2. 中国农业大学动物科技学院, 北京 100193;
3. 天津市农业科学院畜牧兽医研究所, 天津 300192
2. College of Animal Science and Technology, China Agricultural University, Beijing 100193, China;
3. Institute of Animal Husbandry and Veterinary, Tianjin Academy of Agricultural Sciences, Tianjin 300192, China
褪黑素(melatonin, MT)是一种主要由松果体分泌的神经激素,它的合成和分泌均遵循昼夜节律和季节变化,参与机体生殖及免疫调控[1-2]。褪黑素作为强抗氧化剂,可以清除机体活性氧与活性氮,延缓衰老,抑制凋亡基因表达[3],改善体外由于氧化应激引起的各类细胞损伤[3-5]。AANAT及ASMT是褪黑素合成过程中的限速酶,其中,AANAT的活力受到光周期的调控,表现出昼夜节律[6-8]。褪黑素动物模型和临床试验已取得显著成果,临床上逐渐使用褪黑素用于改善人类的内源性昼夜节律失衡,调节睡眠、体温、皮质醇水平和免疫功能失衡问题[9]。褪黑素具有改善卵母细胞质量、保护线粒体功能、提高体外胚胎质量等生物功效[10]。婴儿在4月龄前不分泌褪黑素,主要依靠摄入母乳中的褪黑素来调节生物节律及功能[11]。研究发现,新生儿吮用含褪黑素的牛奶有助于早期昼夜节律的建立,促进免疫系统成熟,增强夜晚睡眠质量和机体抵抗力。老年人补充褪黑素也有助于睡眠,增强机体抵抗力[9, 12-14]。
Yang等[15]研究发现,褪黑素可以显著提高牛奶的乳脂率和乳蛋白率,降低体细胞数量,缓解母畜乳房炎症状,促进幼仔的免疫系统发育并提高其成活率。已有研究运用转基因技术建立了生物反应器,借助血液、膀胱、乳腺和蛋黄、蛋清生产具有生物活性的医用蛋白质类药物,其市场经济价值潜力巨大[16]。运用生物反应器生产褪黑素功能乳或褪黑素功能产品已为今后的研究热点与发展趋势,已有生产乳腺过表达褪黑素合成酶基因绵羊的文献报道[17]。自1980年,Palmiter等[18]将大鼠源生长激素基因显微注入小鼠原核胚中,生产出世界上第一只转基因小鼠后,兔子、猪、小型猪[19]、梅山猪[20]、绵羊[21, 16]、山羊[22]和牛[23]等转基因家畜也相继问世。运用转基因技术建立了人类癌症、老年痴呆和肝炎等遗传性疾病的转基因动物模型。借助转基因技术研究疾病发病机理,进行异种细胞移植治疗,转基因技术变成了研究与治疗人类疾病的技术工具,利用转基因猪肾和其他器官进行人体移植已经发展进入临床试验阶段[24]。与此同时,转基因家畜及农作物新品种的培育、推广与应用也使得转基因生物自身及其产品安全性倍受关注,世界各国相继出台了一系列法律法规来规范转基因生物的研究与应用。
本研究基于前期利用CRISPR/Cas9技术建立的乳腺过表达褪黑素合成酶基因AANAT/ASMT转基因绵羊模型的基础上[17],对转基因绵羊的健康、环境释放及产品安全性进行观测与评估,为AANAT/ASMT转基因绵羊新品种的培育与生产应用奠定数据支撑与安全保障。
1 材料与方法 1.1 试验动物试验绵羊为天津市畜牧兽医研究所试验站的2岁经产转基因杜泊母羊,体重为80~85 kg。所涉及转基因试验样本均为褪黑素合成酶基因特异性乳腺表达的转基因绵羊(n=4)(简称“阳性羊”),非转基因试验样本均为与转基因绵羊同龄的绵羊(n=4)(简称“对照羊”)。
1.2 主要试剂、耗材与仪器PCRMIX、DNA提取试剂盒购自北京天根生物科技公司;琼脂粉购自英国OXOID公司;11.5和15 mL离心管购自Axygen公司;10 mL注射器购自中国杰瑞医用制品公司;4℃离心机购自德国Eppendorf公司;PCR仪购自德国Roche公司;引物由上海生工生物工程有限公司合成;基因测序由北京三博远志生物技术有限责任公司完成。
1.3 试验方法1.3.1 转基因阳性羊的鉴定 剪取1 cm×1 cm大小羊耳组织块,使用试剂盒提取样品DNA,用PCR扩增测序检测转基因后代,PCR反应体系为20 μL:cDNA 0.5 μL,上、下游引物各1 μL,Mix 10 μL,ddH2O 7.5 μL。PCR扩增步骤:95 ℃预变性10 min;95 ℃变性30 s,58 ℃退火30 s,72℃延伸60 s,共35个循环;72 ℃ 10 min后,4 ℃保存。扩增引物序列(由AANAT/ASMT线性载体构建)信息见表 1。
|
|
表 1 PCR扩增引物信息 Table 1 Primer information of PCR amplification |
1.3.2 乳汁、血液收集 母羊产仔后第1天开始收集乳汁,对颜色、气味、粘度进行判断,乳汁是否正常。第15天后连续3 d每天分别于4:00和16:00收集乳汁(每次5 mL),颈部静脉采血(5 mL抗凝集管收集分离血浆,-20℃,避光保存)。血液生化4项及血常规相关指标由天津农学院附属动物医院负责检测并出具检测报告。分别按照阳性羊(n=4)和对照羊(n=4)在不同时间段(4:00和16:00)采集的乳汁和血液进行分组:阳性羊4:00、阳性羊16:00、对照羊4:00、对照羊16:00,每组3个重复12个样本量,用高效液相色谱法检测褪黑素含量。
