2. 甘肃农业大学动物科学技术学院, 兰州 730070
2. College of Animal Science and Technology, Gansu Agricultural University, Lanzhou 730070, China
表观遗传学是指基因核苷酸序列不发生改变,而基因表达可以遗传并造成表型差异的一门遗传学分支学科。表观遗传修饰不仅发生在基因组层面,同样在转录组层面也存在,并且在转录后调控过程中发挥重要作用。目前,已经在RNA中发现100多种不同的化学修饰,其中大部分为甲基化修饰,最常见的是N6-甲基腺苷(N6-methyladenosine,m6A)修饰。早在20世纪70年代,科学家们就在RNA中发现了m6A修饰,但一直由于技术制约其功能未能被很好的揭示。直到2012年,科学家们使用特异性结合m6A抗体富集高度甲基化的mRNA片段,并结合高通量测序技术对富集到的mRNA片段进行全转录组范围内测序,这才使得m6A修饰的神秘面纱被逐渐揭开[1-2]。此外,RNA去甲基化酶的发现打破了人们一直认为m6A修饰是静态的这一观念,m6A修饰的动态可逆为其功能的研究开启了新的局面[3]。
1 mRNA中m6A甲基化的分布与表达以往的研究揭示了各种动态的mRNA修饰,包括m6A、m1A(N1-甲基腺苷,N1-methyladenosine)、m5C(5-甲基胞嘧啶,5-methylcytosine)和ψ(假尿苷,pseudouridine)等,其中m6A修饰是真核生物细胞内最丰富的mRNA修饰[4]。哺乳动物mRNA中分离的m6A约占所有腺苷核苷酸的0.1%~0.4%,平均每个mRNA分子中存在约3~5个m6A甲基化位点[5-6]。同样,m6A修饰也在植物、病毒以及单细胞生物(古细菌、细菌和酵母等)中广泛存在且保守。m6A修饰不是随机分布在成熟转录本中的,而是在特定转录本界标处富集,例如在mRNA内部长的外显子、3′UTR(3′ untranslated region)和终止密码子附近富集,其分布的保守基序为G(m6A)C(70%)或A(m6A)C(30%),表现出一定的序列特异性[1-2, 7-8]。m6A在动物组织中的表达具有特异性和偏好性,例如在大脑、肝和肾中高表达[2]。小鼠小脑和大脑中,m6A表达水平和模式在不同的脑区域显示出高度多样性,小脑中m6A水平高于大脑皮层,脑干、嗅球、皮层、小脑和丘脑中表达水平也不同,这可能与脑的复杂性结构有关[9-10]。m6A的表达也具有一定的保守性,人胚胎干细胞中约70%的m6A修饰同样在小鼠中被检测到,其中约46%的m6A修饰完全一样[1, 11]。此外,m6A修饰存在于tRNA、rRNA和SnRNA中[12-13]。
2 mRNA中m6A修饰的检测方法为了揭示m6A的生物学功能,首先需要确定其在mRNA转录本中的位置。由于mRNA中m6A修饰不影响Watson-Crick碱基配对原则,这使得基于逆转录的方法检测不到其存在,致使难度增加。其次,由于在转录组范围内缺乏检测表观转录组学标记的敏感测序技术,也使得表观转录组学研究落后于表观基因组学。虽然科学家们通过薄层层析(two-dimensional thin layer chromatography, 2D-TLC)、斑点杂交(dot-blot)和液相色谱串联质谱(liquid chromatography-tandem mass spectrometry, LC-MS/MS)等一些传统方法也能检测到转录组中m6A的分布,但是这些方法具有很大的局限性,在很大程度上依赖于物理化学技术,无法满足整个转录组水平上m6A分布的研究要求。直到2012年,两个研究小组独立开发了高通量测序与抗体免疫沉淀相结合的方法(m6A-Seq或MeRIP-Seq,methylated RNA immunoprecipitation and sequencing),这样才能够在整个转录组范围内检测m6A的分布[1-2]。该方法是近年来mRNA修饰研究历程中的一大进步,但同样也存在一定的局限性,例如对原始材料的质量和数量要求高、受抗体影响大、分辨率低等。随后科学家们又开发了SCARLET(site-specific cleavage and radioactive-labeling followed by ligation-assisted extraction and thin-layer chromatography)、PA-m6A-Seq(photo-crosslinking-assisted m6A sequencing)、mi-CLIP-Seq(m6A individual-nucleotide resolution crosslinking and immunoprecipitation sequencing)和m6A-LAIC-Seq(m6A level and isoform characterization sequencing)等方法来检测m6A的分布[7, 14-17]。如表 1所示,本文对这些方法进行了简单描述,并分析了其存在的优缺点。通过试验的方法来鉴定m6A位点既费时费力,又需要大量资金投入。因此,急需开发一些算法或程序来系统的分析鉴定m6A位点。例如RFAthM6A、m6aViewer、TargetM6A、SRAMP、RNA-MethylPred、iRNA-Methyl和pRNAm-PC等算法或程序已被应用于动植物的m6A位点预测[18-24]。这些算法或程序的使用将有助于更加准确、快速鉴定m6A位点,以期更好的解释其生物学功能。
|
|
表 1 mRNA中m6A检测方法的比较 Table 1 Comparison of detection methods of m6A in mRNA |
动物mRNA中的m6A修饰可以被一系列甲基转移酶(writers)、脱甲基酶(erasers)和m6A结合蛋白(readers)动态沉积、去除和识别。目前,已发现的甲基转移酶主要有METTL3(methyltransferase-like 3)、METTL14(methyltransferase-like 14)、WTAP(wilms tumor 1-associated protein)、KIAA1429、RBM15(RNA-binding motif protein 15)、RBM15B(RNA binding motif protein 15B)、METTL16(methyltransferase-like protein 16)和Zc3h13(zinc finger CCCH domain-containing protein 13)等;脱甲基酶主要有FTO(fat mass and obesity-associated protein)和ALKBH5(alkB homologue 5)等;m6A结合蛋白主要有YTHDF1-3(YTH domain family proteins)、YTHDC1-2(YTH domain-containing proteins)、eIF3(eukaryotic initiation factor 3)、HNRNPA2B1((heterogeneous nuclear ribonucleoprotein A1B1)、HNRNPC(heterogeneous nuclear ribonucleoprotein C)和IGF2BP(insulin-like growth factor-2 mRNA-biding proteins)等[28-29]。这些m6A修饰相关酶参与mRNA稳定性、剪接加工、转运以及翻译等一系列代谢过程,对RNA的命运决定发挥重要作用(表 2)。
|
|
表 2 m6A调节器及其在RNA代谢中的作用 Table 2 m6A regulators and their roles in RNA metabolism |
mRNA中m6A修饰是由甲基转移酶介导进行的,研究这些甲基转移酶的结构和机制是深入研究m6A修饰的重点之一。由METTL3、METTL14和WTAP组成的甲基转移酶复合物催化m6A修饰的机制已经清楚。在体外,METTL3和METTL14以1:1化学计量形式形成稳定的异源二聚体,并且两种蛋白协同作用以促进细胞中的mRNA甲基化[26]。其中,METTL3催化S-腺苷甲硫氨酸(S-adenosylmethionine,SAM)介导的甲基转移到腺苷碱基的N6位,而METTL14被认为是催化失活的,具有促进METTL3活性的结构作用[59-60]。WTAP是稳定METTL3和METTL14蛋白之间相互作用必不可少的,可将甲基化酶复合物正确定位于核斑点中,并且识别共有基序RRACH[33, 61]。由于METTL3-METTL14-WTAP复合物只能使一部分位点被甲基化,所以科学家们又发现了其它一些甲基化酶在指导特定靶位点甲基化中起作用,如KIAA1429、RBM15、RBM15B、METTL16和Zc3h13等。KIAA1429是哺乳动物甲基化所必需的,其缺失导致RNA中m6A丰度降低,但其具体的分子机制还不清楚[27]。RBM15及其旁系RBM15B是甲基转移酶复合物的组分,它们可以将甲基化复合物靶向RRACH基序以促进甲基化,研究表明,lncRNA中X非活性特异转录物(X inactive specific transcript,XIST)是RBM15/15B定向甲基化的靶点[34-35]。METTL16是一种靶向前体mRNA和各种非编码RNA的m6A甲基转移酶,其作用方式与METTL3和METTL14明显不同[36, 62]。在脊椎动物中,METTL16作为一个保守的U6 snRNA甲基转移酶,可以通过甲硫氨酸腺苷转移酶2A(methionine adenosyltransferase 2A,MAT2A)mRNA内部的发夹结构来调节选择性剪接进而控制SAM稳态[37, 63]。最近,研究者报道称,Zc3h13是调控m6A修饰的新成员[64]。Zc3h13对m6A的调节是通过控制复合物成员WTAP/Virilizer/Hakai的细胞定位而发挥作用的[38]。抑制Zc3h13表达导致复合物成员WTAP、Virilizer和Hakai蛋白由细胞核向细胞质转移,同时伴随METTL3和METTL14蛋白核内组分的减少,从而抑制m6A形成。
