禽致病性大肠杆菌(avian pathogenic Escherichia coli, APEC)属于肠外致病性大肠杆菌(extraintestinal pathogenic E. coli, ExPEC)[1],可引起禽类急、慢性传染病,常见临床病症以大肠杆菌性败血症、心包炎等为主[2]。随着抗生素的大量使用,禽致病性大肠杆菌耐药现状日益严重,寻找新的药物靶标是防治禽致病性大肠杆菌的重要途径[3]。生物被膜被认为是持续感染及出现抗生素抗性的主要原因,它是附着在组织细胞表面复杂微生物的群落,或者形成聚集体牢固地埋藏在细胞外基质中,设计或筛选能有效减少和根除生物被膜的抗生物被膜分子至关重要[4-7]。
大肠杆菌Ⅲ型分泌系统2(Escherichia coli type Ⅲ secretion system 2, ETT2)是研究者在分析E. coli O157:H7的基因组序列时被首次发现的一种新的Ⅲ型分泌系统[8-9],2014年首次在国内检出禽致病性大肠杆菌ETT2+,结果表明ETT2在禽源大肠杆菌中广泛流行分布[10];整个基因簇的缺失导致雏鸡死亡率、鞭毛相关蛋白表达量和生物被膜形成能力均有显著差异[11],但目前在APEC中有关ETT2的深入研究并不完备。ETT2致病岛编码五种转录因子:YqeI、YgeH、YgeK、EtrA和EivF[12],其中YqeI(Ecs_3704)证实与YgeK共同在QseA调控下参与ler调节区相互作用以激活LEE表达,且LEE毒力岛和QseA对生物被膜形成有一定的影响[13-15]。因此,作者拟研究生物被膜形成是否受YqeI的调控,并研究YqeI作为转录调节子对禽致病性大肠杆菌生物被膜的调控作用。
1 材料与方法 1.1 试验材料禽致病性大肠杆菌AE81野毒株,为实验室保存的临床分离株;Red同源重组所需质粒:pKD46(同源重组的协助质粒,氨苄青霉素抗性);pKD3(携带可被FLP重组酶识别的FRT位点);pCP20(42 ℃可表达FLP重组酶,用于消除FLP位点间的氯霉素抗性基因);pstv28(回复构建质粒),以上质粒均为本实验室保存。
1.2 禽致病性大肠杆菌中yqeI与E. coli O157:H7序列比对以AE81基因组为模板,扩增yqeI[12-13]并进行测序,测序结果与Escherichia coli O157:H7 str. Sakai(GenBank: BA000007.3)中yqeI序列进行比对,检测在APEC中该基因的完整性。并通过软件http://www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi? id=index预测其蛋白结构及功能。
1.3 AE81 yqeI缺失株AE81△YqeI及其回复株AE81-C-YqeI的构建利用Red同源重组的方法将同源臂片段转化到含有pKD46质粒的AE81感受态细胞中进行敲除。筛选阳性重组子并以pCP20质粒42 ℃进行消抗,用yqeI-out-F、yqeI-out-R引物进行PCR验证。将含酶切位点的yqeI基因片段连接到pSTV28载体上,转化到DH5α中,提取阳性株质粒电转导入基因缺失株感受态细胞中。通过yqeI-BamHⅠ/yqeI-HindⅢ引物鉴定测序比对,引物序列见表 1。鉴定成功的回复株命名为AE81-C-YqeI。
