2. 中江县农产品质量安全检验检测站, 德阳 618100;
3. 四川省畜牧科学研究院, 成都 610066
2. Agricultural Product Quality and Safety Inspection and Testing Station of Zhongjiang County, Deyang 618100, China;
3. Sichuan Academy of Animal Science, Chengdu 610066, China
屎肠球菌(Enterococcus faecium)属于革兰阳性细菌,常定植在人或动物的肠道中,为肠道正常菌群。然而近年来由于屎肠球菌抗万古霉素菌株的出现,该细菌导致的疾病案例也越来越多,已经跃居成为院内感染的病原菌第三名[1-3]。该菌在临床上可引起重要感染包括泌尿道感染、菌血症、心内膜炎、憩室炎和脑膜炎等[4-5]。同时该菌对多种药物如β-内酰胺类、氨基糖苷类抗生素等天然耐药,因此研究屎肠球菌耐药性的演变对该菌的药物防控及新药研发可提供理论支撑[6]。
大环内酯类、林可酰胺类和链阳霉抗生素B抗生素(MLSB表型)通常作为耐甲氧西林葡萄球菌(MRSA)感染首选用药,特别适用于皮肤和体表软组织感染及针对青霉素过敏患者[7-8]。然而临床上药物滥用已导致MLSB抗性的金黄色葡萄球菌菌株不断增加,其耐药基因ErmA在临床上也不断被检出,关于该耐药基因传播方式主要以转座子Tn554介导,这使得该耐药基因极易在葡萄球菌种内传播[9-12]。然而关于该耐药基因在屎肠球菌中的传播鲜有报道,目前已开始出现相关研究,若该耐药基因传递到屎肠球菌中,将会对感染屎肠球菌治疗带来巨大的挑战,尤其是耐万古霉素屎肠球菌的出现,因此研究该基因种间传播机制具有极大的意义,同时对屎肠球菌专用抗生素开发也提出了一定的要求,以避免这种肠道常驻菌演变成超级细菌而无有效的治疗药物[13-14]。
基因岛是不同种细菌间进行水平转移的大片段基因组区域,通常这些大片段基因区域参与细菌对环境的适应性变化,包含细菌定植(致病性)、抗生素抗性、共生及新陈代谢等,因此基因岛预测对于细菌进化研究具有极大的意义[15-17]。本研究通过对送检的死亡林麝内脏进行细菌学分离鉴定,得到一株屎肠球菌,命名为屎肠球菌LS170308。之后对该菌进行全基因组测序,并对该基因组进行基因岛预测、结构及亲缘性分析。
1 材料与方法 1.1 样本来源样品来源于陕西镇坪县某林麝养殖场多头死亡林麝内脏组织。
1.2 细菌分离与鉴定规范采集内脏组织进行表面消毒,使用灭菌手术刀片无菌操作切取约1 cm3心、肝等内脏组织,切块接种于10 mL添加有5%无菌血清的TSB液体培养基中,37 ℃分别有氧及厌氧环境下培养16 h。将培养物划线接种于无菌脱纤维绵羊血平板上恒温培养16 h,挑取单个菌落进行扩大培养,提取DNA进行16S rDNA PCR扩增并测序分析。
1.3 药物敏感性试验按照CLSI(2017)指导的K-B药敏扩散法对分离菌进行药物敏感性试验。
1.4 全基因组测序及组装抽提分离菌DNA,选用第三代测序技术PacBio RS Ⅱ测序系统进行de nove测序。将测序返回的原始数据进行处理后利用SMRT Analysis软件2.3.0(Pacific Biosciences)中的HGAP.3进行基因组装配。将组装后的结果上传至NCBI,获得GenBank登录号。