3.2 m6A脱甲基酶(Erasers)m6A脱甲基酶的发现进一步促进了RNA甲基化的研究进展,同时,也使得m6A修饰在细胞中动态调节成为可能。FTO是第一个被鉴定的m6A脱甲基酶,FTO通过N6-羟甲基腺苷(hm6A)和N6-甲酰腺苷(fm6A)中间体才能将m6A催化成A(腺嘌呤,adenine),且该反应速度较快[3, 65]。ALKBH5与FTO同样可促进体内和体外的mRNA脱甲基化。ALKBH5可直接将m6A转化为A,但该反应速度比FTO慢[43]。
3.3 m6A结合蛋白(Readers)对m6A生理功能认识的主要贡献是鉴定了许多m6A结合蛋白。m6A结合蛋白可以通过直接和间接两种模式与m6A位点结合。直接结合即m6A结合蛋白与mRNA中的m6A位点进行选择性结合。间接结合即m6A修饰改变RNA二级结构,从而使RNA可以与RNA结合蛋白相连接。研究表明,含有YTH结构域的YTHDF1-3可以在细胞质中结合m6A修饰的mRNA,其中YTHDF2的结合能力比YTHDF1和YTHDF3强。YTHDF2选择性结合m6A修饰的mRNA,并将其募集到mRNA降解位点,从而调节mRNA稳定性[48]。而YTHDF1和YTHDF3协同作用促进蛋白质合成,并影响YTHDF2介导的mRNA降解[45, 49-50]。存在于细胞核中的YTHDC1影响mRNA剪接,同时也能与YTHDF1-3协同作用快速调控细胞核中mRNA代谢[51]。此外,YTHDC1还可以优先识别XIST上的m6A残基,并且是XIST发挥功能所需的[35]。位于细胞质中的YTHDC2功能目前还不清楚。eIF3也被鉴定为直接结合m6A的蛋白,mRNA 5′UTR中的m6A能够直接结合eIF3,不需要帽子结合因子eIF4E就能招募43S复合体并起始翻译,抑制这种RNA甲基化会选择性减少这类mRNA的翻译[54]。HNRNPA2B1是一种核RNA结合蛋白,能够特异性识别转录本上的m6A修饰,这些发生m6A修饰的位点所在的基序与METTL3基序相匹配,同时HNRNPA2B1能够直接与发生m6A修饰的pri-miRNA相结合,并介导其加工和成熟[55]。HNRNPC是另外一种核RNA结合蛋白,m6A修饰改变mRNA和lncRNA的结构来促进HNRNPC与其结合,进而介导pre-mRNA的加工[57]。IGF2BP是最近发现的一种新的m6A结合蛋白,通过识别共有GG(m6A)C基序靶向数千个mRNA转录物[58]。此外,IGF2BP与YTHDF2功能相反,IGF2BP以m6A依赖性方式促进其靶mRNA稳定和储存。
4 动物mRNA中m6A修饰的生理功能mRNA中m6A修饰几乎参与所有的RNA代谢过程,进而在动物生长发育、生殖过程、脂肪代谢、神经发育、免疫反应、细胞分化与重编程、响应应激和昼夜节律等生理过程中发挥重要作用(图 1)。
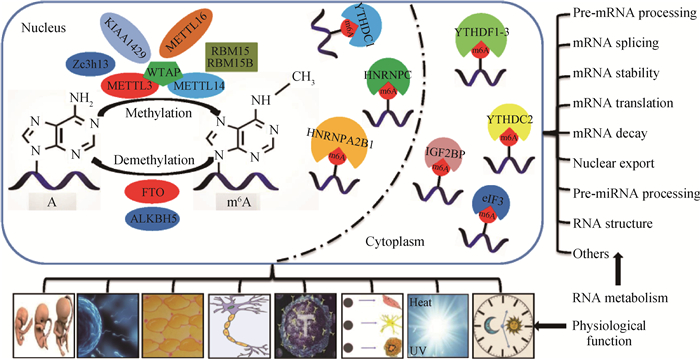
|
图 1 m6A修饰动态调节RNA代谢和动物生理功能示意图 Figure 1 The schematic diagram of m6A modification dynamically regulating RNA metabolism and animal physiological functions |
mRNA中m6A修饰对植物生长发育至关重要,例如在拟南芥生长后期m6A水平降低导致其生长模式改变和顶端优势降低,甲基化复合物组分缺失导致m6A丧失以致于胚胎致死等[25, 66-67]。同样,m6A修饰对维持动物的正常生长发育也是不可缺少的。mRNA中m6A修饰对胚胎发育至关重要,斑马鱼胚胎中甲基化酶复合物组分缺失导致多种发育缺陷,包括较小的头部和眼睛、较小的脑室和弯曲的脊索等[33, 68]。此外,斑马鱼发育早期胚胎中超过三分之一的母源mRNA上存在m6A修饰,而YTHDF2将这些母源mRNA进行清除后才会使合子基因组激活[68]。YTHDF2缺失减慢了m6A修饰的母体mRNA清除,阻碍合子基因组激活,导致细胞周期发生中止从而使斑马鱼子代发育缓慢。在成肌细胞中,METTL3介导的m6A修饰影响了MyoD(myoblast determination protein D)mRNA水平,METTL3是骨骼肌分化的关键调节剂,有助于肌肉生长发育[69]。通过对野猪、长白猪和荣昌猪的肌肉和脂肪组织进行全转录组m6A甲基化图谱分析,发现m6A甲基化程度与转录水平之间存在一定的关系并调节基因的表达,m6A修饰在猪肌肉和脂肪发育过程中发挥作用[70]。猪不同月龄肝中,m6A修饰水平与修饰基因表达之间呈高度负相关,m6A修饰所调节的基因主要参与生长和发育过程,这进一步表明mRNA中m6A修饰对动物生长发育至关重要[71]。
4.2 m6A调节动物生殖过程METTL3和METTL14介导的m6A甲基化调节雄性动物精子发生[72]。METTL3缺失严重抑制精原细胞分化并阻止减数分裂的开始[31]。METTL3和m6A水平升高与精子活力之间存在相关性,弱精子症患者精子中m6A含量、METTL3和METTL14的mRNA表达水平显著高于正常人[73]。METTL3和METTL14介导的m6A甲基化可改变精子发生过程中起重要作用基因的选择性剪接,进而抑制性激素合成和促性腺激素信号传导过程中相关基因的表达[31, 74]。m6A去甲基化酶ALKBH5也在精子发生中发挥重要作用,ALKBH5缺失的雄性小鼠mRNA中m6A增加,精子发生受损[43]。已知ALKBH5调节mRNA剪接和稳定性,在精母细胞和圆形精子细胞核中,ALKBH5介导的m6A可以擦除较长3′UTR使得正常拼接以及减少较短转录物产生[44]。YTHDF2和YTHDC2对于哺乳动物卵母细胞发育和减数分裂过程至关重要。YTHDF2在卵泡发生过程中的卵母细胞和颗粒细胞中均有表达,并且维持MII期卵母细胞发育[75]。YTHDC2可以选择性结合含有m6A的基序,增强其靶标翻译和降解。YTHDC2缺失导致雄性和雌性小鼠不育,睾丸和卵巢明显缩小,生殖细胞发育不会超过偶线期[53, 76]。减数分裂特异性蛋白质MEIOC以不依赖RNA的方式与YTHDC2相互作用以稳定减数分裂前期转录物,从而正确诱导减数分裂过程[77-78]。在减数分裂前期之后发生的转录很少,因此在晚期减数分裂阶段转录,并且不再需要的转录物可能被清除[79-80]。而YTHDC2可以更有效地靶向这些剩余转录物,翻译完成后使转录物不稳定,从而使减数分裂前期适当进展。
4.3 m6A调节动物脂肪代谢mRNA中m6A修饰在脂肪代谢过程中发挥了重要作用。m6A修饰通过调控甲基化相关代谢酶、转录因子和脂肪组织特异性基因表达等参与脂肪代谢,其中FTO作为脱甲基化酶和脂肪特异性表达基因在脂肪代谢过程中的作用不可忽视[81-82]。mRNA中m6A甲基化水平与动物肥胖表型负相关,过表达METTL3可增加m6A水平,抑制脂肪沉积,过表达FTO会降低m6A水平,促进脂肪沉积[42, 81, 83-84]。FTO的脱甲基化酶是前脂肪细胞分化所必需的,FTO缺失阻断脂肪前体细胞分化,具有催化活性的FTO通过控制m6A水平来调节转录因子RUNX1T1的可变剪切,进而调控脂肪前体细胞分化[42, 85]。在骨骼肌中,m6A甲基化水平与骨骼肌脂质含量也呈负相关,其通过影响脂质代谢相关基因的表达而增加骨骼肌细胞中脂质积累[86]。此外,一些甲基供体(甜菜碱)和甲基化抑制剂(环亮氨酸)通过控制mRNA中m6A水平来调节肝脂肪代谢,从而为调控肝脂肪代谢提供了新的靶点[87-89]。
4.4 m6A调节动物神经系统发育神经系统正常发育受多种因子的精确调控,其发育过程中存在高水平的m6A修饰,这些m6A修饰对神经系统功能的发挥非常关键。m6A产生受到抑制将导致果蝇神经元功能受损进而造成其行为异常,例如不能飞行、不能正确折叠翅膀和无法定向等[90-91]。m6A修饰也可以控制哺乳动物神经发生,胚胎小鼠皮层m6A特异性表达基因被富集到神经发生、细胞周期和神经元分化等过程,敲除胚胎期小鼠大脑中METTL14基因可导致m6A丢失,放射状神经胶质细胞的细胞周期延长,并将皮质神经发生延伸到出生后阶段,敲除METTL3的结果与之相同[11]。此外,具有脱甲基酶活性的FTO也参与神经系统发育,其在人神经干细胞和神经元中表达,并且动态调控出生后神经发育过程。FTO缺失导致脑部质量减少,降低神经干细胞增殖和神经元分化,最终导致记忆障碍[92]。
4.5 m6A调节动物免疫反应T细胞具有多种生物学功能,如杀伤靶细胞、调节机体免疫反应和辅助B细胞产生抗体等,体内T细胞稳态对自身免疫至关重要。已经发现,mRNA中m6A修饰在许多细胞过程中起着重要作用,m6A修饰是否对T细胞命运也起重要作用?研究表明,HIV-1感染CD4 T细胞会触发宿主和病毒mRNA中m6A水平显著增加并且帮助其复制,这些被甲基化的基因显著富集到病毒基因表达的功能中[93]。m6A通过METTL3/METTL14和ALKBH5帮助HIV-1复制,敲除METTL3/METTL14导致复制减少,而敲除ALKBH5与之相反[93]。此外,m6A还影响了HIV-1 Rev蛋白与体内RRE的结合,并影响RNA核输出[93]。这项研究确定了控制HIV-1复制及其与宿主免疫系统相互作用的新机制,同样,其它RNA病毒的复制也可能受到m6A的调控,这还需进一步的研究。在小鼠中,m6A修饰通过靶向IL-7/STAT5/SOCS途径来控制CD4 T细胞稳态,METTL3缺失会破坏T细胞稳态和分化,从而抑制naïve CD4 T细胞过继转输诱导肠炎模型中肠炎的发生[94]。在此研究基础上,研究者进一步发现,离乳3~4周的小鼠调节性T细胞中缺失METTL3导致淋巴结肿大、脾肿大、炎症因子表达上调和毛发脱落等严重的自身免疫性疾病,这些现象表明,mRNA中m6A修饰对维持T细胞抑制性功能至关重要[95]。而在幼鼠中,METTL3介导的m6A修饰并不影响T细胞发育过程,离乳后小鼠开始接触外界抗原,METTL3缺失导致T细胞数显著降低且基因表达紊乱,抑制功能显著降低。此外,m6A修饰维持T细胞的抑制功能也受到IL-2-STAT5信号通路的调节,这些结果为T细胞介导的免疫性疾病治疗提供了新的途径。Ⅰ型干扰素在抵抗病毒感染的先天防御中也具有重要作用,研究发现,DDX(DEAD-box)家族成员DDX46在病毒感染后抑制Ⅰ型干扰素的产生[96]。而这一过程主要通过DDX46招募m6A脱甲基化酶ALKBH5诱导核内天然抗病毒基因表达的mRNA滞留,并抑制干扰素产生,进而负调节抗病毒天然免疫反应。
