2. 山东农业大学动物科技学院/动物医学院基础兽医学系, 泰安 271018;
3. 山东农业大学山东省动物生物技术与疾病预防控制重点实验室, 泰安 271018
2. Department of Basic Veterinary Medicine, College of Animal Science and Veterinary Medicine, Shandong Agricultural University, Tai'an 271018, China;
3. Shandong Provincial Key Laboratory of Animal Biotechnology and Disease Control and Prevention, Shandong Agricultural University, Tai'an 271018, China
A型流感病毒是流感病毒中最常见的,可以感染人类、猪、狗、马等动物[1-2]。根据血凝素(HA)和神经氨酸酶(NA)的抗原性可将A型流感病毒分为不同的亚型,已知A型流感病毒HA有18种亚型,NA有11种亚型[3-6]。猪呼吸道中同时含有类禽样唾液酸(sialic acid, SA)-α-2, 3Gal受体和类人样SA-α-2, 6-Gal受体[7-9],可以同时被禽源、人源流感病毒感染。不同种属来源的病毒在猪体内产生重组病毒,这些重组病毒可以导致跨种感染,对人类及动物产生危害。
日前,在世界各地猪群中主要传播H1N1、H1N2和H3N2 3种猪流感病毒(swine influenza virus,SIV)亚型,这些SIV亚型的起源以及抗原和遗传特征不同[10-12]。经典H1N1 SIV被认为是随着1918年西班牙流感大流行进入猪群[13]。中国大陆于1991年发现有经典H1N1病毒[14-15]。欧亚类禽H1N1病毒2001年在中国香港购入的猪中检测到,经典H1N1和欧亚类禽H1N1病毒间重排的毒株出现,并在临床上流行[16]。引起2009年的大流行病毒为3源重排病毒,之后由2009年大流行病毒片段和其他来源的SIV的重排病毒也已从不同国家分离[17],给公共卫生带来巨大的威胁。
不同类型流感病毒在猪体内重排产生的新的毒株,具有潜在的引起猪流感大流行的可能性,需要对SIV进行实时监控。因此,本实验室于2018年4月份在河南省某猪场采集鼻拭子样品150份,分离到1株H1N1亚型SIV,进行了分离病毒全基因组的序列测定与分析及小鼠的致病性研究,研究结果丰富了我国猪流感分子流行病学调查内容,为进一步开展SIV的防控提供重要的理论依据。
1 材料与方法 1.1 临床样品及病毒分离2018年4月,从河南省南阳市猪场采集150份鼻拭子样品,将样品上清液经尿囊腔接种于10日龄SPF鸡胚(0.2 mL·胚-1),置于37 ℃孵化箱孵育72 h,收获尿囊液并通过HA试验检测其血凝效价及使用鉴定引物扩增流感病毒NP节段。将分离的病毒用SPF鸡胚纯化,用Reed-Muench方法测定病毒的EID50。
1.2 病毒亚型的鉴定及测序SIV分型引物及各基因片段扩增引物均由本实验室设计、上海生工生物工程技术服务有限公司合成。采用TRIzol法提取病毒RNA,利用通用引物Uni12(5′-AGCAAAAGG-3′)将RNA反转录为cDNA,以反转录产物为模板进行亚型鉴定及8个基因片段的扩增。扩增产物送上海生工生物工程技术服务有限公司测序。
1.3 病毒的遗传进化与分子特征分析应用DNASTAR软件包中的Seqman软件进行拼接;Megalign软件进行同源性比较和氨基酸特征分析;用MEGA7软件绘制进化树;利用NetNGlyc 1.0 Server糖基化位点预测器(http://www.healthtech.dtu.dk in the future),对氨基酸潜在糖基化位点进行预测分析。
1.4 病毒对小鼠的致病性试验将16只6周龄BALB/c雌性小鼠随机分成感染组和对照组,每组8只。每只小鼠经鼻腔接种106EID50病毒,对照组用相同的方法接种PBS。每天观察小鼠临床症状并称量体重。感染后第3天每组随机各剖杀3只小鼠,无菌采集各组织器官备用。