1.5 基因岛预测使用GIPSy与IslandViewer 4预测软件,以Enterococcus faecium DO作为参考菌株进行基因岛预测,将两者预测结果进行比对分析[18]。
2 结果 2.1 菌株鉴定本研究通过对送检的死亡林麝内脏进行细菌学分离鉴定,在厌氧及需氧环境培养的心、肝组织中分别得到多株屎肠球菌(生化鉴定结果见表 1),16S rDNA测序比对表明上述多株屎肠球菌16S rDNA序列相似性为100%,证实为同一株菌,结果如图 1所示,命名为屎肠球菌LS170308。之后对该菌进行全基因组测序,组装成一条染色体序列(GenBank登录号:CP025077)及一条质粒序列(GenBank登录号:CP025078),进一步对该染色体及质粒进行生物信息学分析。
|
|
表 1 屎肠球菌分离株的生化鉴定 Table 1 Bacterial biochemical identification of isolated Enterococcus faecium strains |

|
图 1 屎肠球菌分离菌株及相关菌株基于16S rDNA的系统发育树 Fig. 1 Phylogenetic tree of isolated and reference Enterococcus faecium strains based on 16S rDNA |
对屎肠球菌LS170308菌株按照CLSI指导的K-B药敏纸片法进行药敏试验结果见表 2。该菌株存在多重耐药性,对阿莫西林、替卡西林、头孢菌素、多西环素、庆大霉素、卡那霉素、妥布霉素、复方新诺明、诺氟沙星、甲氧苄氨嘧啶完全耐药,值得注意的是该菌株对青霉素及链霉素存在一定程度的敏感性;对环丙沙星、丁胺卡那、克林霉素、红霉素等抗生素中介;对氯霉素、氟苯尼考、呋喃妥因抗生素敏感,药物浓度均为30 μg·片-1。
|
|
表 2 屎肠球菌LS170308株药敏试验 Table 2 Drug resistance test results of Enterococcus faecium LS170308 strain |
由于基因岛预测方法各不相同,预测结果也会有所偏差,故本研究同时采用IslandViewer 4和GIPSy进行基因岛预测,将两者的结果进行对比分析。其中IslandViewer 4预测出13个基因岛,GIPSy预测出12个基因岛,通过两者预测结果比较分析确定4个岛为耐药基因岛,如表 3所示。
|
|
表 3 屎肠球菌LS170308株耐药基因岛的GIPSy预测情况 Table 3 GIPSy predicted Enterococcus faecium LS170308 strain resistant island results |
进一步对上述4个耐药基因岛进行逐个分析,仅发现PRI1和PRI4基因岛上携带有耐药基因,其中PRI1携带有ANT(9)、ErmA和AAC(6′)-le-APH(2″)-la,PRI4携带有DfrG耐药基因;PRI2和PRI3携带有与耐药相关的调节基因,如多重药物外排蛋白家族。同时针对上述4个基因岛进行亲缘性分析(图 2~5),结果表明PRI1、PRI4与葡萄球菌亲缘性最高,结果如图 2、5所示;PRI2、PRI3与屎肠球菌亲缘性最高,结果见图 3、4,值得注意的是NCBI数据库中携带有PRI3基因岛的菌株仅有5株,且均为屎肠球菌。