|
|
表 1 根据GenBank中已发布的APEC O1型基因序列设计的缺失和回复引物 Table 1 Deletion and recovery primers designed according to the published APEC O1 gene sequence in GenBank |
从平板上挑取三株菌单菌落接种到LB液体培养基过夜培养。次日1:100转接LB液体培养基中,37 ℃ 150 r·min-1振荡培养至对数期,每隔1 h取200 μL至96孔板中用酶标仪测定OD620 nm,根据OD620 nm数值绘制生长曲线。
1.5 AE81、AE81△YqeI缺失株和AE81-C-YqeI回复株生物被膜成膜能力的检测按1:100转接LB液体培养基中,将三株菌培养至对数生长期,调整OD600 nm至0.03,以LB作阴性对照组,每组3个重复,37 ℃分别于玻璃试管和96孔板培养72 h。玻璃试管中弃培养基并用无菌PBS缓冲液冲洗3次,固定15 min后,晾干观察;96孔板同样PBS洗涤固定,并用结晶紫染色10 min后,用33%乙酸溶解,测量其OD405 nm计算统计学差异。
1.6 AE81、AE81△YqeI缺失株和AE81-C-YqeI回复株生物被膜扫描电镜的观察方法同上。按1:100转接LB液体培养基中,将三株菌培养至对数生长期,将菌液转移到放有爬片的6孔板中,37 ℃静置培养48 h后,取出爬片,PBS清洗3遍,晾干,2.5%戊二醛固定2 h,PBS缓冲液清洗3遍,3%戊二醛4 ℃再固定6 h,用无菌PBS缓冲液漂洗3次,每次20 min;乙醇梯度脱水,每个梯度4 ℃ 20 min;最后置换到100%丙酮,4 ℃重复2次,每次20 min。将处理好的样本浸泡在100%丙酮中, 真空冷冻干燥,用导电胶将干燥后的样品固定于样品台上,在不导电的样品表面喷镀一层10 nm左右的贵金属,进样观察,使用日本日立S-4800冷场发射扫描电镜。
1.7 AE81和AE81△YqeI缺失株转录组学测序 1.7.1 建库测序流程取AE81和AE81△YqeI OD600 nm=1.0的菌液提取total RNA,并通过测量浓度和跑胶验证其质量后,将提取好的total RNA使用生物素标记的特异性探针去除核糖体RNA(rRNA),再对RNA进行纯化和片段化。随后,使用Illumina TruSeq Stranded Kit进行处理,然后双链cDNA加A尾并连接测序接头,连接产物经过纯化扩增得到最终的cDNA文库,最后将构建好的测序文库使用HiSeq[16]测序平台进行测序。
1.7.2 信息分析流程测序所得数据称为raw reads或rawdata,过滤掉低质量、接头污染以及未知碱基N含量过高的reads,得到clean reads。然后将clean reads比对到参考基因组(Escherichia coli O157:H7 str. Sakai, GenBank: BA000007.3)上[17-18],之后进行新转录本预测、SNP & INDEL检测、操纵子等结构分析。对于多个样品根据需求检测不同样品之间的差异表达基因,并对差异表达基因做深入的聚类分析和功能富集分析等。