2 结果 2.1 病毒分离鉴定及同源性分析分离出1株SIV,通过RT-PCR鉴定为H1N1亚型,命名为A/swine/Henan/NY20/2018(H1N1)。利用Megalign软件将参考毒株分别与分离病毒进行核苷酸同源性比较,结果显示,PB2、PB1、NA和NS均与A/swine/Guangxi/NNXD2023/2013(H1N1)相似性最高,在98.4%以上,PA与A/swine/Guangxi/1874/2012(H3N2)相似性最高,为98.8%,M和A/swine/Guangdong/NS2883/2012(H3N2)相似性最高,为98.9%,HA与A/swine/Guangxi/3858/2011(H1N1)相似性最高,为98.2%,NP与A/swine/Guangxi/BB1/2013(H1N1)相似性最高,为99%。
2.2 遗传进化分析对分离病毒8个基因片段进行遗传进化分析。结果显示,分离病毒H1基因属于欧亚类禽H1N1分支,N1基因与H1基因具有相同的分支(图 1)。相比于表面基因,内部基因聚集成相同的聚类模式,除了NS基因属于经典H1N1分支,剩余内部基因全部属于2009年甲型H1N1分支。
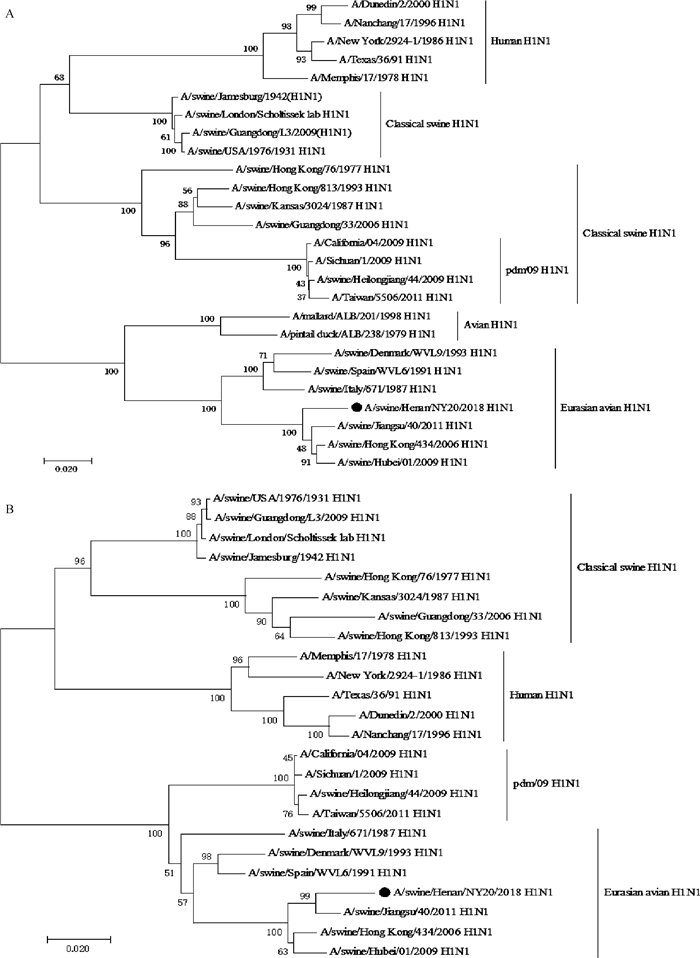
|
图 1 A/swine/Henan/NY20/2018(H1N1)的HA(A)和NA(B)基因与参考序列构建的基因进化树 Fig. 1 Phylogenetic relationship of HA and NA gene of A/swine/Henan/NY20/2018(H1N1) with reference sequences |
对分离病毒A/swine/Henan/NY20/2018(H1N1)基因上的关键氨基酸位点进行了分析,HA蛋白的裂解位点对流感病毒的毒力和致病性具有决定性的作用[18-20]。HA蛋白的裂解位点为PSIQSR↓ GL,具备低致病性流感病毒的分子特征。HA蛋白的受体结合位点发生了T155V(对应H3 HA基因编码位点),T159N、G225E位的突变,T155V和T159N位的突变是禽流感适应猪群的必需突变[21]。HA蛋白225位能够提高病毒颗粒的组装和出芽效率,且225E的病毒比携带225G的病毒复制得更快[22]。分离株的225位为E,对流感病毒的传播起到重要作用。研究发现,HA蛋白226 Q和228 G容易与SA-α-2, 3-Gal受体结合,L226和S228则容易与SA-α-2, 6-Gal受体结合[23]。分离株226位和228位分别为Q和G,表明该病毒容易与SA-α-2, 3-Gal受体结合。HA蛋白糖基化位点分别为14NST16、26NVT28、277NCT279、484NGT486和543NGS545。NA蛋白糖基化位点分别为57NNT59、62NQT64、67NVS69、87NSS89、145NGT146和234NGS236。PB2蛋白的627和701位的被认为是哺乳动物适应性的突变,单个E627K或D701N突变可能会增加病毒的致病性[24]。有相关报道表明,PB2蛋白的591R对于2009甲型H1N1流感病毒对哺乳动物适应很重要[25]。PB2蛋白的627位和701位均未发生突变,但是591位出现了突变。最常见的NA蛋白耐药性的突变存在于N1亚型的H275Y和N295S[26-27],N1蛋白的275位和295位未发生突变,表明对神经氨酸酶抑制剂类药物敏感。有相关报道表明,2009甲型H1N1流感病毒在M2蛋白的31N已经发生突变,对金刚烷胺药物具有耐药性[28]。M2蛋白31位出现S到N的突变,表明具有金刚烷胺类药物的耐药性。
2.4 小鼠致病性试验感染组小鼠第3天开始表现出活动量和采食量减少、扎堆、皮毛皱纹、体重下降等临床症状,体重于感染后的第8天降到最低,最大下降比19.67%,随后体重逐渐恢复。
对照组保持健康且体重保持平稳状态(图 2A)。脏器滴定结果显示,感染组小鼠在肺中检测到的病毒含量为4.25 lg EID50·mL-1,在鼻甲中检测到的病毒含量为2.67 lg EID50·mL-1,在脑、脾和肾中均未检测到病毒复制。对照组未检测到病毒(图 2B)。对小鼠肺HE染色,组织病理变化主要表现为肺泡壁毛细血管充血、出血,肺泡壁增厚,炎性细胞浸润等。对照组未见明显的病理变化(图 2C)。
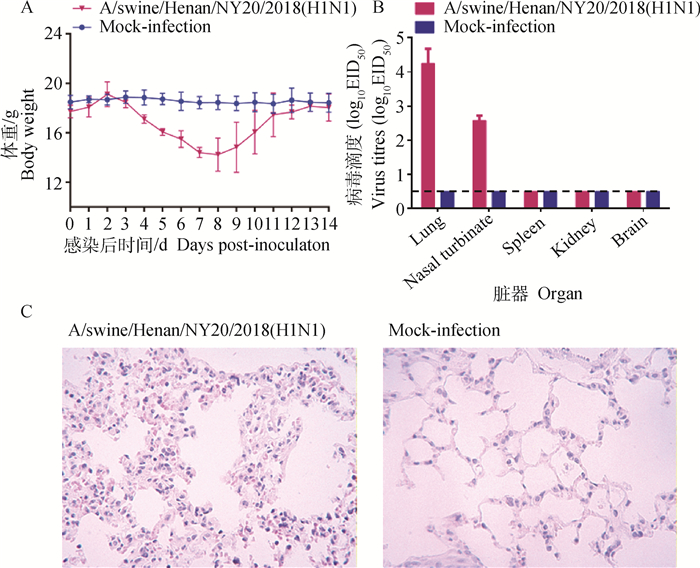
|