1.3.3 尿液、粪样收集 保定羊只后,清理尿道口和肛门附近,并在肛门处涂抹凡士林。等待羊自然排尿,用15 mL离心管接3~5 mL尿液,冰上保存,尿常规相关指标由天津农学院附属动物医院负责检测并出具检测报告。在手套上涂抹凡士林,手指于肛门进入直肠,取直肠段粪样,用液氮速冻后干冰运输,由金唯智生物科技有限公司负责对肠道微生物菌群进行16S rRNA测序并出具检测报告。
1.3.4 高效液相色谱法测定褪黑素浓度取 采集的血清或乳汁,使用高效液相色谱法测定待检样品褪黑素浓度。
1.4 统计分析所有获得的原始数据利用Excel整理,SPSS软件统计分析,采用LSD检验进行两两比较,以P < 0.05表示差异显著,试验结果以“平均数±SEM”表示。
2 结果 2.1 转基因绵羊PCR鉴定以转基因羊DNA为模板进行PCR扩增,扩增产物经1%琼脂糖凝胶电泳检测,结果见图 1,阳性羊AANAT基因扩增片段大小为487 bp,ASMT基因阳性扩增片段大小为742 bp,检测片段的大小与预期目标片段的大小一致。目标片段经切胶回收后测序,测序结果与GenBank数据库中AANAT mRNA(KC290949.1)和ASMT mRNA(KC290950.1)收录的序列一致。PCR鉴定共获得4只阳性转基因杜泊羊,其中乳腺特异表达AANAT蛋白的阳性羊2只,ASMT阳性羊1只,AANAT和ASMT共表达阳性羊1只。

|
A、B. AANAT基因鉴定结果; C、D. ASMT基因鉴定结果 A, B. PCR result of AANAT gene; C, D. PCR result of ASMT gene 图 1 AANAT和ASMT基因的PCR鉴定结果 Fig. 1 Detection of AANAT and ASMT genes by PCR amplification |
乳腺过表达褪黑素合成酶绵羊分别于0、6和12月龄测定体重、体长、身高和胸围等体尺数据。结果见2图,图 2A-D分别是体重、体长、身高和胸围变化趋势,乳腺过表达褪黑素合成酶转基因绵羊(n=4)的生长性能与非转基因绵羊(n=4)均无显著性差异(P>0.05)。

|
图 2 转基因羊生长性能统计 Fig. 2 Statistics of growth performance of transgenic sheep |
2.3.1 转基因绵羊血液生化4项 采集转基因绵羊(n=4)和非转基因对照组绵羊(n=4)血液,统计分析发现,检测绵羊血液总蛋白(P=0.608>0.05)、白蛋白(P=0.057>0.05)、球蛋白(P=0.186>0.05)和胆固醇(P=0.065>0.05)4项指标在转基因与非转基因绵羊间均没有显著性差异,见图 3。

|
图 3 转基因羊体血液生化指标统计 Fig. 3 Statistics of blood biochemical indicators of transgenic sheep |
2.3.2 转基因绵羊血液生理指标 采集绵羊全血用于血液生理指标测定,结果见表 2,转基因阳性绵羊血液中各类细胞组分与对照羊一致,白细胞、红细胞、血小板各项生理指标均在参考范围之内,白细胞系统中除单核细胞含量略高于对照羊之外,其余指标均低于对照羊但差异未达到显著水平。红细胞系统中除平均血红蛋白量其余指标均低于对照羊但差异未达到显著水平;阳性羊与对照羊之间血液生理指标均无显著差异(P>0.05)。
|
|
表 2 转基因绵羊血液生理指标 Table 2 Blood physiological indexes of transgenic sheep |
2.3.3 转基因绵羊尿液生理指标 采集转基因绵羊(n=4)和对照羊(n=4)尿液进行尿常规检测。结果见图 4和表 3,转基因阳性羊与同批次对照羊在相同的饲养条件下,尿液SG比重无显著性差异。正常的尿液为弱酸性,但受饲粮结构影响,转基因阳性羊与对照羊检测结果均为弱碱性。尿液中只存在微量的白细胞、红细胞亚硝酸盐、酮体、胆红素、尿胆元、蛋白质和葡萄糖,若定性检测结果显示阳性则是机体感染疾病的信号,定性检测为阴性说明绵羊机体无炎症反应。转基因绵羊与对照羊尿液各项生理指标均正常且无显著差异(P>0.05)。
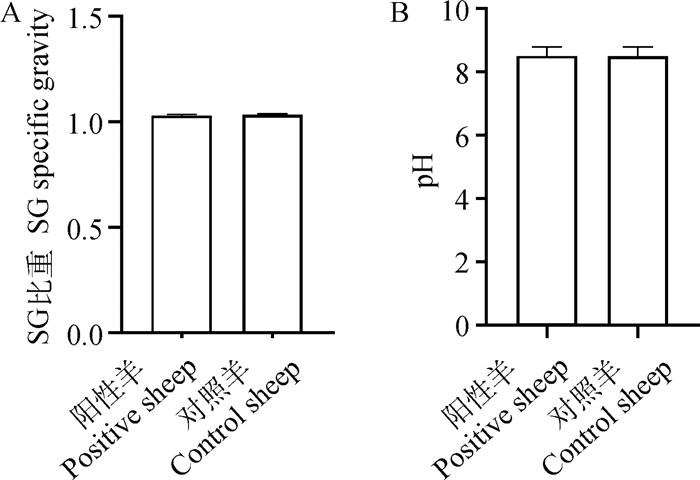
|
图 4 转基因羊尿液生理指标统计 Fig. 4 Statistics of urine physiological indicators of transgenic sheep |
|
|
表 3 转基因绵羊尿液生理指标测定结果 Table 3 Determination results of urine physiological indicators of transgenic sheep |
分别采集转基因绵羊(n=4)和对照羊(n=3)直肠段粪便,液氮速冻之后干冰运输,16S rRNA测量粪便微生物群落组成。对两组样品进行分类,E组为对照羊(n=3),F组为转基因阳性羊(n=4)。对阳性羊和对照羊粪样菌群在属的分类水平下的物种分布进行分析(图 5)。图 5A为在属水平上对两组菌群进行比较,对照羊和转基因羊粪样菌属组成无明显差异,优势菌群差异不显著。图 5B为物种聚类进化树,比较菌群丰度差异。红色背景区域和绿色背景区域表示不同的分组,树枝中红色节点表示在E组中起到重要作用的微生物类群,绿色节点表示在F组中起到重要作用的微生物类群,黄色节点表示的是在两组中均没有起到重要作用的微生物类群。