4.6 m6A调节动物细胞分化和重编程细胞分化是一个复杂的过程,在这个过程中细胞形态、结构和功能向不同方向转变。而表观遗传修饰如RNA甲基化可以改变细胞命运,使分化的细胞逆转恢复全能性或从一种分化细胞转化为另一种分化细胞[97-99]。在造血干/祖细胞、胚胎干细胞和神经干细胞中缺失METTL3或METTL14导致增殖减少,过早分化;而过表达METTL3或METTL14会抑制分化并增加细胞增殖[100-104]。但也有一些研究表明,缺失METTL3的小鼠胚胎干细胞不能分化并且显示胚胎性致死,这与其它研究结果相互矛盾[105]。进一步的研究发现,m6A通过影响促进多能性基因(转录因子)mRNA水平与稳定性,对胚胎干细胞多能性进行调控[106]。例如SMAD2/3与METTL3-METTL14-WTAP复合物直接互作,在干细胞分化时,SMAD2/SMAD3调控下游靶基因(nanog)mRNA中m6A水平升高,使得干细胞迅速跳出多能性,向特定方向分化[107]。Zc3h13与WTAP、Virilizer和Hakai之间的相互作用对于m6A修饰和胚胎干细胞多能性调节是必需的[38]。Zc3h13缺失导致胚胎干细胞形态改变和自我更新能力降低,这主要是通过Zc3h13复合物来调节分化基因的表达进而影响多能性。近期研究发现,具有m6A修饰的LincRNA 1281在胚胎干细胞中特异性高表达,并且对于胚胎干细胞正常分化是必须的[108]。LincRNA 1281作为内源竞争性RNA与miRNA(let-7)相互作用,从而维持胚胎干细胞正常的分化潜能,这种新的RNA调控机制为探索mRNA中m6A修饰在细胞分化中提供了基础。总之,可以通过控制m6A甲基化水平以及多能性基因表达来调节细胞分化或细胞转换,这对细胞分化和发育过程中塑造细胞状态至关重要。
4.7 m6A响应动物应激反应无论热休克、缺氧和紫外线应激的起源如何,机体和细胞必须以增加其存活机会或使其凋亡的方式改变其代谢和基因表达。而mRNA水平的变化可以迅速发生并且对于适应快速变化的环境是必需的。越来越多的证据表明,mRNA中m6A修饰响应于应激而改变,并且可以将外部应激与复杂的转录、转录后和翻译过程网络连接起来。在小鼠胚胎成纤维细胞中,为了响应热休克应激,5′UTR中腺苷会被优先甲基化[109]。5′UTR中m6A修饰峰的增加是由于应激诱导后YTHDF2核定位所致。在热应激情况下,核YTHDF2通过限制FTO阻止脱甲基化维持应激诱导转录物5′UTR甲基化。此外,热休克诱导的HSP70翻译是由5′UTR介导的,HSP70在5′UTR内含有m6A位点,热休克后HSP70的5′UTR中m6A峰显著增加,使其以不依赖帽子的方式翻译[54]。多项研究将缺氧与mRNA中m6A修饰联系起来,m6A脱甲基化酶ALKBH5是缺氧诱导因子1α(HIF-1α)的直接靶点[110-111]。缺氧环境使ALKBH5表达增加而促进m6A脱甲基化,同时增强了一些多能性因子(如NANOG)的稳定性,这是肿瘤形成和转移所必需的[112]。mRNA中m6A修饰还可以调节紫外线诱导的DNA损伤应答反应[113]。紫外线诱导的DNA损伤导致m6A水平升高,METTL3和METTL14定位于DNA损伤部位。RNA中m6A相关酶Poly-ADP核糖聚合酶(PARP)、METTL3/14、FTO和DNA聚合酶κ(polκ)形成一种新的动态通路,可在紫外线诱导的DNA损伤中快速应答。此外,mRNA中m6A修饰还可以响应饥饿、毒素、干扰素和氧化应激等刺激,这些结果为机体或细胞响应应激的研究提供了一个新的途径。
4.8 m6A调节动物昼夜节律昼夜节律是机体适应环境周期性变化的一种精细机制,在代谢稳态和细胞周期等多种生理过程中发挥作用。像DNA甲基化一样,mRNA中m6A修饰在哺乳动物昼夜节律调节中也发挥着重要作用,其丰度与昼夜节律成反比[114]。哺乳动物昼夜节律也可以通过时钟基因(隐花色素蛋白)的反转录-翻译反馈环调节,隐花色素蛋白缺失导致m6A水平降低,进而影响m6A介导的昼夜节律[115]。已知mRNA中m6A修饰可以调节RNA加工,而时钟基因具有多个m6A甲基化位点,m6A缺乏导致RNA加工延迟,从而影响时钟基因表达的速度和稳定性。
5 展望动物mRNA中m6A修饰是最丰富的RNA修饰,其可以被甲基转移酶、脱甲基酶和m6A结合蛋白动态沉积、去除和识别,一旦打破这种平衡将会引起严重的生理失调。近年来,随着基于抗体的测序技术不断发展,m6A与mRNA代谢之间的关系不断被阐明。mRNA中m6A修饰几乎影响RNA代谢的每个步骤,包括前体mRNA加工、选择性剪接、核输出、翻译和降解等。目前,已经构建出人、小鼠、猪和斑马鱼等动物的全转录组m6A修饰图谱,揭示其在动物生长发育、生殖过程、脂肪代谢、神经发育、免疫反应、细胞分化与重编程、响应应激和昼夜节律等生理过程中发挥重要作用。mRNA中m6A修饰研究已经取得了实质性进展,但仍然存在一些问题或挑战。第一,目前在转录组范围内检测m6A的方法主要依赖于m6A抗体富集,与其质量密切相关,抗体选择不当将造成假阳性;第二,MeRIP-Seq方法只能将m6A残基定位在100~200 nt的转录本区域中,无法在全转录组水平上鉴定m6A的精确位置;第三,像其它RNA结合蛋白免疫沉淀方法一样,MeRIP-Seq测序需要至少300 μg或高达mg级别的总RNA作为原始材料,对于一些小样本或珍贵样品不适用;第四,为什么m6A甲基化酶只对一些mRNA起作用而不是全部,是否还存在其它甲基化相关酶以及这些酶之间是如何协调作用的;第五,其它RNA修饰与m6A之间是否存在联系,它们是否一起调节某种转录物的生物学过程。这些问题有待进一步研究。
| [1] | DOMINISSINI D, MOSHITCH-MOSHKOVITZ S, SCHWARTZ S, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq[J]. Nature, 2012, 485(7397): 201–206. DOI: 10.1038/nature11112 |
| [2] | MEYER K D, SALETORE Y, ZUMBO P, et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons[J]. Cell, 2012, 149(7): 1635–1646. DOI: 10.1016/j.cell.2012.05.003 |
| [3] | JIA G F, FU Y, ZHAO X, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO[J]. Nat Chem Biol, 2011, 7(12): 885–887. DOI: 10.1038/nchembio.687 |
| [4] | DESROSIERS R C, FRIDERICI K H, ROTTMAN F M. Characterization of Novikoff hepatoma mRNA methylation and heterogeneity in the methylated 5′ terminus[J]. Biochemistry, 1975, 14(20): 4367–4374. DOI: 10.1021/bi00691a004 |
| [5] | WEI C M, GERSHOWITZ A, MOSS B. Methylated nucleotides block 5′ terminus of HeLa cell messenger RNA[J]. Cell, 1975, 4(4): 379–386. DOI: 10.1016/0092-8674(75)90158-0 |
| [6] | ROTTMAN F, SHATKIN A J, PERRY R P. Sequences containing methylated nucleotides at the 5′ termini of messenger RNAs:possible implications for processing[J]. Cell, 1974, 3(3): 197–199. DOI: 10.1016/0092-8674(74)90131-7 |
| [7] | KE S D, ALEMU E A, MERTENS C, et al. A majority of m6A residues are in the last exons, allowing the potential for 3′ UTR regulation[J]. Genes Dev, 2015, 29(19): 2037–2053. DOI: 10.1101/gad.269415.115 |
| [8] | BATISTA P J. The RNA modification N6-methyladenosine and its implications in human disease[J]. Genomics Proteom Bioinf, 2017, 15(3): 154–163. DOI: 10.1016/j.gpb.2017.03.002 |
| [9] | ENGEL M, CHEN A. The emerging role of mRNA methylation in normal and pathological behavior[J]. Genes Brain Behav, 2018, 17(3): e12428. DOI: 10.1111/gbb.2018.17.issue-3 |