A.攻毒后14 d内小鼠体重变化; B.攻毒后第3天小鼠脏器内病毒含量; C.攻毒后第3天小鼠肺部病理切片(200×) A. The body weight changes of mice over 14 days post infection; B. Viral titer in organs of mice at 3 days post infection; C. Histopathological of the mice lung tissue at 3 days post infection (200×) 图 2 A/swine/Henan/NY20/2018(H1N1)对小鼠的致病性 Fig. 2 Pathogenicity of A/swine/Henan/NY20/2018(H1N1) in mice. |
由于猪具有A型流感病毒“混合容器”的作用,所以当多种流感病毒亚型的共同循环可导致基因重排新病毒的产生[29-33]。2009年,在墨西哥和美国出现人感染2009甲型H1N1,随后的一段时间内迅速传播至多个国家[34]。在发生人感染2009甲型H1N1之后,在加拿大发现猪感染2009甲型H1N1的病例[35-36]。在我国香港发现第1个重配的H1N1病毒,其基因片段来源于2009甲型H1N1(NA),欧亚类禽H1N1(HA),6个内部基因3重重组SIV[37]。目前,猪群中经常分离到含有2009甲型H1N1基因的新型重配病毒[32]。本研究中,A/swine/Henan/NY20/2018(H1N1)为1株3源重排H1N1病毒,其基因片段来源于欧亚类禽H1N1 (HA和NA)、2009甲型H1N1(PB2、PB1、PA、NP、M)和经典H1N1(NS)。此种基因型在2013年广西省曾报道过[25]。此次分离的病毒5个内部基因来源于2009甲型H1N1,再次验证了含有2009甲型H1N1基因已经相对稳定的存在猪群中。
为了评估分离株在哺乳动物物种中的致病性,笔者将BALB/c小鼠作为感染模型,研究结果发现分离株在小鼠肺和鼻甲能够有效的复制,肺出现明显的病理学变化,没有出现小鼠的死亡。说明分离株对小鼠的有一定的致病性,但致病力不强,其具体的致病性还有待于到猪体内验证。
2009甲型H1N1可以从猪群传染给人类,也可以从人类传播到猪群[38]。含有2009甲型H1N1基因片段的重配病毒已在全世界的猪中广泛传播[39-40]。含有2009甲型H1N1基因的流感病毒是否会引发下一次的流感大流行,将是关乎公共卫生学意义的重要事情。因此,需要加强SIV流行病学的监测。
4 结论在河南省流行病学调查中分离到1株H1N1亚型SIV,该分离株属于1株3源重排H1N1病毒,对小鼠呈现一定的致病力。提示需要进一步加强对SIV监测,为流感的防控及流感大流行的预测提供重要的理论依据。
| [1] | FONI E, GARBARINO C, CHIAPPONI C, et al. Epidemiological survey of swine influenza A virus in the wild boar population of two Italian provinces[J]. Influenza Other Respir Viruses, 2013, 7(S4): 16–20. |
| [2] | WEBSTER R G, BEAN W J, GORMAN O T, et al. Evolution and ecology of influenza A viruses[J]. Microbiol Rev, 1992, 56(1): 152–179. DOI: 10.1128/MMBR.56.1.152-179.1992 |
| [3] | MUKHERJEE T R, AGRAWAL A S, CHAKRABARTI S, et al. Full genomic analysis of an influenza A (H1N2) virus identified during 2009 pandemic in Eastern India: evidence of reassortment event between co-circulating A(H1N1)pdm09 and A/Brisbane/10/2007-like H3N2 strains[J]. Virol J, 2012, 9: 233. DOI: 10.1186/1743-422X-9-233 |
| [4] | FOUCHIER R A M, MUNSTER V, WALLENSTEN A, et al. Characterization of a novel influenza A virus hemagglutinin subtype (H16) obtained from black-headed gulls[J]. J Virol, 2005, 79(5): 2814–2822. DOI: 10.1128/JVI.79.5.2814-2822.2005 |