|
图 2 屎肠球菌LS170308和相关参考株基于耐药基因岛PRI1的系统发育树 Fig. 2 Phylogenetic tree of Enterococcus faecium LS170308 and related reference strains based on the drug-resistant genomic island PRI1 |
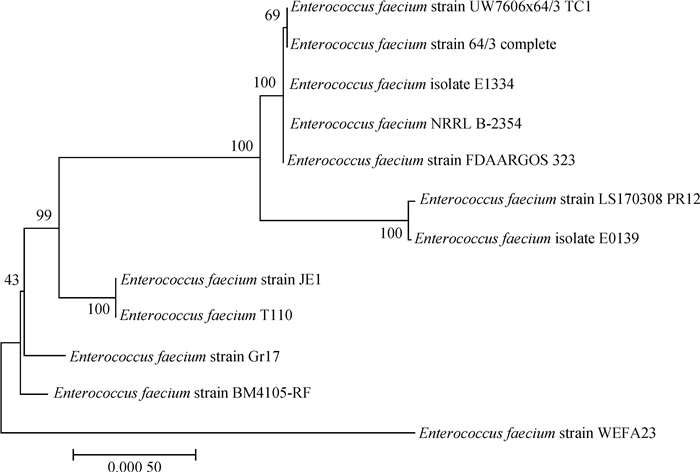
|
图 3 屎肠球菌LS170308和相关参考株基于耐药基因岛PRI2的系统发育树 Fig. 3 Phylogenetic tree of Enterococcus faecium LS170308 and related reference strains based on the drug-resistant genomic island PRI2 |
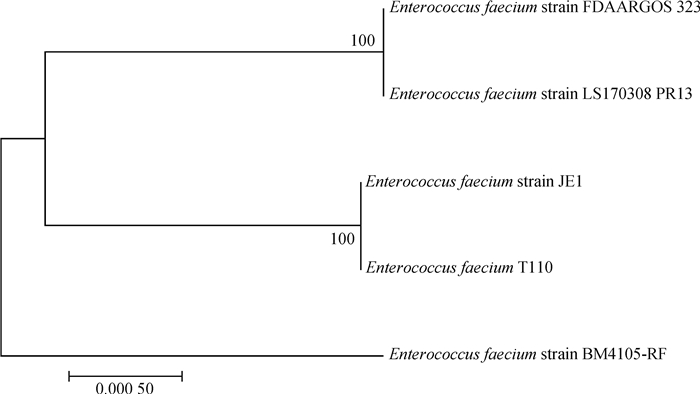
|
图 4 屎肠球菌LS170308和相关参考株基于耐药基因岛PRI3的系统发育树 Fig. 4 Phylogenetic tree of Enterococcus faecium LS170308 and related reference strains based on the drug-resistant genomic island PRI3 |
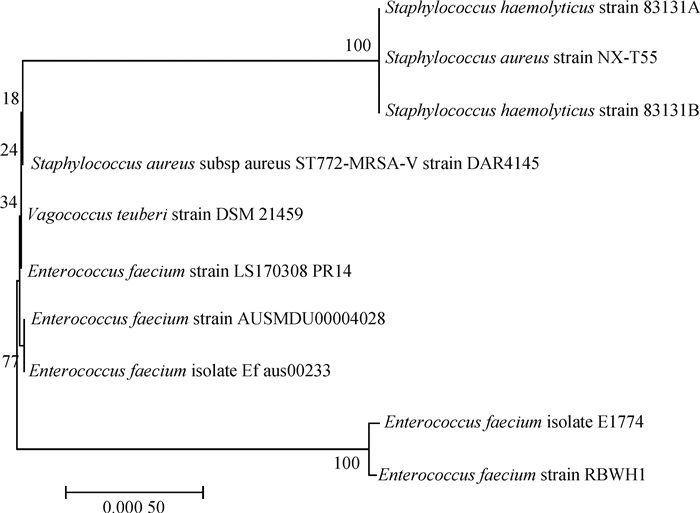
|
图 5 屎肠球菌LS170308和相关参考株基于耐药基因岛PRI4的系统发育树 Fig. 5 Phylogenetic tree of Enterococcus faecium LS170308 and related reference strains based on the drug-resistant genomic island PRI4 |
在屎肠球菌LS170308染色体上找到PRI1位置,并在其前后截取1 000 bp序列合计13 552 bp进行序列分析,使用ORFfinder工具查找这段序列的开放阅读框,筛选后对这段序列进行基因功能注释,注释结果见图 6。
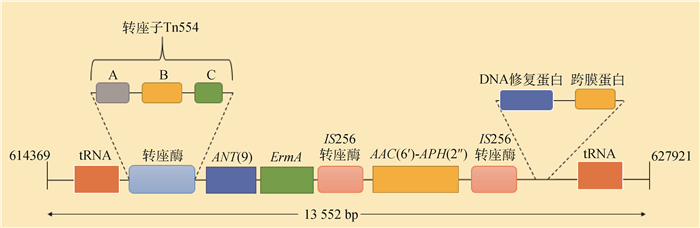
|
图 6 屎肠球菌LS170308耐药基因岛PRI1结构 Fig. 6 Structure of the drug-resistant genomic island PRI1 of Enterococcus faecium LS170308 |
将基因岛PRI1序列上传至NCBI进行blast比对,建立进化树分析其亲缘关系,表明该基因岛与金黄色葡萄球菌亲缘性最近,进一步分别对ANT(9)、ErmA、AAC(6′)-APH(2″)、转座酶Tn554、IS256插入元件等进行亲缘性分析,发现其与金黄色葡萄球菌Mu50株亲缘关系最近,详细结果见表 4。
|
|
表 4 耐药基因岛PRI1上部分基因亲缘关系 Table 4 The genetic relationship of part genes from the drug-resistant genomic island PRI1 |