| [10] | CHANG M Q, LV H Y, ZHANG WL, et al. Region-specific RNA m6A methylation represents a new layer of control in the gene regulatory network in the mouse brain[J]. Open Biol, 2017, 7(9): 170166. DOI: 10.1098/rsob.170166 |
| [11] | YOON K J, RINGELING F R, VISSERS C, et al. Temporal control of mammalian cortical neurogenesis by m6A methylation[J]. Cell, 2017, 171(4): 877–889. DOI: 10.1016/j.cell.2017.09.003 |
| [12] | CHANDOLA U, DAS R, PANDA B. Role of the N6-methyladenosine RNA mark in gene regulation and its implications on development and disease[J]. Brief Funct Genomics, 2015, 14(3): 169–179. DOI: 10.1093/bfgp/elu039 |
| [13] | ALARCÓN C R, LEE H, GOODARZI H, et al. N6-methyladenosine marks primary microRNAs for processing[J]. Nature, 2015, 519(7544): 482–485. DOI: 10.1038/nature14281 |
| [14] | LIU N, PARISIEN M, DAI Q, et al. Probing N6-methyladenosine RNA modification status at single nucleotide resolution in mRNA and long noncoding RNA[J]. RNA, 2013, 19(12): 1848–1856. DOI: 10.1261/rna.041178.113 |
| [15] | CHEN K, LU Z K, WANG X, et al. High-resolution N6-methyladenosine (m6A) map using photo-crosslinking-assisted m6A sequencing[J]. Angew Chem Int Ed, 2015, 54(5): 1587–1590. DOI: 10.1002/anie.201410647 |
| [16] | LINDER B, GROZHIK A V, OLARERIN-GEORGE A O, et al. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome[J]. Nat Methods, 2015, 12(8): 767–772. DOI: 10.1038/nmeth.3453 |
| [17] | MOLINIE B, WANG J K, LIM K S, et al. m6A-LAIC-seq reveals the census and complexity of the m6A epitranscriptome[J]. Nat Methods, 2016, 13(8): 692–698. DOI: 10.1038/nmeth.3898 |
| [18] | WANG X F, YAN R X. RFAthM6A:a new tool for predicting m6A sites in Arabidopsis thaliana[J]. Plant Mol Biol, 2018, 96(3): 327–337. DOI: 10.1007/s11103-018-0698-9 |
| [19] | ANTANAVICIUTE A, BAQUERO-PEREZ B, WATSON C M, et al. m6aViewer:software for the detection, analysis, and visualization of N6-methyladenosine peaks from m6A-seq/ME-RIP sequencing data[J]. RNA, 2017, 23(10): 1493–1501. DOI: 10.1261/rna.058206.116 |
| [20] | LI G Q, LIU Z, SHEN H B, et al. TargetM6A:identifying N6-methyladenosine sites from RNA sequences via position-specific nucleotide propensities and a support vector machine[J]. IEEE Trans Nanobioscience, 2016, 15(7): 674–682. DOI: 10.1109/TNB.2016.2599115 |
| [21] | ZHOU Y, ZENG P, LI Y H, et al. SRAMP:prediction of mammalian N6-methyladenosine (m6A) sites based on sequence-derived features[J]. Nucleic Acids Res, 2016, 44(10): e91. DOI: 10.1093/nar/gkw104 |
| [22] | JIA C Z, ZHANG J J, GU W Z. RNA-MethylPred:a high-accuracy predictor to identify N6-methyladenosine in RNA[J]. Anal Biochem, 2016, 510: 72–75. DOI: 10.1016/j.ab.2016.06.012 |
| [23] | CHEN W, FENG P M, DING H, et al. iRNA-Methyl:identifying N6-methyladenosine sites using pseudo nucleotide composition[J]. Anal Biochem, 2015, 490: 26–33. DOI: 10.1016/j.ab.2015.08.021 |
| [24] | LIU Z, XIAO X, YU D J, et al. pRNAm-PC:predicting N6-methyladenosine sites in RNA sequences via physical-chemical properties[J]. Anal Biochem, 2016, 497: 60–67. DOI: 10.1016/j.ab.2015.12.017 |
| [25] | ZHONG S L, LI H Y, BODI Z, et al. MTA is an Arabidopsis messenger RNA adenosine methylase and interacts with a homolog of a sex-specific splicing factor[J]. Plant Cell, 2008, 20(5): 1278–1288. DOI: 10.1105/tpc.108.058883 |
| [26] | LIU J Z, YUE Y A, HAN D L, et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation[J]. Nat Chem Biol, 2014, 10(2): 93–95. DOI: 10.1038/nchembio.1432 |
| [27] | SCHWARTZ S, MUMBACH M R, JOVANOVIC M, et al. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5′ sites[J]. Cell Rep, 2014, 8(1): 284–296. DOI: 10.1016/j.celrep.2014.05.048 |
| [28] | YANG Y, HSU P J, CHEN Y S, et al. Dynamic transcriptomic m6A decoration:writers, erasers, readers and functions in RNA metabolism[J]. Cell Rep, 2018, 28(6): 616–624. DOI: 10.1038/s41422-018-0040-8 |
| [29] | DENG X L, SU R, WENG H Y, et al. RNA N6-methyladenosine modification in cancers:current status and perspectives[J]. Cell Rep, 2018, 28(5): 507–517. DOI: 10.1038/s41422-018-0034-6 |
| [30] | DU Y Z, HOU G F, ZHANG H L, et al. SUMOylation of the m6A-RNA methyltransferase METTL3 modulates its function[J]. Nucleic Acids Res, 2018, 46(10): 5195–5208. DOI: 10.1093/nar/gky156 |