| [5] | TONG S X, ZHU X Y, LI Y, et al. New world bats harbor diverse influenza A viruses[J]. PLoS Pathog, 2013, 9(10): e1003657. DOI: 10.1371/journal.ppat.1003657 |
| [6] | TONG S X, LI Y, RIVAILLER P, et al. A distinct lineage of influenza A virus from bats[J]. Proc Natl Acad Sci U S A, 2012, 109(11): 4269–4274. DOI: 10.1073/pnas.1116200109 |
| [7] | YANG H L, CHEN Y, QIAO C L, et al. Prevalence, genetics, and transmissibility in ferrets of Eurasian avian-like H1N1 swine influenza viruses[J]. Proc Natl Acad Sci U S A, 2016, 113(2): 392–397. DOI: 10.1073/pnas.1522643113 |
| [8] | ZHAO N, MARTIN B E, YANG C K, et al. Association analyses of large-scale glycan microarray data reveal novel host-specific substructures in influenza A virus binding glycans[J]. Sci Rep, 2015, 5: 15778. DOI: 10.1038/srep15778 |
| [9] | SUZUKI Y, ITO T, SUZUKI T, et al. Sialic acid species as a determinant of the host range of influenza A viruses[J]. J Virol, 2000, 74(24): 11825–11831. DOI: 10.1128/JVI.74.24.11825-11831.2000 |
| [10] | VAN REETH K, BROWN I H, DVRRWALD R, et al. Seroprevalence of H1N1, H3N2 and H1N2 influenza viruses in pigs in seven European countries in 2002-2003[J]. Influenza Other Respir Viruses, 2008, 2(3): 99–105. DOI: 10.1111/j.1750-2659.2008.00043.x |
| [11] | CHUTINIMITKUL S, THIPPAMOM N, DAMRONGWATANAPOKIN S, et al. Genetic characterization of H1N1, H1N2 and H3N2 swine influenza virus in Thailand[J]. Arch Virol, 2008, 153(6): 1049–1056. DOI: 10.1007/s00705-008-0097-7 |
| [12] | KONG W L, HUANG L Z, QI H T, et al. Genetic characterization of H1N2 influenza a virus isolated from sick pigs in Southern China in 2010[J]. Virol J, 2011, 8: 469. DOI: 10.1186/1743-422X-8-469 |
| [13] | VINCENT A L, PEREZ D R, RAJAO D, et al. Influenza A virus vaccines for swine[J]. Vet Microbiol, 2017, 206: 35–44. DOI: 10.1016/j.vetmic.2016.11.026 |