进一步分析发现PRI1与其他菌株耐药岛差异在于PRI1插入了两个IS256插入元件,并携带AAC(6′)-APH(2″)耐药基因,见图 7。
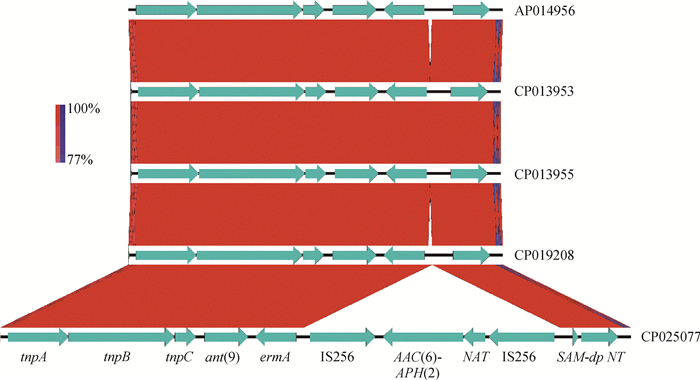
|
图 7 屎肠球菌LS170308耐药基因岛PRI1结构比对 Fig. 7 Structural comparison of the drug-resistant genomic island PRI1 among Enterococcus faecium LS170308 and other strains |
从死亡林麝内脏组织中分离到一株屎肠球菌,命名为屎肠球菌LS170308株。尽管目前研究尚不能说明该菌株感染是导致林麝出现严重败血症的主要病因,但该菌株具有较强的机体侵袭力,同时药敏试验结果证实该菌株出现多重耐药,耐药程度严重。通过全基因测序发现该菌染色体及质粒上携带efmA、ErmA、dfrG、AAC(6′)-Ie-APH(2″)-Ia、AAC(6′)-Ii、eatAv、liaS 7、cat等耐药基因,与临床耐药表征较符合。该菌株携带有cat基因,但药敏结果证实该菌株对氯霉素类抗生素敏感,根据文献报道分析原因可能是cat基因包含cat A型和cat B型,需要同时存在上述基因才会表现出氯霉素抗性[19]。尽管屎肠球菌基因组不存在相关β-内酰胺类药物失活酶,但该菌对此类药物几乎完全耐药,分析可能是青霉素结合蛋白(PBPS)靶位点突变或被替换所致,资料显示屎肠球菌PBP5不能与β-内酰胺类抗生素结合,且PBP5可能参与了屎肠球菌分裂过程中细胞壁合成的重要部分,因而屎肠球菌对β-内酰胺类抗生素具有天然的耐药优势[20]。该菌株携带多个针对氨基糖苷类药物的抗性基因包括AAC(6′)-le-APH(2″)-la、ANT(6)-la、AAC(6′)-li,其中AAC(6′)-le-APH(2″)-la为氨基糖苷类药物双功能修饰酶,属于革兰阳性菌中一种非常重要的耐药酶,由于其有强大的底物谱,几乎可使所有氨基糖苷类抗生素失效[21-22]。资料显示ErmA基因起源于金黄色葡萄球菌抗大环内酯-林可酰胺-链霉抗生物蛋白B核苷酸,因此间接证实屎肠球菌LS170308株多重耐药基因岛PRI1与金黄色葡萄球菌中类似基因岛的亲缘性较高[23]。
通过对屎肠球菌LS170308株使用多种方法进行基因岛预测,分析出一个多重耐药基因岛PRI1,该耐药基因岛上携带AAC(6′)-le-APH(2″)-la、ErmA、ANT(9)耐药基因,同时该基因岛上的包含两种转座酶分别为转座子Tn554和Tn4001(IS256插入元件)。研究表明Tn554是金黄色葡萄球菌的一种转座元件,具有与溶原性噬菌体相似的整合特性,其包含三个转座基因,分别为转座酶A,转座酶B和转座酶C,Tn554主要携带ANT(9)和ErmA耐药基因[24-25]。IS256是一种高度活跃的插入序列,作为复合型转座子Tn4001的一个组成部分,在金黄色葡萄球菌抵抗氨基糖苷类药物时发挥了关键作用[26]。有文献报道,转座子Tn4001携带的两侧IS256元件能够产生头对尾串联拷贝或自由的圆形形式,表明这两个插入元件都是活跃的[27-28]。本研究通过对屎肠球菌LS170308株PRI1耐药基因岛结构分析,转座子Tn4001的两个IS256元件携带AAC(6′)-le-APH(2″)-la基因,而Tn554主要携带ANT(9)和ErmA耐药基因,推测这两个来自于金黄色葡萄球菌携带有耐药基因的转座子通过水平转移或自然转化插入到屎肠球菌LS170308株染色体上,构成耐药基因岛,之后经过DNA修复蛋白等功能蛋白进行修饰使该基因岛稳定遗传。本试验结果有助于对屎肠球菌耐药演变的研究,也给屎肠球菌临床治疗提供更多的思路。但针对该基因岛是否由金黄色葡萄球菌水平转移而来,且该基因岛在屎肠球菌中是否具有水平传播能力,目前尚不可知,需要进一步试验验证。