|
图A中,不同面积代表菌属的数量。粪样中的5种优势菌群排列为:瘤胃球菌(Ruminococcaceae)、理研菌(Rikenellaceae)、毛螺旋菌(Lachnospiraceae)、厚壁菌门中的克里斯滕森菌(Christenella)、拟杆菌(Bacteeoides)。瘤胃球菌、理研菌和毛螺旋菌是瘤胃液的主要菌群,克里斯滕森菌是将糖发酵产生挥发性脂肪酸的主要菌群,拟杆菌则是降解消化纤维,提高宿主免疫系统的主要菌群。图B显示,在对照组绵羊肠道中起重要作用的2种菌群为厚壁菌,而在转基因阳性绵羊肠道中起重要作用的3种菌群均为拟杆菌 In figure A, different areas represent the number of genera. The 5 dominant bacterial groups in stool are: Ruminococcaceae, Rikenellaceae, Lachnospiraceae, Christenella, Bacteeoides. Ruminococcaceae, Rikenellaceae and Lachnospiraceae are the main flora in rumen fluid, Christenella is the main flora of fermenting sugar to produce volatile fatty acids, Bacteeoides is the main flora that degrades digestive fiber and improves the host's immune system. In figure B, the two bacterial groups that play an important role in the sheep intestine of the control group are Chlamydia, while the 3 bacterial groups that play an important role in the intestine of the transgenic sheep are Bacteroides 图 5 菌群物种结构差异分析 Fig. 5 Analysis of species structure differences in the flora |
产后第15天分别采集转基因绵羊(n=4)和对照组绵羊(n=4)4:00和16:00时的羊乳及血液各5 mL,连续3 d,测定乳样和血清褪黑素含量。转基因绵羊4:00时奶中褪黑素含量均显著高于其16:00时褪黑素含量(P<0.05),转基因羊和对照羊4:00时血清褪黑素含量显著高于16:00时褪黑素含量(P<0.05)。不同绵羊在同一时间点,阳性羊乳中褪黑素含量均极显著高于对照羊(P<0.05),但血中褪黑素含量在两组羊间无显著差异(P>0.05,图 6)。
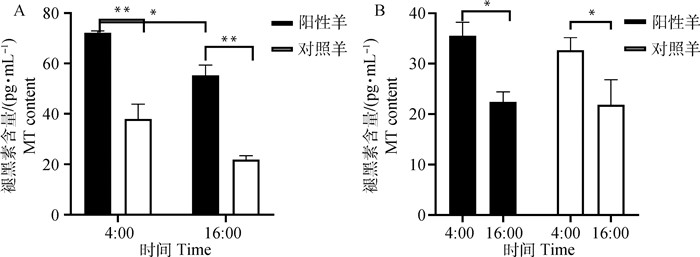
|
A.不同时间段奶样褪黑素含量检测;B.不同时间段血清褪黑素含量检测。*.P < 0.05;**. P < 0.01,下同 A. Detection of melatonin content in milk at different periods; B. Detection of melatonin content in serum at different periods.*.P < 0.05;**. P < 0.01, the same as below 图 6 绵羊乳和血清中褪黑素含量检测结果 Fig. 6 Detection results of melatonin content in milk and serum of sheep |
采集转基因阳性羊(n=4)和对照羊(n=4)产后第15天羊乳5 mL,连续3 d。对乳脂率、乳蛋白、乳糖和尿素氮含量等指标进行测定。其中,两组间乳脂率、乳蛋白、非脂乳固形物、总干物质含量和尿素氮浓度均无显著差异(P>0.05),转基因阳性羊乳糖含量显著高于对照羊(P<0.05),体细胞数量极显著低于对照羊(P<0.01,图 7)。
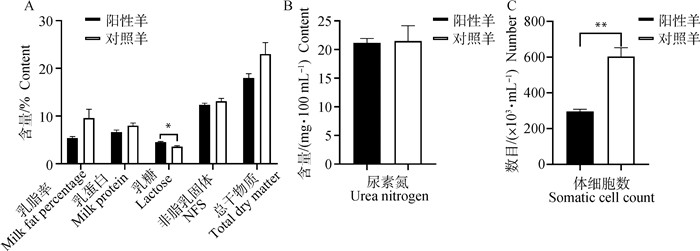
|
A.奶样乳脂率、乳蛋白、乳糖、非脂乳固形物和总干物质含量;B.奶样尿素氮含量;C.奶样体细胞数 A. Milk fat percentage, milk protein, lactose, non-fat solids and total dry matter contents in milk; B. Urea nitrogen content in milk; C. Somatic cell count in milk 图 7 转基因羊乳成分测定结果 Fig. 7 Determination results of milk composition of transgenic sheep |