1.8 荧光定量PCR检测生物被膜相关基因转录量细菌总RNA的提取和cDNA的合成:参照Trizol法提取三组细菌的总RNA。核酸蛋白浓度测定仪检测RNA浓度,并用1%琼脂糖凝胶电泳检测RNA的完整性。采用反转录试剂盒对RNA进行反转录。将合成的cDNA保存于-20 ℃。
qRT-PCR反应,采用诺唯赞AceQ qPCR SYBR Green Master Mix荧光定量试剂盒,用实时荧光定量PCR对五种与生物被膜形成相关的差异基因转录量进行相对定量的检测,体系程序参照试剂盒使用说明。测试样品重复三管;熔解曲线分析并鉴定引物特异性。引物序列见表 2。
|
|
表 2 生物被膜相关荧光定量引物列表 Table 2 Biofilm-related fluorescence quantitative primer list |
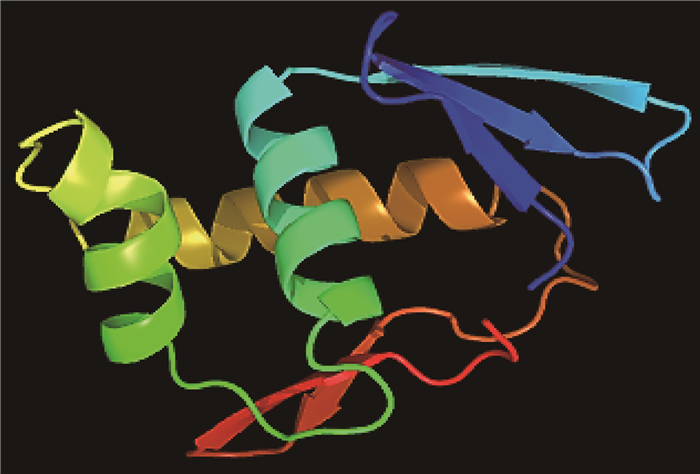
经测序比对(如图 1 A),证实在AE81中检测到的序列与肠出血性大肠杆菌E. coli O157:H7(GenBank: BA000007.3)中公布的ETT2 yqeI序列一致,两者所编码的实为同一种蛋白,并通过软件预测其蛋白结构功能为转录因子,为后续在APEC中研究其作为ETT2组分发挥的作用提供最基础的理论保障。
|
图像以彩虹色序由N端向C端着色 Image coloured by rainbow N inbow N bow 图 1 APEC中YqeI蛋白功能预测图 Fig. 1 Function prediction of YqeI protein in APEC |
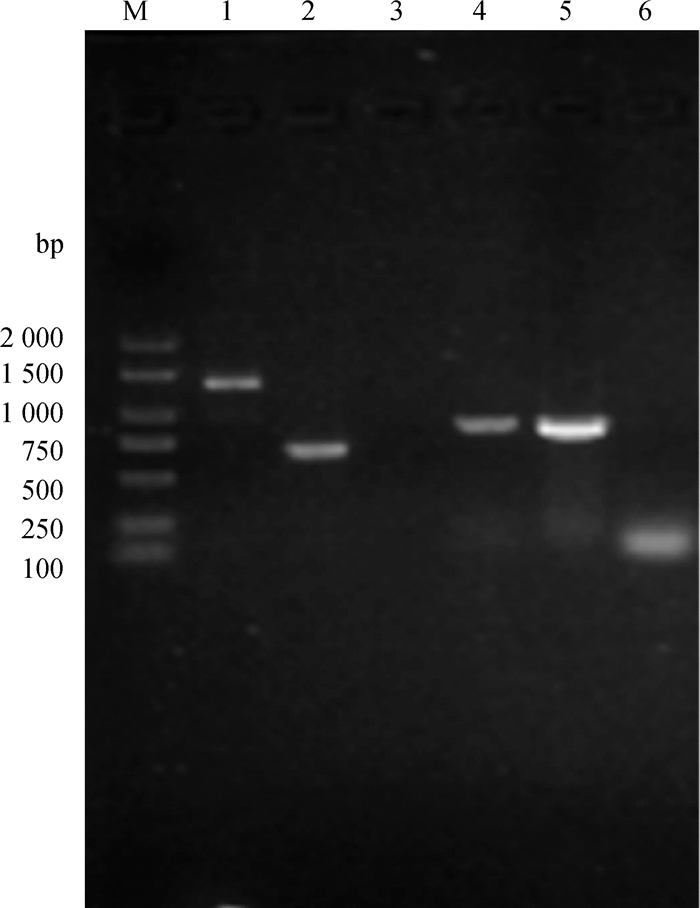
验证结果显示缺失株较原始株的外侧引物片段减小了732 pb,说明810 pb的yqeI片段成功缺失大部分碱基而丧失其蛋白能力,成功构建AE81△YqeI缺失株(图 2);经回复后,能够检测到该基因完整片段,成功构建AE81-C-YqeI回复株。
|
M. DNA相对分子质量标准;1. AE81-YqeI-OUT;2. △YqeI-YqeI-OUT;3. YqeI-OUT阴性对照;4. AE81-yqeI-BamHⅠ/yqeI-HindⅢ;5. C-YqeI-yqeI-BamHⅠ/yqeI-HindⅢ;6. yqeI-BamHⅠ/yqeI-HindⅢ阴性对照 M. DNA marker; 1. AE81-YqeI-OUT; 2. △ YqeI-YqeI-OUT; 3. YqeI-OUT negative control; 4. AE81-yqeI-BamHⅠ/yqeI-HindⅢ; 5. C-YqeI-yqeI-BamHⅠ/yqeI-HindⅢ; 6. yqeI-BamHⅠ/yqeI-HindⅢ negative control 图 2 AE81△YqeI和AE81-C-YqeI的验证 Fig. 2 Verification of AE81△YqeI and AE81-C-YqeI |
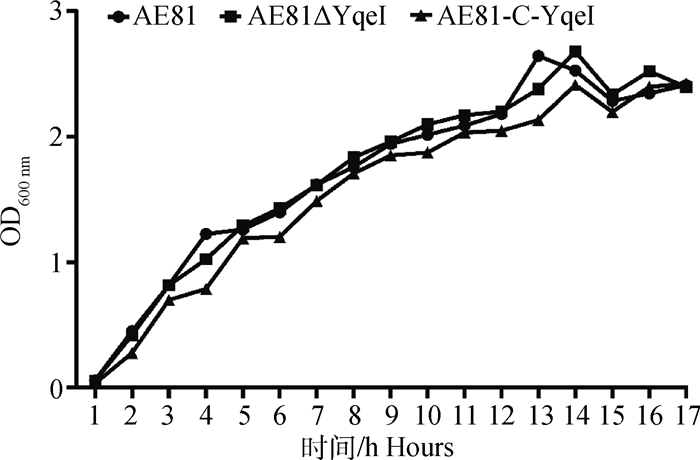
缺失yqeI后,菌株AE81的生长状况未发生显著变化(如图 3)。原始株AE81、缺失株AE81△YqeI、AE81-C-YqeI生长状态基本趋于一致。以P < 0.05为有差异的标准,与原始株AE81相比AE81△YqeI和AE81-C-YqeI的P值分别为0.964 1和0.607 8,差异均不显著。
