.61,进行单场估计EBV确准性为0.47,其中第9、28、29场估计EBV准确性分别为0.59、0.51和0.47。关联组2中,联合估EBV的准确性为0.56,进行单场估计EBV确准性为0.45,其中第36、37、38场估计EBV准确性分别为0.46、0.54和0.46。关联组1和关联组2联合估计较单场估计准确性分别提高了29.8%和24.4%。
|
|
表 5 单场和联合评估估计育种值排名 Table 5 The ranking of estimated breeding value from single herd and joint genetic evaluation |
|
|
表 6 单场和联合评估估计育种值准确性比较 Table 6 The accuracy comparison of estimated breeding value from single herd and joint genetic evaluation |
本研究计算了中国肉用西门塔尔牛的场间关联率,根据Mathur等[10]关联组划分的原则,划分出两个关联组。中国肉用西门塔尔牛关联率处于较低水平,平均关联率仅有1.91%,而两个关联组的关联率分别为11.23%和12.54%,组内关联率明显提高。张锁宇等[8, 21]对全国大白、长白、杜洛克猪场间的关联进行了分析,全国平均关联率约为0.4%,关联水平较低,对各品种分别划分了3个关联组,其中长白猪的一个关联组平均关联率最大,为3.53%。
这表明,我国仅能实现大白、长白、杜洛克部分猪场之间的联合估计。我国肉用西门塔尔牛场间关联率较低可能包括几种原因:1)地理、经济等因素导致中国肉用西门塔尔牛在全国各场间遗传物质的交流较少,较多的遗传交流是提高场间关联率的关键。本研究划分的关联组2中大部分场间的地理距离较近,可能更易于遗传物质交流,导致场间关联率较高;2)部分场对牛号、系谱的登记不规范,系谱信息直接关系到场间遗传关联计算的准确度,缺乏完整的系谱信息导致忽视了很多场之间的遗传联系,总体关联率较低导致全国范围内的遗传关联无法建立[22]。
本研究采用非求导约束最大似然法估计了中国肉用西门塔尔牛初生重性状的遗传力, 属于中高遗传力性状,所估计的遗传力处于文献报道的范围之内[23-24]。对于初生重遗传力估计值,Bullock等[25]估计的海福特牛的遗传力为0.49,Jeyaruban等[26]估计澳大利亚西门塔尔牛的遗传力为0.36,汪春乾等[27]估计的中国西门塔尔牛遗传力为0.32。有研究认为,遗传进展与遗传变异、选择强度、准确度和世代间隔、分析方法及统计模型密切相关[28]。当群体规模扩大,群体内的遗传变异随之增多,选择强度随之增大,遗传参数估计准确度也随之提高[1]。遗传参数估计的准确程度主要受群体规模、分析方法和模型的影响[1, 10, 29-31]。单一育种场规模较小,而场间联系紧密的各场形成关联组后可以视为同一群体,扩大了群体规模,增大了遗传参数估计的准确性。本研究分别对单场和关联组进行遗传力估计,关联组1数据量远大于关联组2,遗传力估计标准误更小。在单场估计中也出现同样规律,规模较大、数据分布较均匀的场遗传力估计的标准误更小,遗传力估计准确性更高。把遗传关联紧密的场划分成一个关联组相当于扩大了群体规模,提高了遗传参数估计的准确性。
对单场和联合估计EBV的大小进行比较,发现两组EBV的排序有所不同(表 5),在单场内排名较低的个体在联合估计时仍有被选择的机会。通过联合遗传估计,可以跨场对种公牛进行比较和选择,从而为种公牛提供更加公平的评估机会,筛选出更优个体。本研究中,相对于单场评估,两关联组的EBV准确性均得到了明显提高,今后可以通过人工授精等方式传递优秀种公牛遗传物质,加强关联组内遗传交流。在全国范围内,加强种公牛站建设可更有效地传递优秀遗传物质[32]。同时应完善肉牛登记和记录系统,使系谱信息记录完整。
4 结论本研究通过对中国肉用西门塔尔牛的场间遗传关联分析,发现全国牛场间的关联较弱。依据场间关联率可划分出两个关联组,关联组内场间关联率明显高于全国平均水平,关联组内计算遗传力和估计EBV的准确性均高于单场。因此,依据关联率划分关联组进行联合育种有利于加快中国肉用西门塔尔牛的育种进程,为实现全国范围内中国肉用西门塔尔牛的联合育种奠定基础。
| [1] |
张勤. 家畜育种值和遗传参数估计方法的发展及现状[J]. 国外畜牧学(草食家畜), 1990(6): 1–4, 46.
ZHANG Q. The development and status of methods of estimated breeding value and genetic parameters[J]. Grass-Feeding Livestock, 1990(6): 1–4, 46. (in Chinese) |
| [2] |
李娅兰, 刘珍云, 刘敬顺, 等. 世界种猪育种体系及对我国种猪育种借鉴[J]. 中国畜牧业, 2013(6): 52–54.