| [31] | XU K, YANG Y, FENG G H, et al. Mettl3-mediated m6A regulates spermatogonial differentiation and meiosis initiation[J]. Cell Res, 2017, 27(9): 1100–1114. DOI: 10.1038/cr.2017.100 |
| [32] | LI Q, LI X, TANG H, et al. NSUN2-Mediated m5C methylation and METTL3/METTL14-mediated m6A methylation cooperatively enhance p21 translation[J]. J Cell Biochem, 2017, 118(9): 2587–2598. DOI: 10.1002/jcb.v118.9 |
| [33] | PING X L, SUN B F, WANG L, et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase[J]. Cell Res, 2014, 24(2): 177–189. DOI: 10.1038/cr.2014.3 |
| [34] | HORIUCHI K, KAWAMURA T, IWANARI H, et al. Identification of Wilms' tumor 1-associating protein complex and its role in alternative splicing and the cell cycle[J]. J Biol Chem, 2013, 288(46): 33292–33302. DOI: 10.1074/jbc.M113.500397 |
| [35] | PATIL D P, CHEN C K, PICKERING B F, et al. m6A RNA methylation promotes XIST-mediated transcriptional repression[J]. Nature, 2016, 537(7620): 369–373. DOI: 10.1038/nature19342 |
| [36] | WARDA A S, KRETSCHMER J, HACKERT P, et al. Human METTL16 is a N6-methyladenosine (m6A) methyltransferase that targets pre-mRNAs and various non-coding RNAs[J]. EMBO Rep, 2017, 18(11): 2004–2014. DOI: 10.15252/embr.201744940 |
| [37] | SHIMA H, MATSUMOTO M, ISHIGAMI Y, et al. S-adenosylmethionine synthesis is regulated by selective N6-adenosine methylation and mRNA degradation involving METTL16 and YTHDC1[J]. Cell Rep, 2017, 21(12): 3354–3363. DOI: 10.1016/j.celrep.2017.11.092 |
| [38] | WEN J, LV R T, MA H H, et al. Zc3h13 regulates nuclear RNA m6A methylation and mouse embryonic stem cell self-renewal[J]. Mol Cell, 2018, 69(6): 1028–1038. DOI: 10.1016/j.molcel.2018.02.015 |
| [39] | BARTOSOVIC M, MOLARES H C, GREGOROVA P, et al. N6-methyladenosine demethylase FTO targets pre-mRNAs and regulates alternative splicing and 3′-end processing[J]. Nucleic Acids Res, 2017, 45(19): 11356–11370. DOI: 10.1093/nar/gkx778 |
| [40] | MAUER J, LUO X B, BLANJOIE A, et al. Reversible methylation of m6Am in the 5′ cap controls mRNA stability[J]. Nature, 2017, 541(7637): 371–375. DOI: 10.1038/nature21022 |
| [41] | YU J, CHEN M X, HUANG H J, et al. Dynamic m6A modification regulates local translation of mRNA in axons[J]. Nucleic Acids Res, 2018, 46(3): 1412–1423. DOI: 10.1093/nar/gkx1182 |
| [42] | ZHAO X, YANG Y, SUN B F, et al. FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis[J]. Cell Res, 2014, 24(12): 1403–1419. DOI: 10.1038/cr.2014.151 |
| [43] | ZHENG G Q, DAHL J A, NIU Y M, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility[J]. Mol Cell, 2013, 49(1): 18–29. DOI: 10.1016/j.molcel.2012.10.015 |
| [44] | TANG C, KLUKOVICH R, PENG H Y, et al. ALKBH5-dependent m6A demethylation controls splicing and stability of long 3′-UTR mRNAs in male germ cells[J]. Proc Natl Acad Sci U S A, 2018, 115(2): E325–E333. DOI: 10.1073/pnas.1717794115 |
| [45] | WANG X, ZHAO B S, ROUNDTREE I A, et al. N6-methyladenosine modulates messenger RNA translation efficiency[J]. Cell, 2015, 161(6): 1388–1399. DOI: 10.1016/j.cell.2015.05.014 |
| [46] | LI J F, MENG S, XU M J, et al. Downregulation of N6-methyladenosine binding YTHDF2 protein mediated by miR-493-3p suppresses prostate cancer by elevating N6-methyladenosine levels[J]. Oncotarget, 2018, 9(3): 3752–3764. |
| [47] | YANG Z, LI J, FENG G X, et al. microRNA-145 modulates N6-methyladenosine levels by targeting the 3′-untranslated mRNA region of the N6-methyladenosine binding YTH domain family 2 protein[J]. J Biol Chem, 2017, 292(9): 3614–3623. DOI: 10.1074/jbc.M116.749689 |
| [48] | WANG X, LU Z K, GOMEZ A, et al. N6-methyladenosine-dependent regulation of messenger RNA stability[J]. Nature, 2014, 505(7481): 117–120. DOI: 10.1038/nature12730 |
| [49] | LI A, CHEN Y S, PING X L, et al. Cytoplasmic m6A reader YTHDF3 promotes mRNA translation[J]. Cell Res, 2017, 27(3): 444–447. DOI: 10.1038/cr.2017.10 |
| [50] | SHI H L, WANG X, LU Z K, et al. YTHDF3 facilitates translation and decay of N6-methyladenosine-modified RNA[J]. Cell Res, 2017, 27(3): 315–328. DOI: 10.1038/cr.2017.15 |
| [51] | XIAO W, ADHIKARI S, DAHAL U, et al. Nuclear m6A reader YTHDC1 regulates mRNA splicing[J]. Mol Cell, 2016, 61(4): 507–519. DOI: 10.1016/j.molcel.2016.01.012 |
| [52] | ROUNDTREE I A, LUO G Z, ZHANG Z J, et al. YTHDC1 mediates nuclear export of N6-methyladenosine methylated mRNAs[J]. eLife, 2017, 6: e31311. DOI: 10.7554/eLife.31311 |