|
图 3 各菌株的生长曲线 Fig. 3 Growth curve of each strain |
玻璃试管结合结晶紫的数据,原始株AE81菌体表面形成的生物被膜致密完整且较为厚实;缺失株表面无法形成完整被膜;回复株虽没有原始株的完整度高,但回复株生物被膜能力得到一定的回补(图 4)。

|
与AE81相比,* *.P < 0.01 Compared with AE81, * *.P < 0.01 图 4 生物被膜形成(A)及结晶紫数据(B) Fig. 4 Biofilm formation (A) and crystal violet data (B) |
借助扫描电镜观察结合结晶紫数据结果发现(如图 5),原始株AE81形成的生物被膜由多层菌体搭建而成,具有丰富的立体层次结构,菌体之间形成较为明显的黏连现象;而AE81△YqeI缺失株的生物被膜由单层菌体构成,菌体间不黏连;在缺失株的基础上回补yqeI后,生物被膜形态与原始株基本相似,呈立体多层黏连构架。

|
A. AE81,8 000倍放大;B. AE81△YqeI,8 000倍放大;C. AE81-C-YqeI,8 000倍放大;D. AE81,10 000倍放大;E. AE81△YqeI,10 000倍放大;F. AE81-C-YqeI,10 000倍放大 A. AE81, 8 000 times magnification; B. AE81△YqeI, 8 000 times magnification; C. AE81-C-YqeI, 8 000 times magnification; D. AE81, 10 000 times magnification; E. AE81△YqeI; 10 000 times magnification; F. AE81-C-YqeI, 10 000 times magnification 图 5 生物被膜扫描电镜图像 Fig. 5 Scanning electron micrograph of biofilm |
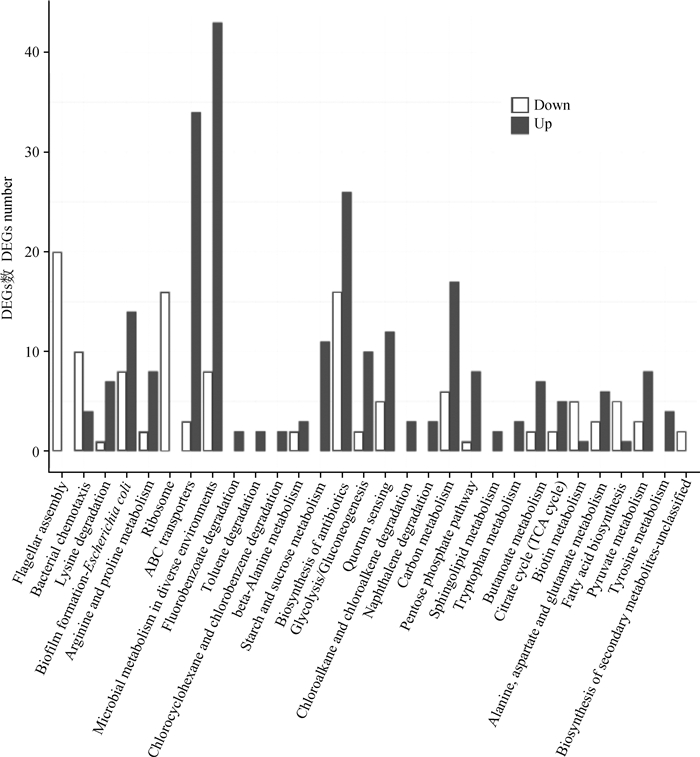
转录组学测序结果中yqeI影响了587个差异基因(DEGs),其中上调391个,下调196个;上调的基因中最多有43个富集在不同环境中微生物的代谢上,34个涉及ABC转运蛋白,22个影响碳代谢,21个参与抗生素的合成;下调的基因中最多下调20个的鞭毛装置基因且差异最为显著(图 6)。
|
横坐标为显著差异表达基因富集的Pathway(按富集显著程度排列,从左至右富集显著性依次降低),纵坐标表示对应Pathway中显著变化差异表达基因数目 The abscissa is a Pathway with significant differentially expressed genes enriched (according to the significant degree of enrichment, from left to right, the signigicance of enrichment decreased in turm). The ordinate