转基因是近年来用于探索生殖、疾病、遗传等科学奥秘的热门课题。用CRISPR建立了人类癌症、神经病学、心血管系统、免疫缺陷疾病模型[25]。使用基因编辑技术使生物机体产生可遗传和适应性免疫变化,已获得了能够稳定遗传的基因编辑的山羊、绵羊、小鼠等动物[26-29]。随后,这一技术在家畜育种中广泛应用,相继获得突变beta-lactoglobulin (BLG)基因的转基因牛[30],突变myostatin(MSTN)基因的梅山猪[20],突变SP110基因的抗结核病的牛[31],过表达AANAT基因的转基因绵羊[17]。
由于克隆动物重编程过程会出现异常状态,所以能成功分娩并存活的克隆动物也存在安全隐患。本研究中,对0、6和12月龄转基因绵羊测定体重、体长、身高和胸围等体尺数据,生长期与成年期的生长曲线证明,转基因绵羊生长性能与对照组绵羊差异不显著。对生长期的体细胞克隆牛与非克隆牛进行体重、体高、胸厚、腰围监测,结果发现,这些指标的增长速度和成年胴体重均无显著差异[32],体细胞克隆绵羊的出生体重和臀长、腹围、胸围、体重等生长指标与对照组绵羊相比没有显著差异[33],对体细胞核移植山羊和猪的生长性能检测也得到了相同的结论[34]。
尿液生理生化指标与肾功能相关,血液生理生化参数反映了机体代谢、营养和疾病情况,血液是生理学、病理学以及毒理学的重要参考指标[35]。本研究对转基因绵羊血液和尿液生理生化指标进行了检测,结果表明,转基因绵羊的血液生化组分没有发生改变,各组分含量与对照组无显著差异。体细胞核移植牛和猪血液中红细胞、白细胞、血细胞比容等生理生化指标与对照组相比也未显示出显著性差异[36-38]。本研究对转基因阳性羊尿液组分及生化指标的检测结果与对照组羊之间无显著差异。肠道菌群与机体吸收代谢息息相关,菌群群落结构和组成丰度是反映生物体健康状况的重要指标,Xu等[39]通过16S rDNA测序对转化编码人类溶菌酶(hLZ)、乳铁蛋白和人α乳清蛋白基因的转基因奶牛进行了牛肠道微生物群落的研究,结果表明,转基因对牛肠道的微生物群落没有显著影响。通过对转基因绵羊的粪便进行微生物菌群测序结果显示,转基因绵羊与正常羊肠道微生物群落组成和优势菌群无显著差异。
对克隆动物产品进行安全检测也是生物安全评价研究的重要环节,研究者对不同品种克隆奶牛及不同泌乳周期的牛奶成分进行分析后发现,体细胞核移植荷斯坦奶牛的牛乳总化学成分(总固体、脂肪、乳糖、蛋白质含量等)与对照牛乳无显著差异[40-42],且各个成分之间的比率与非克隆牛牛乳差别不大[42]。本研究对转基因绵羊乳成分分析发现,两组间除乳糖含量有显著差异外,其余组分未显示出显著差异。有研究对克隆牛进行乳腺健康检测,结果显示,奶中体细胞数均在正常值范围内[43]。本研究中,转基因绵羊乳体细胞数显著低于对照组羊则可能与试验转基因绵羊中乳腺过表达褪黑素合成酶基因有关,与褪黑素具有抗炎的结论相符[44]。体细胞核移植的奶牛具有正常的生长、生殖与泌乳特性,但总产奶量与对照组有明显差异[45]。使用体细胞核移植牛乳与牛肉进行大鼠90 d饲喂试验,结果与对照组无显著差异[46]。本试验因条件限制没有检测总产奶量和大鼠饲喂试验。
为了确保转基因生物的安全性,需要对转基因动物、后代及可能影响其性能的指标进行全面分析,Mehrotra和Goyal[47]认为,全面的表型和分子检测是真正检验转基因生物安全的措施之一。本研究对转基因绵羊表型生长性状、血液尿液生理生化指标、粪便微生物群落及乳成分进行了全面细致的分析,结果显示,转基因绵羊与对照羊间在各指标均无显著差异,可推测乳腺过表达褪黑素合成酶AANAT及ASMT基因绵羊是健康安全的。
4 结论乳腺过表达AANAT及ASMT基因阳性绵羊与对照组羊之间在生长性状、血液和尿液生理生化指标以及肠道微生物组成等均无显著差异。两组间血清褪黑素水平无显著差异,而乳中褪黑素含量阳性转基因羊显著高于对照组羊;阳性绵羊乳组分与对照组羊基本保持一致,乳糖含量显著高于对照绵羊,体细胞数含量极显著降低,验证了褪黑素的抗炎作用,其余组分含量与对照组绵羊相当。
| [1] | ROBINSON J J, WIGZELL S, AITKEN R P, et al. Daily oral administration of melatonin from March onwards advances by 4 months the breeding season of ewes maintained under the ambient photoperiod at 57°N[J]. Anim Reprod Sci, 1992, 27(2-3): 141–160. DOI: 10.1016/0378-4320(92)90054-H |
| [2] | BROWN A J, PENDERGAST J S, YAMAZAKI S. Peripheral circadian oscillators[J]. Yale J Biol Med, 2019, 92(2): 327–335. |