LI Y L, LIU Z Y, LIU J S, et al. World swine breeding system and reference for breeding swine breding in China[J]. China Animal Industry, 2013(6): 52–54. DOI: 10.3969/j.issn.2095-2473.2013.06.025 (in Chinese) |
| [3] |
李冉. 国外畜禽良种繁育发展及经验借鉴[J]. 世界农业, 2014(3): 30–33, 37.
LI R. Development and experience of livestock breeding in foreign countries[J]. World Agriculture, 2014(3): 30–33, 37. DOI: 10.3969/j.issn.1002-4433.2014.03.005 (in Chinese) |
| [4] |
王爱国, 李力, 梅克义, 等. 加强联合育种提高种猪质量[J]. 中国畜禽种业, 2005(10): 8–9.
WANG A G, LI L, MEI K Y, et al. Enhance joint breeding and improve the quality of swine[J]. The Chinese Livestock Breeding, 2005(10): 8–9. (in Chinese) |
| [5] |
朱波, 李姣, 汪聪勇, 等. 我国肉用西门塔尔牛群体生长发育性状遗传参数估计及其遗传进展[J]. 畜牧兽医学报, 2020, 51(8): 1833–1844.
ZHU B, LI J, WANG C Y, et al. Genetic parameter and genetic gain estimation for growth and development traits in Chinese Simmental beef cattle[J]. Acta Veterinaria et Zootechnica Sinica, 2020, 51(8): 1833–1844. (in Chinese) |
| [6] |
牛红, 宝金山, 吴洋, 等. 中国西门塔尔牛肉用群体重要经济性状遗传参数估计[J]. 畜牧兽医学报, 2016, 47(9): 1817–1823.
NIU H, BAO J S, WU Y, et al. Estimation of genetic parameters for economic important traits in Chinese Simmental beef cattle[J]. Acta Veterinaria et Zootechnica Sinica, 2016, 47(9): 1817–1823. (in Chinese) |
| [7] |
王雅春, 陈幼春. 世界西门塔尔牛育种现状及其在中国的应用方向[J]. 中国牛业科学, 2008, 34(6): 1–4.
WANG Y C, CHEN Y C. Breeding progress of Simmental world-wide and the principle of its application in China[J]. China Cattle Science, 2008, 34(6): 1–4. DOI: 10.3969/j.issn.1001-9111.2008.06.001 (in Chinese) |
| [8] |
张金鑫, 张锁宇, 邱小田, 等. 我国杜洛克、长白和大白猪场间遗传联系分析[J]. 畜牧兽医学报, 2017, 48(9): 1591–1601.
ZHANG J X, ZHANG S Y, QIU X T, et al. The genetic connectedness of Duroc, Landrace and Yorkshire pigs in China[J]. Acta Veterinaria et Zootechnica Sinica, 2017, 48(9): 1591–1601. (in Chinese) |
| [9] | KENNEDY B W, TRUS D. Considerations on genetic connectedness between management units under an animal model[J]. J Anim Sci, 1993, 71(9): 2341–2352. DOI: 10.2527/1993.7192341x |
| [10] | MATHUR P K, SULLIVAN B P, CHESNAIS J P.Measuring connectedness: concept and application to a large industry breeding program[C]//Proceedings of the 7th World Congress on Genetics Applied to Livestock Production.Montpellier, France, 2002. |
| [11] | FOUILLOUX M N, LALOË D.A method for measuring connectedness among herds in mixed linear models[C]//Proceedings of the 8th WCGALP, 2006. |
| [12] | ÅKESSON M, BENSCH S, HASSELQUIST D, et al. Estimating heritabilities and genetic correlations: comparing the 'animal model' with parent-offspring regression using data from a natural population[J]. PLoS One, 2008, 3(3): e1739. DOI: 10.1371/journal.pone.0001739 |
| [13] | KRUUK L E B. Estimating genetic parameters in natural populations using the 'animal model'[J]. Philos Trans R Soc B: Biol Sci, 2004, 359(1446): 873–900. DOI: 10.1098/rstb.2003.1437 |
| [14] | PEGOLO N T, LALOË D, DE OLIVEIRA H N, et al. Trends of the genetic connectedness measures among Nelore beef cattle herds[J]. J Anim Breed Genet, 2012, 129(1): 20–29. DOI: 10.1111/j.1439-0388.2011.00934.x |
| [15] |
陈军, 孙东晓, 王雅春, 等. 中国荷斯坦牛不同群体间遗传联系性研究[J]. 畜牧兽医学报, 2009, 40(1): 129–132.
CHEN J, SUN D X, WANG Y C, et al. Measures of genetic connectedness among herds in Chinese Holstein[J]. Acta Veterinaria et Zootechnica Sinica, 2009, 40(1): 129–132. DOI: 10.3321/j.issn:0366-6964.2009.01.022 (in Chinese) |
| [16] | MADSEN P, SORENSEN P, SU G, et al.DMU-a package for analyzing multivariate mixed models[C]//Proceedings of the 8th World Congress on Genetics Applied to Livestock Production.2006. |
| [17] | HENDERSON C R, QUAAS R L. Multiple trait evaluation using relatives' records[J]. J Anim Sci, 1976, 43(6): 1188–1197. DOI: 10.2527/jas1976.4361188x |
| [18] | ZHANG L C, NING Z H, XU G Y, et al. Heritabilities and genetic and phenotypic correlations of egg quality traits in brown-egg dwarf layers[J]. Poult Sci, 2005, 84(8): 1209–1213. DOI: 10.1093/ps/84.8.1209 |
| [19] |
叶健, 傅金銮, 张锁宇, 等. 安徽省猪育种核心群场间联系性和遗传参数估计[J]. 中国畜牧杂志, 2015, 51(18): 62–67.