| [14] | KONG W L, HUANG Y M, CAO N, et al. Isolation and phylogenetic analysis of H1N1 swine influenza virus from sick pigs in Southern China[J]. Indian J Virol, 2011, 22(1): 66–71. DOI: 10.1007/s13337-011-0035-2 |
| [15] | ZHU W F, YANG S, GUO Y J, et al. Imported pigs may have introduced the first classical swine influenza viruses into Mainland China[J]. Infect Genet Evol, 2013, 17: 142–146. DOI: 10.1016/j.meegid.2013.03.007 |
| [16] | ZHU H, WEBBY R, LAM T T, et al. History of Swine influenza viruses in Asia[J]. Curr Top Microbiol Immunol, 2013, 370: 57–68. |
| [17] | FAN X H, ZHU H C, ZHOU B P, et al. Emergence and dissemination of a swine H3N2 reassortant influenza virus with 2009 pandemic H1N1 genes in pigs in China[J]. J Virol, 2012, 86(4): 2375–2378. DOI: 10.1128/JVI.06824-11 |
| [18] | WEBSTER R G, ROTT R. Influenza virus a pathogenicity:the pivotal role of hemagglutinin[J]. Cell, 1987, 50(5): 665–666. DOI: 10.1016/0092-8674(87)90321-7 |
| [19] | KLENK H D, ROTT R. The molecular biology of influenza virus pathogenicity[J]. Adv Virus Res, 1988, 34: 247–281. DOI: 10.1016/S0065-3527(08)60520-5 |
| [20] | HORIMOTO T, KAWAOKA Y. Pandemic threat posed by avian influenza A viruses[J]. Clin Microbiol Rev, 2001, 14(1): 129–149. DOI: 10.1128/CMR.14.1.129-149.2001 |
| [21] |
刘清政, 王宏宇, 杜以军, 等. 一株猪流感病毒的遗传进化分析及其致病力研究[J]. 中国预防兽医学报, 2017, 39(12): 1022–1025.
LIU Q Z, WANG H Y, DU Y J, et al. Genetic evolution analysis and pathogenicity of a swine influenza virus[J]. Chinese Journal of Preventive Veterinary Medicine, 2017, 39(12): 1022–1025. (in Chinese) |
| [22] | WANG Z, YANG H L, CHEN Y, et al. A single-amino-acid substitution at position 225 in hemagglutinin alters the transmissibility of Eurasian avian-like H1N1 swine influenza virus in guinea pigs[J]. J Virol, 2017, 91(21): e00800–17. |
| [23] |
李婷, 朱启运, 冉多良, 等. 猪流感病毒分离株A/Swine/Fujian/F1/2001(H5N1)表面蛋白基因序列分析[J]. 新疆农业大学学报, 2006, 29(3): 1–4.