| [3] | ACUÑA-CASTROVIEJO D, ESCAMES G, VENEGAS C, et al. Extrapineal melatonin:sources, regulation, and potential functions[J]. Cell Mol Life Sci, 2014, 71(16): 2997–3025. DOI: 10.1007/s00018-014-1579-2 |
| [4] | DE CASTRO T B, BORDIN-JUNIOR N A, DE ALMEIDA E A, et al. Evaluation of melatonin and AFMK levels in women with breast cancer[J]. Endocrine, 2018, 62(1): 242–249. DOI: 10.1007/s12020-018-1624-2 |
| [5] | PENG W, LEI M T, ZHANG J, et al. The protective effect of melatonin on the in vitro development of yak embryos against hydrogen peroxide-induced oxidative injury[J]. Zygote, 2019, 27(3): 118–125. DOI: 10.1017/S0967199418000412 |
| [6] | COELHO L A, ANDRADE-SILVA J, MOTTA-TEIOXEIRA L C, et al. The absence of pineal melatonin abolishes the daily rhythm of Tph1(Tryptophan Hydroxylase 1), Asmt (Acetylserotonin O-Methyltransferase), and Aanat (Aralkylamine N-Acetyltransferase) mRNA expressions in rat testes[J]. Mol Neurobiol, 2019, 56(11): 7800–7809. DOI: 10.1007/s12035-019-1626-y |
| [7] | YAMANAKA Y, YAMADA Y, HONMA K I, et al. Cryptochrome deficiency enhances transcription but reduces protein levels of pineal Aanat[J]. J Mol Endocrinol, 2018, 61(4): 219–229. DOI: 10.1530/JME-18-0101 |
| [8] | CAUSTON H C. Metabolic rhythms:A framework for coordinating cellular function[J]. Eur J Neurosci, 2020, 51(1): 1–12. |
| [9] | ARENDT J, SKENE D J. Melatonin as a chronobiotic[J]. Sleep Med Rev, 2005, 9(1): 25–39. DOI: 10.1016/j.smrv.2004.05.002 |
| [10] | ZOU H J, CHEN B L, DING D, et al. Melatonin promotes the development of immature oocytes from the COH cycle into healthy offspring by protecting mitochondrial function[J]. J Pineal Res, 2020, 68(1): e12621. |
| [11] | JALDO-ALBA F, MUÑOZ-HOYOS A, MOLINA-CARBALLO A, et al. Light deprivation increases plasma levels of melatonin during the first 72 h of life in human infants[J]. Acta Endocrinol, 1993, 129(5): 442–445. DOI: 10.1530/acta.0.1290442 |
| [12] | MCKENNA H, REIES I K M. The case for a chronobiological approach to neonatal care[J]. Early Hum Dev, 2018, 126: 1–5. DOI: 10.1016/j.earlhumdev.2018.08.012 |
| [13] | JAN J E, WASDELL M B, FREEMAN R D, et al. Evidence supporting the use of melatonin in shortgestation infants[J]. J Pineal Res, 2007, 42(1): 22–27. DOI: 10.1111/j.1600-079X.2006.00398.x |