YE J, FU J L, ZHANG S Y, et al. Estimation of genetic parameters and connectedness in pig breeding nucleus herds of AnHui province[J]. Chinese Journal of Animal Science, 2015, 51(18): 62–67. DOI: 10.3969/j.issn.0258-7033.2015.18.013 (in Chinese) |
| [20] | TARRÉS J, FINA M, PIEDRAFITA J. Connectedness among herds of beef cattle bred under natural service[J]. Genet Sel Evol, 2010, 42(1): 6. DOI: 10.1186/1297-9686-42-6 |
| [21] |
张锁宇, 邱小田, 丁向东, 等. 利用关联组估计中国大白、长白及杜洛克猪繁殖性状的遗传参数[J]. 畜牧兽医学报, 2016, 47(3): 429–438.
ZHANG S Y, QIU X T, DING X D, et al. Estimation of genetic parameters by connecting groups for reproductive traits of Yorkshire, Landrace and Duroc pigs in China[J]. Acta Veterinaria et Zootechnica Sinica, 2016, 47(3): 429–438. (in Chinese) |
| [22] |
赵玉民, 李俊雅. 肉牛联合育种方案设计与实施[J]. 现代畜牧兽医, 2010(3): 8–9.
ZHAO Y M, LI J Y. Design and implementation of joint breeding on meet cattle[J]. Modern Journal of Animal Husbandry and Veterinary Medicine, 2010(3): 8–9. DOI: 10.3969/j.issn.1672-9692.2010.03.004 (in Chinese) |
| [23] | KAUSE A, MIKKOLA L, STRANDÉN I, et al. Genetic parameters for carcass weight, conformation and fat in five beef cattle breeds[J]. Animal, 2015, 9(1): 35–42. DOI: 10.1017/S1751731114001992 |
| [24] | BENYSHEK L L. Heritabilities for growth and carcass traits estimated from data on Herefords under commercial conditions[J]. J Anim Sci, 1981, 53(1): 49–56. DOI: 10.2527/jas1981.53149x |
| [25] | BULLOCK K D, BERTRAND J K, BENYSHEK L L. Genetic and environmental parameters for mature weight and other growth measures in Polled Hereford cattle[J]. J Anim Sci, 1993, 71(7): 1737–1741. DOI: 10.2527/1993.7171737x |
| [26] | JEYARUBAN M G, JOHNSTON D J, TIER B, et al. Genetic parameters for calving difficulty using complex genetic models in five beef breeds in Australia[J]. Anim Prod Sci, 2015, 56(5): 927–933. |
| [27] |
汪春乾, 许尚忠, 陈宏权, 等. 中国西门塔尔牛生长性状遗传参数估计[J]. 安徽农业大学学报, 2004, 31(4): 417–420.
WANG C Q, XU S Z, CHEN H Q, et al. Estimating parameters growth traits of Chinese Simmental[J]. Journal of Anhui Agricultural University, 2004, 31(4): 417–420. DOI: 10.3969/j.issn.1672-352X.2004.04.008 (in Chinese) |
| [28] | MISZTAL I. Genetics and breeding: restricted maximum likelihood estimation of variance components in animal model using sparse matrix inversion and a supercomputer[J]. J Dairy Sci, 1990, 73(1): 163–172. DOI: 10.3168/jds.S0022-0302(90)78660-2 |
| [29] | DARFOUR-ODURO K A, NAAZIE A, AHUNU B K, et al. Genetic parameter estimates of growth traits of indigenous pigs in Northern Ghana[J]. Livestock Sci, 2009, 125(2-3): 187–191. DOI: 10.1016/j.livsci.2009.04.007 |
| [30] | GROENEVELD E, KOVAC M. A note on multiple solutions in multivariate restricted maximum like-lihood covariance component estimation[J]. J Dairy Sci, 1990, 73(8): 2221–2229. DOI: 10.3168/jds.S0022-0302(90)78902-3 |
| [31] | JALIL-SARGHALE A, KHOLGHI M, SHAHREBABAK M M, et al. Model comparisons and genetic parameter estimates of growth traits in Baluchi sheep[J]. Slovak J Anim Sci, 2014, 47(1): 12–18. |
| [32] |
陈伟生. 实施全国畜禽遗传改良计划应处理好几个关系[J]. 中国畜牧杂志, 2013, 49(6): 3–5, 13.
CHEN W S. Several relations should be handled well in implementing the national livestock and poultry genetic improvement plan[J]. Chinese Journal of Animal Science, 2013, 49(6): 3–5, 13. (in Chinese) |