LI T, ZHU Q Y, RAN D L, et al. Sequence analysis on the surface glycoprotein genes of a swine influenza virus isolate A/Swine/Fujian/F1/2001(H5N1)[J]. Journal of Xinjiang Agricultural University, 2006, 29(3): 1–4. DOI: 10.3969/j.issn.1007-8614.2006.03.001 (in Chinese) |
| [24] | ZHU W F, LI L, YAN Z G, et al. Dual E627K and D701N mutations in the PB2 protein of A(H7N9) influenza virus increased its virulence in mammalian models[J]. Sci Rep, 2015, 5: 14170. DOI: 10.1038/srep14170 |
| [25] | HE P, WANG G J, MO Y N, et al. Novel triple-reassortant influenza viruses in pigs, Guangxi, China[J]. Emerg Microbes Infect, 2018, 7(1): 85. |
| [26] | LI X D, LIAO H, LIU Y, et al. Drug-resistant and genetic evolutionary analysis of influenza virus from patients during the 2013 and 2014 influenza season in Beijing[J]. Microb Drug Resist, 2017, 23(2): 253–260. DOI: 10.1089/mdr.2015.0297 |
| [27] | IKEMATSU H, KAWAI N, IWAKI N, et al. In vitro neuraminidase inhibitory activity of four neuraminidase inhibitors against influenza virus isolates in the 2011-2012 season in Japan[J]. J Infect Chemother, 2014, 20(2): 77–80. |
| [28] | GARTEN R J, DAVIS C T, RUSSELL C A, et al. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans[J]. Science, 2009, 325(5937): 197–201. DOI: 10.1126/science.1176225 |
| [29] | ITO T, COUCEIRO J N, KELM S, et al. Molecular basis for the generation in pigs of influenza A viruses with pandemic potential[J]. J Virol, 1998, 72(9): 7367–7373. DOI: 10.1128/JVI.72.9.7367-7373.1998 |
| [30] | GAMMELIN M, MANDLER J, SCHOLTISSEK C. Two subtypes of nucleoproteins (NP) of influenza A viruses[J]. Virology, 1989, 170(1): 71–80. |
| [31] | VAN POUCKE S G, NICHOLLS J M, NAUWYNCK H J, et al. Replication of avian, human and swine influenza viruses in porcine respiratory explants and association with sialic acid distribution[J]. Virol J, 2010, 7: 38. DOI: 10.1186/1743-422X-7-38 |
| [32] | NONTHABENJAWAN N, CHANVATIK S, CHAIYAWONG S, et al. Genetic diversity of swine influenza viruses in Thai swine farms, 2011-2014[J]. Virus Genes, 2015, 50(2): 221–230. DOI: 10.1007/s11262-014-1153-x |
| [33] | SONG Y F, ZHANG Y, ZHANG B, et al. Identification, genetic analysis, and pathogenicity of classical swine H1N1 and human-swine reassortant H1N1 influenza viruses from pigs in China[J]. Viruses, 2020, 12(1): 55. |
| [34] | SMITH G J D, VIJAYKRISHNA D, BAHL J, et al. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic[J]. Nature, 2009, 459(7250): 1122–1125. DOI: 10.1038/nature08182 |
| [35] | HOWDEN K J, BROCKHOFF E J, CAYA F D, et al. An investigation into human pandemic influenza virus (H1N1) 2009 on an Alberta swine farm[J]. Can Vet J, 2009, 50(11): 1153–1161. |
| [36] | NElSON M I, VINCENT A L, KITIKOON P, et al. Evolution of novel reassortant A/H3N2 influenza viruses in North American swine and humans, 2009-2011[J]. J Virol, 2012, 86(16): 8872–8878. DOI: 10.1128/JVI.00259-12 |
| [37] | VIJAYKRISHNA D, POON L L M, ZHU H C, et al. Reassortment of pandemic H1N1/2009 influenza A virus in swine[J]. Science, 2010, 328(5985): 1529. DOI: 10.1126/science.1189132 |
| [38] | NELSON M I, VINCENT A L. Reverse zoonosis of influenza to swine:new perspectives on the human-animal interface[J]. Trends Microbiol, 2015, 23(3): 142–153. |
| [39] | WATSON S J, LANGAT P, REID S M, et al. Molecular epidemiology and evolution of influenza viruses circulating within European swine between 2009 and 2013[J]. J Virol, 2015, 89(19): 9920–9931. DOI: 10.1128/JVI.00840-15 |
| [40] | LEWIS N S, RUSSELL C A, LANGAT P, et al. The global antigenic diversity of swine influenza A viruses[J]. eLife, 2016, 5: e12217. DOI: 10.7554/eLife.12217 |