| [14] | BAGHBAN R S, MOHEBBI A, VAKILZADEH G, et al. Enhancement of therapeutic DNA vaccine potency by melatonin through inhibiting VEGF expression and induction of antitumor immunity mediated by CD8+ T cells[J]. Arch Virol, 2018, 163(3): 587–597. DOI: 10.1007/s00705-017-3647-z |
| [15] | YANG M H, SHI J M, TIAN J H, et al. Exogenous melatonin reduces somatic cell count of milk in Holstein cows[J]. Sci Rep, 2017, 7(1): 43280. |
| [16] | WANG L, ZHAO Y, REITER R J, et al. Changes in melatonin levels in transgenic 'Micro-Tom' tomato overexpressing ovine AANAT and ovine HIOMT genes[J]. J Pineal Res, 2014, 56(2): 134–142. DOI: 10.1111/jpi.12105 |
| [17] | MA T, TAO J L, YANG M H, et al. An AANAT/ASMT transgenic animal model constructed with CRISPR/Cas9 system serving as the mammary gland bioreactor to produce melatonin-enriched milk in sheep[J]. J Pineal Res, 2017, 63(1): e12406. DOI: 10.1111/jpi.12406 |
| [18] | PALMITER R D, BRINSTER R L, HAMMER R E, et al. Dramatic growth of mice that develop from eggs microinjected with metallothionein-growth hormone fusion genes[J]. Nature, 1982, 300(5893): 611–615. DOI: 10.1038/300611a0 |
| [19] | UCHIDA M, SHIMATSU Y, ONOE K, et al. Production of transgenic miniature pigs by pronuclear microinjection[J]. Transgenic Res, 2001, 10(6): 577–582. DOI: 10.1023/A:1013059917280 |
| [20] | QIAN L L, TANG M X, YANG J Z, et al. Targeted mutations in myostatin by zinc-finger nucleases result in double-muscled phenotype in Meishan pigs[J]. Sci Rep, 2015, 5(1): 14435. DOI: 10.1038/srep14435 |
| [21] | TIAN X Z, LV D Y, MA T, et al. AANAT transgenic sheep generated via OPS vitrified-microinjected pronuclear embryos and reproduction efficiency of the transgenic offspring[J]. PeerJ, 2018, 6: e5420. DOI: 10.7717/peerj.5420 |
| [22] | WANG B, BALDASSARE H, TAO T, et al. Transgenic goats produced by DNA pronuclear microinjection of in vitro derived zygotes[J]. Mol Reprod Dev, 2002, 63(4): 437–443. DOI: 10.1002/mrd.10199 |
| [23] | WANG M, SUN Z L, ZOU Z Y, et al. Efficient targeted integration into the bovine Rosa26 locus using TALENs[J]. Sci Rep, 2018, 8(1): 10385. |
| [24] | COOPER D K C, GASTON R, ECKHOFF D, et al. Xenotransplantation-the current status and prospects[J]. Br Med Bull, 2018, 125(1): 5–14. DOI: 10.1093/bmb/ldx043 |