indicates the number of differentially expressed genes corresponding to significant changes in Pathway 图 6 显著变化差异表达基因富集的Pathway分类 Fig. 6 Pathway classification map of the enrichment of significantly changed differentially expressed genes |
转录组学差异基因中与生物被膜相关的基因有22个,其中上调的14个,下调的8个;而有4个上调的基因(csrA、yciR、adrB和gcvA)对生物被膜的形成起到负调控作用[19-21]。这22个基因中有涉及到鞭毛、菌毛、纤维素和多糖代谢基因(表 3),变化幅度最大的是下调的多糖生物合成、输出蛋白wzA[22],下调幅度2.38;而上调最明显的纤维素合成蛋白bcsA的变化幅度1.79;生物膜PGA合成蛋白PgaA上调幅度1.01、PgaD下调幅度1.05,且后者变化幅度大[23];鞭毛蛋白flhC、flhD、flgM和ycgR均显著下调。这些基因共同对生物被膜的形成起到直接或间接的影响,形成一个完整的生物被膜调控网络(图 7)。
|
|
表 3 生物被膜差异基因信息表 Table 3 Biofilm differential gene information table |

|
cAMP/CRP信号通路影响细菌运动性和氨基酸缺乏,氨基酸缺乏调控菌毛和纤维素合成影响生物被膜,而运动性间接调控生物被膜的形成,此外下方图框中描绘生物被膜中涉及的环境信号和sRNA调控机制 cAMP/CRP signaling pathway affects bacterial motility and amino acid starvation, amino acid starvation regulates pili and cellulose synthesis to affect biofilm, while motility indirectly regulates biofilm formation, and the Environment signals and sRNA regulatory involved in biofilm is depicted in the lower panel 图 7 大肠杆菌生物被膜调控通路 Fig. 7 Biofilm regulatory pathway map of Escherichia coli |
荧光定量验证涉及鞭毛、菌毛等与生物被膜相关基因的检测结果与转录组学差异基因结果一致,均有不同程度的下调(图 8),进一步证实了yqeI的缺失在转录水平上显著降低了生物被膜相关基因的表达。

|
*P < 0.05, **.P < 0.01;** *.P < 0.001 图 8 生物被膜基因差异转录 Fig. 8 Biofilm gene differential transcription map |
