2. Department of Pathogenic Biology, Chengde Medical University, Chengde 067000, Hebei, China;
3. Department for Viral Diarrhea, National Institute for Viral Disease Control and Prevention, Chinese Center for Disease Control and Prevention, Beijing 102206, China
Objective Newly identified human rhinovirus C (HRV-C) and human bocavirus (HBoV) cannot propagate in vitro in traditional cell culture models; thus obtaining knowledge about these viruses and developing related vaccines are difficult. Therefore, it is necessary to develop a novel platform for the propagation of these types of viruses.
Methods A platform for culturing human airway epithelia in a three-dimensional (3D) pattern using Matrigel as scaffold was developed. The features of 3D culture were identified by immunochemical staining and transmission electron microscopy. Nucleic acid levels of HRV-C and HBoV in 3D cells at designated time points were quantitated by real-time polymerase chain reaction (PCR). Levels of cytokines, whose secretion was induced by the viruses, were measured by ELISA.
Results Properties of bronchial-like tissues, such as the expression of biomarkers CK5, ZO-1, and PCK, and the development of cilium-like protuberances indicative of the human respiration tract, were observed in 3D-cultured human airway epithelial (HAE) cultures, but not in monolayer-cultured cells. Nucleic acid levels of HRV-C and HBoV and levels of virus-induced cytokines were also measured using the 3D culture system.
Conclusion Our data provide a preliminary indication that the 3D culture model of primary epithelia using a Matrigel scaffold in vitro can be used to propagate HRV-C and HBoV.
Non-influenza respiratory viruses such as parainfluenza virus, human rhinovirus, human metapneumovirus, respiratory syncytial virus (RSV), and human coronavirus affect people of all age groups, and they can cause mild to severe respiratory illnesses from common colds to severe respiratory disease[1-2]. The human rhinovirus C (HRV-C) species was discovered in 2006[3], and it is more virulent than HRV-A and HRV-B species, which are associated with severe illnesses such as pneumonia, acute wheezing, and exacerbation of asthma in infancy[4]. Human bocavirus (HBoV) was identified in 2005 from human nasopharyngeal aspirates of patients with respiratory tract illness[5]. Acute wheezing was the most common clinical symptoms observed in HBoV-infected patients[6]. Infections caused by these two viruses pose a substantial disease burden to young children; however, the biological properties and pathogenesis of HRV-C and HBoV have not been fully clarified. The ability of HRV-C and HBoV to propagate in traditional monolayer (2D) cellular culture or animals represents the bottleneck that limits related studies.
Traditional 2D cellular culture is a common platform to study infectious disease[7]; however, this method cannot mimic the physiological complexity of in vivo microenvironments, which might restrict the propagation of HRV-C and HBoV. The development of suitable cell culture models is thus promising for studying the life cycle of infectious viruses and their interactions with the host. Previously, studies have shown that HRV-C15 and HRV-C11 strains propagate in fully differentiated human nasal or bronchial epithelial cells[8], and HRV-C15 and HRV-C41 strains propagate in the differentiated human sinus epithelial cells[9-11]. Additionally, HRV-C2, HRV-C7, HRV-C12, HRV-C15, and HRV-C29 types were all capable of mediating productive infection in reconstituted 3D human airway epithelia[12]. Human trachea epithelial cells were previously cultured in an air-liquid interface (ALI), and they formed a pseudostratified epithelium, in which HBoV propagated[13]. These studies suggest that novel 3D tissue-like cell culture systems are suitable for studying HRV-C and HBoV.
Matrigel basement membrane matrix is a commercial cell culture medium consisting of a gelatinous protein mixture secreted by Engelbreth-Holm-Swarm (EHS) mouse sarcoma cells. It is rich in extracellular matrix (ECM) components, and used widely for 3D cell culture[14]. Cells cultured in Matrigel show many differences in gene and protein expression, survival, proliferation, differentiation, and metabolism, compared to those cultured by traditional 2D culture methods[15-18]. Cells grown in 2D and 3D cultures also respond differently to chemical drugs or radiation[19-20]. Thus, 3D cell culture is the third model that bridges the gap between traditional cell culture and animal models[21-23].
Although it has been demonstrated that viruses (HBoV, infectious bronchitis virus, avian influenza viruses, RSV, and HRV-C) propagate well in 3D cultured cells using ALI methodologies[24-27], this strategy is time-consuming (approximately 15 days). In the current study, we constructed a 3D cell culture model using Matrigel as the scaffold and measured the propagation abilities of HRV-C and HBoV using this platform. The results of this study showed that both HRV-C and HBoV propagate in the 3D Matrigel system, whereas they could not be detected in 2D cultures.