| [25] | RODRÍGUEZ-RODRÍGUEZ D R, RAMÍREZ-SOLÍS R, GARZA-ELIZONDO M A, et al. Genome editing:A perspective on the application of CRISPR/Cas9 to study human diseases (Review)[J]. Int J Mol Med, 2019, 43(4): 1559–1574. |
| [26] | BEHLER J, HESS W R. Approaches to study CRISPR RNA biogenesis and the key players involved[J]. Methods, 2020, 172: 12–26. DOI: 10.1016/j.ymeth.2019.07.015 |
| [27] | KALDS P, ZHOU S W, CAI B, et al. Sheep and goat genome engineering:from random transgenesis to the CRISPR era[J]. Front Genet, 2019, 10: 750. DOI: 10.3389/fgene.2019.00750 |
| [28] | SUN N, PETIWALA S, WANG R, et al. Development of drug-inducible CRISPR-Cas9 systems for large-scale functional screening[J]. BMC Genomics, 2019, 20(1): 225. DOI: 10.1186/s12864-019-5601-9 |
| [29] | KITAMOTO K, TAKETANI Y, FUJII W, et al. Gene-ration of mouse model of TGFBI-R124C corneal dystrophy using CRISPR/Cas9-mediated homology-directed repair[J]. Sci Rep, 2020, 10(1): 2000. |
| [30] | SUN Z L, WANG M, HAN S W, et al. Production of hypoallergenic milk from DNA-free beta-lactoglobulin (BLG) gene knockout cow using zinc-finger nucleases mRNA[J]. Sci Rep, 2018, 8(1): 15430. DOI: 10.1038/s41598-018-32024-x |
| [31] | WU H B, WANG Y S, ZHANG Y, et al. TALE nickase-mediated SP110 knockin endows cattle with increased resistance to tuberculosis[J]. Proc Natl Acad Sci U S A, 2015, 112(13): E1530–E1539. DOI: 10.1073/pnas.1421587112 |
| [32] | SHIGA K, UMEKI H, SHIMURA H, et al. Growth and fertility of bulls cloned from the somatic cells of an aged and infertile bull[J]. Theriogenology, 2005, 64(2): 334–343. DOI: 10.1016/j.theriogenology.2004.12.002 |
| [33] | EDWARDS L, PEURA T, HARTWICH K, et al. Postnatal growth and circulating ACTH and cortisol concentratoins during the first month of life in cloned lambs[J]. Endocrinology, 2002, 143(9): 3699. DOI: 10.1210/en.2002-220500 |
| [34] | CHEN G S, SUI Y. Production, performance, slaughter characteristics, and meat quality of Ziwuling wild crossbred pigs[J]. Trop Anim Health Prod, 2018, 50(2): 365–372. DOI: 10.1007/s11250-017-1441-2 |
| [35] |
乔娜, 干驰, 朱佳琪, 等. 转hLF基因山羊血液生理生化指标检测分析[J]. 南京农业大学学报, 2016, 39(3): 483–487.