4 结论从死亡林麝内脏组织中分离得到一株屎肠球菌LS170308,该菌株携带有一个多重耐药基因组岛PRI1,该PRI1与金黄色葡萄球菌亲缘性最高。
| [1] |
张波, 吕晓玥, 朱武, 等. 2012-2015年肠球菌属临床感染及耐药性变迁[J]. 中华医院感染学杂志, 2017, 27(1): 47–50.
ZHANG B, LYU X Y, ZHU W, et al. Clinical infection caused by Enterococcus spp and change of drug resistance from 2012 to 2015[J]. Chinese Journal of Nosocomiology, 2017, 27(1): 47–50. (in Chinese) |
| [2] | CATTOIR V, GIARD J C. Antibiotic resistance in Enterococcus faecium clinical isolates[J]. Expert Rev Anti-infect Ther, 2014, 12(2): 239–248. DOI: 10.1586/14787210.2014.870886 |
| [3] | MELEGH S, NYUL A, KOVÃCS K, et al. Dissemination of VanA-type Enterococcus faecium isolates in hungary[J]. Microb Drug Resist, 2018, 24(9): 1376–1390. DOI: 10.1089/mdr.2017.0296 |
| [4] | GUZEK A, RYBICKI Z, TOMASZEWSKI D. Changes in bacterial flora and antibiotic resistance in clinical samples isolated from patients hospitalized in the Military Institute of Medicine in Warsaw, Poland, between 2005 and 2012[J]. Przegl Epidemiol, 2017, 71(2): 165–176. |
| [5] | COTTON M J, PACKER C D. Vancomycin-resistant Enterococcus faecium empyema in an asplenic patient[J]. Cureus, 2018, 10(8): e3227. |
| [6] |
方志华, 余素飞. 2011—2013临床分离屎肠球菌和粪肠球菌耐药情况[J]. 现代预防医学, 2015, 42(4): 766–768.