| [53] | HSU P J, ZHU Y F, MA H H, et al. Ythdc2 is an N6-methyladenosine binding protein that regulates mammalian spermatogenesis[J]. Cell Res, 2017, 27(9): 1115–1127. DOI: 10.1038/cr.2017.99 |
| [54] | MEYER K D, PATIL D P, ZHOU J, et al. 5′ UTR m6A promotes cap-independent translation[J]. Cell, 2015, 163(4): 999–1010. DOI: 10.1016/j.cell.2015.10.012 |
| [55] | ALARCÓ C R, GOODARZI H, LEE H, et al. HNRNPA2B1 is a mediator of m6A-dependent nuclear RNA processing events[J]. Cell, 2015, 162(6): 1299–1308. DOI: 10.1016/j.cell.2015.08.011 |
| [56] | CIENIKOVÓ Z, DAMBERGER F F, HALL J, et al. Structural and mechanistic insights into poly (uridine) tract recognition by the hnRNP C RNA recognition motif[J]. J Am Chem Soc, 2014, 136(41): 14536–14544. DOI: 10.1021/ja507690d |
| [57] | LIU N, DAI Q, ZHENG G Q, et al. N6-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions[J]. Nature, 2015, 518(7540): 560–564. DOI: 10.1038/nature14234 |
| [58] | HUANG H L, WENG H Y, SUN W J, et al. Recognition of RNA N6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation[J]. Nat Cell Biol, 2018, 20(3): 285–295. DOI: 10.1038/s41556-018-0045-z |
| [59] | WANG X, FENG J, XUE Y, et al. Structural basis of N6-adenosine methylation by the METTL3-METTL14 complex[J]. Nature, 2016, 534(7608): 575–578. DOI: 10.1038/nature18298 |
| [60] | WANG P, DOXTADER K A, NAM Y S. Structural basis for cooperative function of mettl3 and mettl14 methyltransferases[J]. Mol Cell, 2016, 63(2): 306–317. DOI: 10.1016/j.molcel.2016.05.041 |
| [61] | ŚLEDŹ P, JINEK M. Structural insights into the molecular mechanism of the m6A writer complex[J]. eLife, 2016, 5: e18434. DOI: 10.7554/eLife.18434 |
| [62] | RUSZKOWSKA A, RUSZKOWSKI M, DAUTER Z, et al. Structural insights into the RNA methyltransferase domain of METTL16[J]. Sci Rep, 2018, 8(1): 5311. DOI: 10.1038/s41598-018-23608-8 |
| [63] | PENDLETON K E, CHEN B B, LIU K Q, et al. The U6 snRNA m6A methyltransferase METTL16 regulates SAM synthetase intron retention[J]. Cell, 2017, 169(5): 824–835. DOI: 10.1016/j.cell.2017.05.003 |
| [64] | KNUCKLES P, LENCE T, HAUSSMANN I U, et al. Zc3h13/Flacc is required for adenosine methylation by bridging the mRNA-binding factor Rbm15/Spenito to the m6A machinery component Wtap/Fl(2)d[J]. Genes Dev, 2018, 32(5-6): 415–429. DOI: 10.1101/gad.309146.117 |
| [65] | FU Y, JIA G F, PANG X Q, et al. FTO-mediated formation of N6-hydroxymethyladenosine and N6-formyladenosine in mammalian RNA[J]. Nat Commun, 2013, 4: 1798. DOI: 10.1038/ncomms2822 |
| [66] | BODI Z, ZHONG S L, MEHRA S, et al. Adenosine methylation in arabidopsis mRNA is associated with the 3′ end and reduced levels cause developmental defects[J]. Front Plant Sci, 2012, 3: 48. |
| [67] | VESPA L, VACHON G, BERGER F, et al. The immunophilin-interacting protein AtFIP37 from Arabidopsis is essential for plant development and is involved in trichome endoreduplication[J]. Plant Physiol, 2004, 134(4): 1283–1292. DOI: 10.1104/pp.103.028050 |
| [68] | ZHAO B S, WANG X, BEADELL A V, et al. m6A-dependent maternal mRNA clearance facilitates zebrafish maternal-to-zygotic transition[J]. Nature, 2017, 542(7642): 475–478. DOI: 10.1038/nature21355 |
| [69] | KUDOU K, KOMATSU T, NOGAMI J, et al. The requirement of Mettl3-promoted MyoD mRNA maintenance in proliferative myoblasts for skeletal muscle differentiation[J]. Open Biol, 2017, 7(9): 170119. DOI: 10.1098/rsob.170119 |
| [70] | TAO X L, CHEN J N, JIANG Y Z, et al. Transcriptome-wide N6-methyladenosine methylome profiling of porcine muscle and adipose tissues reveals a potential mechanism for transcriptional regulation and differential methylation pattern[J]. BMC Genomics, 2017, 18(1): 336. DOI: 10.1186/s12864-017-3719-1 |
| [71] | HE S, WANG H, LIU R, et al. mRNA N6-methyladenosine methylation of postnatal liver development in pig[J]. PLoS One, 2017, 12(3): e0173421. DOI: 10.1371/journal.pone.0173421 |
| [72] | LIN Z, HSU P J, XING X D, et al. Mettl3-/Mettl14-mediated mRNA N6-methyladenosine modulates murine spermatogenesis[J]. Cell Res, 2017, 27(10): 1216–1230. DOI: 10.1038/cr.2017.117 |
| [73] | YANG Y, HUANG W, HUANG J T, et al. Increased N6-methyladenosine in human sperm RNA as a risk factor for asthenozoospermia[J]. Sci Rep, 2016, 6: 24345. DOI: 10.1038/srep24345 |