QIAO N, GAN C, ZHU J Q, et al. Hematological and biochemical indices in blood of hLF transgenic goat[J]. Journal of Nanjing Agricultural University, 2016, 39(3): 483–487. (in Chinese) |
| [36] | CHAVATTE-PALMER P, HEYMAN Y, RICHARD C, et al. Clinical, hormonal, and hematologic characteristics of bovine calvesderived from nuclei from somatic cells[J]. Biol Reprod, 2002, 66(6): 1596–1603. DOI: 10.1095/biolreprod66.6.1596 |
| [37] | KIM E Y, SONG D H, PARK M J, et al. Post-death cloning of endangered Jeju black cattle (Korean Native Cattle):fertility and serum chemistry in a cloned bull and cow and their offspring[J]. J Reprod Dev, 2013, 59(6): 536–543. DOI: 10.1262/jrd.2013-047 |
| [38] | WANG M F, FENG S F, MA G J, et al. Whole-genome methylation analysis reveals epigenetic variation in cloned and donor pigs[J]. Front Genet, 2020, 11: 23. DOI: 10.3389/fgene.2020.00023 |
| [39] | XU J X, ZHAO J, WANG J W, et al. Molecular-based environmental risk assessment of three varieties of genetically engineered cows[J]. Transgenic Res, 2011, 20(5): 1043–1054. DOI: 10.1007/s11248-010-9477-3 |
| [40] | WALSH M K, LUCEY J A, GOVINDASAMY-LUCEY S, et al. Comparison of milk produced by cows cloned by nuclear transfer with milk from non-cloned cows[J]. Cloning Stem Cells, 2003, 5(3): 213–219. DOI: 10.1089/153623003769645875 |
| [41] | MONTAZER-TORBATI F, BOUTINAUD M, BRUN N, et al. Differences during the first lactation between cows cloned by somatic cell nuclear transfer and noncloned cows[J]. J Dairy Sci, 2016, 99(6): 4778–4794. DOI: 10.3168/jds.2015-10532 |
| [42] | YONAI M, KANEYAMA K, MIYASHITA N, et al. Growth, reproduction, and lactation in somatic cell cloned cows with short telomeres[J]. J Dairy Sci, 2005, 88(11): 4097–4110. DOI: 10.3168/jds.S0022-0302(05)73094-0 |
| [43] | LAIBLE G, BROPHY B, KNIGHTON D, et al. Compositional analysis of dairy products derived from clones and clonedtransgenic cattle[J]. Theriogenology, 2007, 67(1): 166–177. DOI: 10.1016/j.theriogenology.2006.09.028 |
| [44] | TAO J L, YANG M H, WU H, et al. Effects of AANAT overexpression on the inflammatory responses and autophagy activity in the cellular and transgenic animal levels[J]. Autophagy, 2018, 14(11): 1850–1869. DOI: 10.1080/15548627.2018.1490852 |
| [45] | TAKAHASHI M, TSUCHIYA H, HAMANO S, et al. Clinical study report on milk production in the offspring of a somatic cell cloned Holstein cow[J]. J Reprod Dev, 2013, 59(6): 595–598. DOI: 10.1262/jrd.2013-025 |
| [46] | HEYMAN Y, CHAVATTE-PALMER P, BERTHELOT V, et al. Assessing the quality of products from cloned cattle:An integrative approach[J]. Theriogenology, 2007, 67(1): 134–141. DOI: 10.1016/j.theriogenology.2006.09.020 |
| [47] | MEHROTRA S, GOYAL V. Evaluation of designer crops for biosafety-A scientist's perspective[J]. Gene, 2013, 515(2): 241–248. DOI: 10.1016/j.gene.2012.12.029 |