FANG Z H, YU S F. Drug resistance of Enterococcus faecium and Enterococcus faecalis clinically isolated between 2011 and 2013[J]. Modern Preventive Medicine, 2015, 42(4): 766–768. (in Chinese) |
| [7] | SARROU S, MALLI E, TSILIPOUNIDAKI K, et al. MLSB-resistant Staphylococcus aureus in central greece: rate of resistance and molecular characterization[J]. Microb Drug Resist, 2018. DOI: 10.1089/mdr.2018.0259 |
| [8] | AJANTHA G S, KULKARNI R D, SHETTY J, et al. Phenotypic detection of inducible clindamycin resistance among Staphylococcus aureus isolates by using the lower limit of recommended inter-disk distance[J]. Indian J Pathol Microbiol, 2008, 51(3): 376–378. DOI: 10.4103/0377-4929.42515 |
| [9] | ZEHRA A, GULZAR M, SINGH R, et al. Prevalence, multidrug resistance and molecular typing of methicillin-resistant Staphylococcus aureus (MRSA) in retail meat from punjab, India[J]. J Glob Antimicrob Resist, 2019, 16: 152–158. DOI: 10.1016/j.jgar.2018.10.005 |
| [10] | KHASHEI R, MALEKZADEGAN Y, SEDIGH EBRAHIM-SARAIE H, et al. Phenotypic and genotypic characterization of macrolide, lincosamide and streptogramin B resistance among clinical isolates of staphylococci in southwest of Iran[J]. BMC Res Notes, 2018, 11: 711. DOI: 10.1186/s13104-018-3817-4 |
| [11] | SZEMRAJ M, KWASZEWSKA A, SZEWCZYK E M. New gene responsible for resistance of clinical corynebacteria to macrolide, lincosamide and streptogramin B[J]. Pol J Microbiol, 2018, 67(2): 237–240. DOI: 10.21307/pjm-2018-028 |
| [12] | TOKA ÖZER T. The rate of inducible MLSB resistance in the methicillin-resistant staphylococci isolated from clinical samples[J]. J Clin Lab Anal, 2016, 30(5): 490–493. DOI: 10.1002/jcla.2016.30.issue-5 |
| [13] | CAI J, SCHWARZ S, CHI D, et al. Faecal carriage of optrA-positive enterococci in asymptomatic healthy humans in Hangzhou, China[J]. Clin Microbiol Infect, 2018. DOI: 10.1016/j.cmi.2018.07.025 |
| [14] | ISOGAI N, URUSHIBARA N, KAWAGUCHIYA M, et al. Characterization of Enterococcus faecium with macrolide resistance and reduced susceptibility to quinupristin/dalfopristin in a Japanese hospital:detection of extensive diversity in erm(B)-regulator regions[J]. Microb Drug Resist, 2013, 19(4): 298–307. DOI: 10.1089/mdr.2012.0176 |
| [15] |
周纯葆.基因岛预测与隔离迁移模型并行化[D].长春: 吉林大学, 2012.