| [74] | XIA H, ZHONG C R, WU X X, et al. Mettl3 mutation disrupts gamete maturation and reduces fertility in zebrafish[J]. Genetics, 2018, 208(2): 729–743. DOI: 10.1534/genetics.117.300574 |
| [75] | IVANOVA I, MUCH C, DI GIACOMO M, et al. The RNA m6A reader YTHDF2 is essential for the post-transcriptional regulation of the maternal transcriptome and oocyte competence[J]. Mol Cell, 2017, 67(6): 1059–1067. DOI: 10.1016/j.molcel.2017.08.003 |
| [76] | JAIN D, PUNO M R, MEYDAN C, et al. Ketu mutant mice uncover an essential meiotic function for the ancient RNA helicase YTHDC2[J]. eLife, 2018, 7: e30919. DOI: 10.7554/eLife.30919 |
| [77] | ABBY E, TOURPIN S, RIBEIRO J, et al. Implementation of meiosis prophase I programme requires a conserved retinoid-independent stabilizer of meiotic transcripts[J]. Nat Commun, 2016, 7: 10324. DOI: 10.1038/ncomms10324 |
| [78] | SOH Y Q S, MIKEDIS M M, KOJIMA M, et al. Meioc maintains an extended meiotic prophase I in mice[J]. PLoS Genet, 2017, 13(4): e1006704. DOI: 10.1371/journal.pgen.1006704 |
| [79] | PAGE J, DE LA FUENTE R, MANTEROLA M, et al. Inactivation or non-reactivation:what accounts better for the silence of sex chromosomes during mammalian male meiosis[J]. Chromosoma, 2012, 121(3): 307–326. DOI: 10.1007/s00412-012-0364-y |
| [80] | MONESI V. Ribonucleic acid synthesis during mitosis and meiosis in the mouse testis[J]. J Cell Biol, 1964, 22(3): 521–532. DOI: 10.1083/jcb.22.3.521 |
| [81] | WANG X X, ZHU L N, CHEN J Q, et al. mRNA m6A methylation downregulates adipogenesis in porcine adipocytes[J]. Biochem Biophys Res Commun, 2015, 459(2): 201–207. DOI: 10.1016/j.bbrc.2015.02.048 |
| [82] | WANG X X, SUN B F, JIANG Q, et al. mRNA m6A plays opposite role in regulating UCP2 and PNPLA2 protein expression in adipocytes[J]. Int J Obes, 2018. DOI: 10.1038/s41366-018-0027-z |
| [83] | CHURCH C, MOIR L, MCMURRAY F, et al. Overexpression of Fto leads to increased food intake and results in obesity[J]. Nat Genet, 2010, 42(12): 1086–1092. DOI: 10.1038/ng.713 |
| [84] | FISCHER J, KOCH L, EMMERLING C, et al. Inactivation of the Fto gene protects from obesity[J]. Nature, 2009, 458(7240): 894–898. DOI: 10.1038/nature07848 |
| [85] | ZHANG M Z, ZHANG Y, MA J, et al. The demethylase activity of FTO (fat mass and obesity associated protein) is required for preadipocyte differentiation[J]. PLoS One, 2015, 10(7): e0133788. DOI: 10.1371/journal.pone.0133788 |
| [86] | WU W C, FENG J, JIANG D H, et al. AMPK regulates lipid accumulation in skeletal muscle cells through FTO-dependent demethylation of N6-methyladenosine[J]. Sci Rep, 2017, 7: 41606. DOI: 10.1038/srep41606 |
| [87] | KANG H F, ZHANG Z W, YU L, et al. FTO reduces mitochondria and promotes hepatic fat accumulation through RNA demethylation[J]. J Cell Biochem, 2018, 119(7): 5676–5685. DOI: 10.1002/jcb.v119.7 |
| [88] | ZHOU X H, CHEN J Q, CHEN J, et al. The beneficial effects of betaine on dysfunctional adipose tissue and N6-methyladenosine mRNA methylation requires the AMP-activated protein kinase α1 subunit[J]. J Nutr Biochem, 2015, 26(12): 1678–1684. DOI: 10.1016/j.jnutbio.2015.08.014 |
| [89] | LU N, LI X M, YU J Y, et al. Curcumin attenuates lipopolysaccharide-induced hepatic lipid metabolism disorder by modification of m6A RNA methylation in piglets[J]. Lipids, 2018, 53(1): 53–63. DOI: 10.1002/lipd.2018.53.issue-1 |
| [90] | LENCE T, AKHTAR J, BAYER M, et al. m6A modulates neuronal functions and sex determination in Drosophila[J]. Nature, 2016, 540(7632): 242–247. DOI: 10.1038/nature20568 |
| [91] | HAUSSMANN I U, BODI Z, SANCHEZ-MORAN E, et al. m6A potentiates Sxl alternative pre-mRNA splicing for robust Drosophila sex determination[J]. Nature, 2016, 540(7632): 301–304. DOI: 10.1038/nature20577 |
| [92] | LI L P, ZANG L Q, ZHANG F R, et al. Fat mass and obesity-associated (FTO) protein regulates adult neurogenesis[J]. Hum Mol Genet, 2017, 26(13): 2398–2411. DOI: 10.1093/hmg/ddx128 |
| [93] | LICHINCHI G, GAO S, SALETORE Y, et al. Dynamics of the human and viral m6A RNA methylomes during HIV-1 infection of T cells[J]. Nat Microbiol, 2016, 1(4): 16011. DOI: 10.1038/nmicrobiol.2016.11 |
| [94] | LI H B, TONG J Y, ZHU S, et al. m6A mRNA methylation controls T cell homeostasis by targeting the IL-7/STAT5/SOCS pathways[J]. Nature, 2017, 548(7667): 338–342. DOI: 10.1038/nature23450 |