禽致病性大肠杆菌侵染宿主的过程中常形成生物被膜以抵抗宿主免疫及抗菌制剂的杀伤[24-25]。生物被膜的形成由鞭毛、菌毛、外膜蛋白、脂多糖和胞外多糖等因素介导[26],在抗菌剂的杀伤下仍能顽固地存活[25]。研究发现,二元调控系统中转录因子phoP可通过调节细菌鞭毛和卷曲菌毛的菌毛蛋白的组装而影响APEC生物膜的形成[26-27];转录因子mcbR的缺失通过上调纤维素合成、鞭毛和菌毛相关基因的转录增强APEC生物被膜形成能力[28]。众多转录因子通过调控生物被膜的形成能力而影响细菌本身耐药性。细菌耐药正是目前在利用抗菌药物治疗细菌感染途径中急需解决的研究难题。
目前主要抗生物被膜模式包括干扰群体感应途径、黏附机制、脂多糖、胞外多糖和抑制参与各种信号传导途径的第二信使等[29-30],上述途径均受到转录因子调控的影响,但ETT2毒力岛中是否存在影响生物被膜的调节因子未知。本研究从ETT2的转录因子YqeI入手,探究YqeI对生物被膜形成能力的影响及相关机制。通过检测原始株与缺失株的成膜能力并结合扫描电镜观察生物被膜结构形态,发现转录因子yqeI基因的缺失使细菌胞外聚合物膜基质组分大量减少,细菌无法形成厚实完整的生物被膜。
转录组测序及荧光定量结果显示,生物被膜相关基因中,受YqeI转录因子影响最显著的为外膜脂蛋白基因wzA[22],其缺失使荚膜层无法形成,且对抗不利环境因素的耐受作用减弱[31];生物膜黏附素PGA的孔蛋白PgaA表达量上调,但辅助糖基转移酶PgaC催化UDP-N-乙酰葡糖胺聚合产生PGA的PgaD下调[23];此外,鞭毛通过参与黏附在介质表面的过程而影响生物被膜形成的初始阶段,转录组学数据显示,差异表达基因中鞭毛相关基因数量最多且均为下调[32]。本研究发现,YqeI转录因子从多个途径介导生物被膜的形成,影响生物被膜形成的不同阶段,为以YqeI为方向探寻新的抗耐药机制提供基础。
4 结论以禽致病性大肠杆菌AE81野毒株为亲本株,成功构建yqeI基因缺失株,yqeI的缺失不影响缺失株的生长曲线,但其生物被膜形成能力显著下降,且相关生物被膜基因转录量显著下调。
| [1] |
马兴树. 禽大肠杆菌病疫苗研究进展[J]. 中国畜牧兽医, 2015, 42(1): 234–244.
MA X S. Research progress on vaccines of avian colibacillosis[J]. China Animal Husbandry & Veterinary Medicine, 2015, 42(1): 234–244. (in Chinese) |
| [2] |
张宇曦, 韩先干, 左佳坤, 等. 禽致病性大肠杆菌脂多糖核心型分布与毒力基因的相关性分析[J]. 微生物学通报, 2015, 42(8): 1619–1625.
ZHANG Y X, HAN X G, ZUO J K, et al. Distribution of lipopolysaccharide core type in avian pathogenic Escherichia coli and its correlation with virulence gene[J]. Microbiology China, 2015, 42(8): 1619–1625. (in Chinese) |
| [3] |
范国博, 韩月, 张宇曦, 等. 禽致病性大肠杆菌gspL基因缺失株构建及生物学特性[J]. 微生物学报, 2015, 55(1): 89–95.