ZHOU C B. Detection of genomic islands and parallelization of isolation with migration model[D]. Changchun: Jilin University, 2012. (in Chinese) http://cdmd.cnki.com.cn/Article/CDMD-10183-1012365382.htm |
| [16] | SOLIMAN A M, SHIMAMOTO T, NARIYA H, et al. Emergence of Salmonella genomic island 1 variant SGI1-W in a clinical isolate of Providencia stuartii from Egypt[J]. Antimicrob Agents Chemother, 2019, 63(1). DOI: 10.1128/AAC.01793-18 |
| [17] | ORIESKOVA M, KAJSIK M, SZEMES T, et al. Contribution of the thermotolerance genomic island to increased thermal tolerance in Cronobacter strains[J]. Antonie van Leeuwenhoek, 2016, 109(3): 405–414. DOI: 10.1007/s10482-016-0645-1 |
| [18] | BERTELLI C, LAIRD M R, WILLIAMS K P, et al. IslandViewer 4: expanded prediction of genomic islands for larger-scale datasets[J]. Nucleic Acids Res, 2017, 45(W1): W30–W35. DOI: 10.1093/nar/gkx343 |
| [19] | HUANG L, YUAN H, LIU M F, et al. Type B chloramphenicol acetyltransferases are responsible for chloramphenicol resistance in Riemerella anatipestifer, China[J]. Front Microbiol, 2017, 8: 297. |
| [20] | MONTEALEGRE M C, ROH J H, RAE M, et al. Differential penicillin-binding protein 5 (PBP5) levels in the Enterococcus faecium clades with different levels of ampicillin resistance[J]. Antimicrob Agents Chemother, 2017, 61(1): e02034–16. |
| [21] |
李聪然, 游雪甫, 蒋建东. 氨基糖苷类双功能修饰酶AAC(6')-APH(2″)的研究进展[J]. 中国感染与化疗杂志, 2007, 7(6): 468–471.
LI C R, YOU X F, JIANG J D. Research update of aminoglycoside bifunctional modifying enzyme AAC(6')-APH(2″)[J]. Chinese Journal of Infection and Chemotherapy, 2007, 7(6): 468–471. DOI: 10.3321/j.issn:1009-7708.2007.06.022 (in Chinese) |
| [22] | DE FREITAS A D A R, FARIA A R, DE CASTRO ABREU PINTO T, et al. Distribution of species and antimicrobial resistance among enterococci isolated from the fecal microbiota of captive blue-fronted parrot (Amazona aestiva) in Rio de Janeiro, Brazil[J]. Sci Total Environ, 2018, 615: 1428–1437. DOI: 10.1016/j.scitotenv.2017.09.004 |
| [23] | MURPHY E. Nucleotide sequence of ermA, a macrolide-lincosamide-streptogramin B determinant in Staphylococcus aureus[J]. J Bacteriol, 1985, 162(2): 633–640. |
| [24] | BASTOS M C, MURPHY E. Transposon Tn554 encodes three products required for transposition[J]. EMBO J, 1988, 7(9): 2935–2941. DOI: 10.1002/embj.1988.7.issue-9 |
| [25] | MURPHY E, HUWYLER L, DE FREIRE BASTOS M D C. Transposon Tn554:complete nucleotide sequence and isolation of transposition-defective and antibiotic-sensitive mutants[J]. EMBO J, 1985, 4(12): 3357–3365. DOI: 10.1002/embj.1985.4.issue-12 |
| [26] | SCHREIBER F, SZEKAT C, JOSTEN M, et al. Antibiotic-induced autoactivation of IS256 in Staphylococcus aureus[J]. Antimicrob Agents Chemother, 2013, 57(12): 6381–6384. DOI: 10.1128/AAC.01585-13 |
| [27] | PRUDHOMME M, TURLAN C, CLAVERYS J P, et al. Diversity of Tn4001 transposition products:the flanking IS256 elements can form tandem dimers and IS circles[J]. J Bacteriol, 2002, 184(2): 433–443. DOI: 10.1128/JB.184.2.433-443.2002 |
| [28] | MURUGESAN S, MANI S, KUPPUSAMY I, et al. Role of insertion sequence element is256 as a virulence marker and its association with biofilm formation among methicillin-resistant Staphylococcus epidermidis from hospital and community settings in Chennai, South India[J]. Indian J Med Microbiol, 2018, 36(1): 124–126. DOI: 10.4103/ijmm.IJMM_17_276 |