| [95] | TONG J Y, CAO G C, ZHANG T, et al. m6A mRNA methylation sustains Treg suppressive functions[J]. Cell Res, 2018, 28(2): 253–256. DOI: 10.1038/cr.2018.7 |
| [96] | ZHENG Q L, HOU J, ZHOU Y, et al. The RNA helicase DDX46 inhibits innate immunity by entrapping m6A-demethylated antiviral transcripts in the nucleus[J]. Nat Immunol, 2017, 18(10): 1094–1103. DOI: 10.1038/ni.3830 |
| [97] | CHEN T, HAO Y J, ZHANG Y, et al. m6A RNA methylation is regulated by microRNAs and promotes reprogramming to pluripotency[J]. Cell Stem Cell, 2015, 16(3): 289–301. DOI: 10.1016/j.stem.2015.01.016 |
| [98] | LV J H, ZHANG Y F, GAO S W, et al. Endothelial-specific m6A modulates mouse hematopoietic stem and progenitor cell development via Notch signaling[J]. Cell Res, 2018, 28(2): 249–252. DOI: 10.1038/cr.2017.143 |
| [99] | AGUILO F, WALSH M J. The N6-methyladenosine RNA modification in pluripotency and reprogramming[J]. Curr Opin Genet Dev, 2017, 46: 77–82. DOI: 10.1016/j.gde.2017.06.006 |
| [100] | WENG H Y, HUANG H L, WU H Z, et al. METTL14 inhibits hematopoietic stem/progenitor differentiation and promotes leukemogenesis via mRNA m6A modification[J]. Cell Stem Cell, 2018, 22(2): 191–205. DOI: 10.1016/j.stem.2017.11.016 |
| [101] | ZHANG C X, CHEN Y S, SUN B F, et al. m6A modulates haematopoietic stem and progenitor cell specification[J]. Nature, 2017, 549(7671): 273–276. DOI: 10.1038/nature23883 |
| [102] | WANG Y, LI Y, TOTH J I, et al. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells[J]. Nat Cell Biol, 2014, 16(2): 191–198. DOI: 10.1038/ncb2902 |
| [103] | VU L P, PICKERING B F, CHENG Y M, et al. The N6-methyladenosine (m6A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells[J]. Nat Med, 2017, 23(11): 1369–1376. |
| [104] | WANG Y, LI Y, YUE M H, et al. N6-methyladenosine RNA modification regulates embryonic neural stem cell self-renewal through histone modifications[J]. Nat Neurosci, 2018, 21(2): 195–206. |
| [105] | BATISTA P J, MOLINIE B, WANG J K, et al. m6A RNA modification controls cell fate transition in mammalian embryonic stem cells[J]. Cell Stem Cell, 2014, 15(6): 707–719. DOI: 10.1016/j.stem.2014.09.019 |
| [106] | GEULA S, MOSHITCH-MOSHKOVITZ S, DOMINISSINI D, et al. Stem cells.m6A mRNA methylation facilitates resolution of naive pluripotency toward differentiation[J]. Science, 2015, 347(6225): 1002–1006. DOI: 10.1126/science.1261417 |
| [107] | BERTERO A, BROWN S, MADRIGAL P, et al. The SMAD2/3 interactome reveals that TGFβ controls m6A mRNA methylation in pluripotency[J]. Nature, 2018, 555(7695): 256–259. DOI: 10.1038/nature25784 |
| [108] | YANG D D, QIAO J, WANG G Y, et al. N6-methyladenosine modification of lincRNA 1281 is critically required for mESC differentiation potential[J]. Nucleic Acids Res, 2018, 46(8): 3906–3920. DOI: 10.1093/nar/gky130 |
| [109] | ZHOU J, WAN J, GAO X W, et al. Dynamic m6A mRNA methylation directs translational control of heat shock response[J]. Nature, 2015, 526(7574): 591–594. DOI: 10.1038/nature15377 |
| [110] | FRY N J, LAW B A, ILKAYEVA O R, et al. N6-methyladenosine is required for the hypoxic stabilization of specific mRNAs[J]. RNA, 2017, 23(9): 1444–1455. DOI: 10.1261/rna.061044.117 |
| [111] | THALHAMMER A, BENCOKOVA Z, POOLE R, et al. Human AlkB homologue 5 is a nuclear 2-oxoglutarate dependent oxygenase and a direct target of hypoxia-inducible factor 1α(HIF-1α)[J]. PLoS One, 2011, 6(1): e16210. DOI: 10.1371/journal.pone.0016210 |
| [112] | ZHANG C Z, SAMANTA D, LU H Q, et al. Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m6A-demethylation of NANOG mRNA[J]. Proc Natl Acad Sci U S A, 2016, 113(14): e2047–e2056. DOI: 10.1073/pnas.1602883113 |
| [113] | XIANG Y, LAURENT B, HSU C H, et al. RNA m6A methylation regulates the ultraviolet-induced DNA damage response[J]. Nature, 2017, 543(7646): 573–576. DOI: 10.1038/nature21671 |
| [114] | FUSTIN J M, DOI M, YAMAGUCHI Y, et al. RNA-methylation-dependent RNA processing controls the speed of the circadian clock[J]. Cell, 2013, 155(4): 793–806. DOI: 10.1016/j.cell.2013.10.026 |
| [115] | WANG C Y, YEH J K, SHIE S S, et al. Circadian rhythm of RNA N6-methyladenosine and the role of cryptochrome[J]. Biochem Biophys Res Commun, 2015, 465(1): 88–94. DOI: 10.1016/j.bbrc.2015.07.135 |