FAN G B, HAN Y, ZHANG Y X, et al. Construction and characterization of a gspL mutant of avian pathogenic Escherichia coli[J]. Acta Microbiologica Sinica, 2015, 55(1): 89–95. (in Chinese) |
| [4] | ROY R, TIWARI M, DONELLI G, et al. Strategies for combating bacterial biofilms:a focus on anti-biofilm agents and their mechanisms of action[J]. Virulence, 2018, 9(1): 522–554. DOI: 10.1080/21505594.2017.1313372 |
| [5] | KALI A, BHUVANESHWAR D, CHARLES P M, et al. Antibacterial synergy of curcumin with antibiotics against biofilm producing clinical bacterial isolates[J]. J Basic Clin Pharm, 2016, 7(3): 93–96. DOI: 10.4103/0976-0105.183265 |
| [6] | YU C, LI X N, ZHANG N, et al. Inhibition of biofilm formation by D-tyrosine:effect of bacterial type and D-tyrosine concentration[J]. Water Res, 2016, 92: 173–179. DOI: 10.1016/j.watres.2016.01.037 |
| [7] | WU S M, LIU G, JIN W H, et al. Antibiofilm and anti-infection of a marine bacterial exopolysaccharide against Pseudomonas aeruginosa[J]. Front Microbiol, 2016, 7: 102. |
| [8] | HAYASHI T, MAKINO K, OHNISHI M, et al. Complete genome sequence of enterohemorrhagic Eschelichia coli O157:H7 and genomic comparison with a laboratory strain K-12[J]. DNA Res, 2001, 8(1): 11–22. DOI: 10.1093/dnares/8.1.11 |
| [9] | SHULMAN A, YAIR Y, BIRAN D, et al. The Escherichia coli type Ⅲ secretion system 2 has a global effect on cell surface[J]. mBio, 2018, 9(4): e01070–18. |
| [10] |
李丽丽.禽源大肠杆菌主要毒力因子的分子流行病学分析及F1与P菌毛单克隆抗体的制备[D].扬州: 扬州大学, 2014.
LI L L. Molecular epidemiological analysis of major virulence factors of avian Escherichia coli and preparation of monoclonal antibodies against avian E. coli's F1 and P pili[D]. Yangzhou: Yangzhou University, 2014. (in Chinese) |
| [11] | HUJA S, OREN Y, TROST E, et al. Genomic avenue to avian colisepticemia[J]. mBio, 2015, 6(1): e01681–14. |
| [12] | REN C P, CHAUDHURI R R, FIVIAN A, et al. The ETT2 gene cluster, encoding a second type Ⅲ secretion system from Escherichia coli, is present in the majority of strains but has undergone widespread mutational attrition[J]. J Bacteriol, 2004, 186(11): 3547–3560. DOI: 10.1128/JB.186.11.3547-3560.2004 |
| [13] | LUZADER D H, WILLSEY G G, WARGO M J, et al. The type three secretion system 2-encoded regulator EtrB modulates enterohemorrhagic Escherichia coli virulence gene expression[J]. Infect Immunity, 2016, 84(9): 2555–2565. DOI: 10.1128/IAI.00407-16 |
| [14] | VIKRAM A, JESUDHASAN P R, PILLAI S D, et al. Isolimonic acid interferes with Escherichia coli O157:H7 biofilm and TTSS in QseBC and QseA dependent fashion[J]. BMC Microbiol, 2012, 12: 261. DOI: 10.1186/1471-2180-12-261 |
| [15] | WEIGEL W A, DEMUTH D R. QseBC, a two-component bacterial adrenergic receptor and global regulator of virulence in Enterobacteriaceae and Pasteurellaceae[J]. Mol Oral Microbiol, 2016, 31(5): 379–397. DOI: 10.1111/omi.12138 |
| [16] | KIM D, LANGMEAD B, SALZBERG S L. HISAT:a fast spliced aligner with low memory requirements[J]. Nat Methods, 2015, 12(4): 357–360. DOI: 10.1038/nmeth.3317 |
| [17] | LANGMEAD B, SALZBERG S L. Fast gapped-read alignment with Bowtie 2[J]. Nat Methods, 2012, 9(4): 357–359. DOI: 10.1038/nmeth.1923 |
| [18] | LI B, DEWEY C N. RSEM:accurate transcript quantification from RNA-Seq data with or without a reference genome[J]. BMC Bioinformatics, 2011, 12: 323. DOI: 10.1186/1471-2105-12-323 |
| [19] | LEISTRA A N, GELDERMAN G, SOWA S W, et al. A canonical biophysical model of the CsrA Global regulator suggests flexible regulator-target interactions[J]. Sci Rep, 2018, 8(1): 9892. DOI: 10.1038/s41598-018-27474-2 |
| [20] | LINDENBERG S, KLAUCK G, PESAVENTO C, et al. The EAL domain protein YciR acts as a trigger enzyme in a c-di-GMP signalling cascade in E.coli biofilm contro[J]. EMBO J, 2013, 32(14): 2001–2014. DOI: 10.1038/emboj.2013.120 |
| [21] | STAUFFER L T, STAUFFER G V. Antagonistic roles for GcvA and GcvB in hdeAB expression in Escherichia coli[J]. ISRN Microbiol, 2012, 2012: 697308. |
| [22] | DONG C L, BEIS K, NESPER J, et al. The structure of Wza, the translocon for group 1 capsular polysaccharides in Escherichia coli, identifies a new class of outer membrane protein[J]. Nature, 2006, 444(7116): 226–229. DOI: 10.1038/nature05267 |
| [23] | KANG J M, LI Q Q, LIU L, et al. The specific effect of gallic acid on Escherichia coli biofilm formation by regulating pgaABCD genes expression[J]. Appl Microbiol Biotechnol, 2018, 102(4): 1837–1846. DOI: 10.1007/s00253-017-8709-3 |
| [24] |
苏莉, 王慧. 病原体感染及其免疫逃逸机制研究进展[J]. 生物技术通讯, 2016, 27(4): 582–585.
SU L, WANG H. Advances on mechanisms of pathogen infection and immune escape[J]. Letters in Biotechnology, 2016, 27(4): 582–585. DOI: 10.3969/j.issn.1009-0002.2016.04.028 (in Chinese) |
| [25] | POUDYAL B, SAUER K. The ABC of biofilm drug tolerance:the MerR-like regulator BrlR is an activator of ABC transport systems, with PA1874-77 contributing to the tolerance of Pseudomonas aeruginosa biofilms to tobramycin[J]. Antimicrob Agents Chemother, 2018, 62(2): e01981–17. |
| [26] | FORMOSA-DAGUE C, MICKAЁL C, MARTIN-YKEN H, et al. The role of Glycans in bacterial adhesion to mucosal surfaces:how can single-molecule techniques advance our understanding?[J]. Microorganisms, 2018, 6(2): 39. DOI: 10.3390/microorganisms6020039 |
| [27] | YIN L, LI Q W, XUE M, et al. The role of the phoP transcriptional regulator on biofilm formation of avian pathogenic Escherichia coli[J]. Avian Pathol, 2019, 48(4): 362–370. DOI: 10.1080/03079457.2019.1605147 |
| [28] | YU L M, LI W C, QI K Z, et al. McbR is involved in biofilm formation and H2O2 stress response in avian pathogenic Escherichia coli X40[J]. Poultry Sci, 2019, 98(9): 4094–4103. DOI: 10.3382/ps/pez205 |
| [29] | WU H, MOSER C, WANG H Z, et al. Strategies for combating bacterial biofilm infections[J]. Int J Oral Sci, 2015, 7(1): 1–7. DOI: 10.1038/ijos.2014.65 |
| [30] | JANESCH P, ROUHA H, BADARAU A, et al. Assessing the function of pneumococcal neuraminidases NanA, NanB and NanC in in vitro and in vivo lung infection models using monoclonal antibodies[J]. Virulence, 2018, 9(1): 1521–1538. DOI: 10.1080/21505594.2018.1520545 |
| [31] | YI H B, YUAN B, LIU J B, et al. Identification of a wza-like gene involved in capsule biosynthesis, pathogenicity and biofilm formation in Riemerella anatipestifer[J]. Microb Pathog, 2017, 107: 442–450. DOI: 10.1016/j.micpath.2017.04.023 |
| [32] | DUFRÊNE Y F. Sticky microbes:forces in microbial cell adhesion[J]. Trends Microbiol, 2015, 23(6): 376–382. DOI: 10.1016/j.tim.2015.01.011 |