MATERIALS AND METHODS 2D Cell CulturePrimary human airway epithelial (HAE) cells were obtained from the Chinese Center for Disease Control and Prevention. Cells were cultured in BEGM media (Lonza, Walkersville, MD). HAE cells were used from passage 1 to passage 5 for all experiments, with passage 1 defined as the thawed cells from the nitrogen canister.
Trypsin-EDTA (0.25%) was used to remove cells from culture flasks for sub-culturing. Cells were incubated at 37 ℃ in a humidified atmosphere containing 5% CO2. Two million cells were seeded in 75 cm2 culture flasks and sub-cultured at 80% confluence.
3D CultureThe 3D culture system using Matrigel as a scaffold was prepared according to the manufacturer's protocol. Briefly, the mixture of Matrigel (BD Biosciences, Baltimore, MD) solution, thawed in an ice box, and cell suspension (1 × 104 cells/insert, 200 μL/insert) was added to the insert (12-Transwell plate), or pre-plated cells in the membrane of the insert, using cooled pipette tips, and then covered with a Matrigel layer. These samples and Matrigel only-coated plates, were incubated at 37 ℃ for 30-45 min for solidification. Further, the cells were plated on the surface of the Matrigel layer, for Matrigel only-coated inserts, and then 3D culture medium was added to all inserts and wells; medium was changed every 2 days. Cells in the inserts of 12-well uncoated plates were used as 2D controls. The morphology of cells at day 7 was observed using a reverse phase-contrast microscope.
Immunochemical StainingThe primary antibodies used for immunostaining of 2D and 3D cells included ZO-1 (1:500, Abcam, USA), Cytokeratin 5 (1:500, CST, USA), Pan-cytokeratin (1:500, Abcam, USA), and β-tubulin (1:1, 000, CST, USA). Secondary antibodies (anti-mouse or rabbit conjugated with Alexa Fluor488 and Alexa Fluor594) were purchased from Beyotime (1:2, 000, China).
For 2D cultures, cells were seeded at 1 × 104 cells in each well of 12-well tissue culture plates and cultured for 48 h before detection. Cells were fixed with 4% paraformaldehyde (PFA) for 20 min at room temperature and permeabilized with 0.5% triton X-100 in PBS on ice for 10 min. Non-specific binding was blocked with 5% bovine serum albumin (BSA) in PBS for 60 min at room temperature prior to probing with primary antibodies. The cells were incubated with the primary antibody diluted in 5% BSA/1 × PBS for 1 h at room temperature. After incubation, cells were washed three times with 1 × PBS (10 min each) and then incubated with the appropriate Alexa Fluor secondary antibodies diluted in 5% BSA for 1 h. Cells were then washed three times with PBS for 10 min each. Nuclei were counterstained with DAPI (Beyotime, China).
The 3D structures in Matrigel were immunostained as described previously[28] with the following modifications. The 3D cultures were fixed with 4% PFA for 30 min at room temperature and thereafter washed three times with PBS for 20 min each. Blocking was performed by incubating the structures in immunofluorescence buffer (5% BSA/0.2% triton X-100 in PBS) for 4 h. The 3D structures were incubated with primary antibody diluted in 5% BSA/1 × PBS overnight at room temperature. After incubation, the structures were washed three times with PBS, for 20 min each, and then incubated with the appropriate Alexa Fluor secondary antibodies diluted in 5% BSA for 1 h. The structures were washed three times with PBS for 20 min each. Nuclei were counterstained with DAPI. Analyses were performed with a fluorescence microscope (Axio Imager Z2) at 20× magnification.
Measurement by Transmission Electron Microscopy (TEM)Specimens were post-fixed in 1% osmium tetroxide, en bloc stained with 4% uranyl acetate, dehydrated through a graded series of alcohol and propylene oxide, and embedded in a mixture of Epon substitute and Araldite. Thin sections were stained with 4% uranyl acetate and Reynolds's lead citrate, and the images were obtained using a transmission electron microscope.
Virus and InfectionHRV-C15 and HBoV uncultured specimens were obtained from the Chinese Center for Disease Control and Prevention. They were then identified and quantified as previously described[29-30].
Media used for 3D cultures for a period of 7 days, and those for 2D cultures at exponential growth phase, were removed, and the cells were washed three times with sterile PBS. Viruses were diluted with BEGM to a desired concentration. Briefly, a 100 μL inoculum [approximately 106 copies of viral nucleotides (PCR genome equivalents)] per well was added to either the 3D cultures after culturing for 7 days or the 2D cultures. Cultures were gently shaken for 15 min at room temperature and then incubated with virus at 34 ℃ for 3 h[29]. The inoculum was removed and cells were washed twice with BEGM. Then, 500 μL and 1 mL of BEGM were added to the insert and basolateral chamber of the well, respectively. The media in the insert and basolateral chamber and cells were harvested at the designated time after infection and stored at -80 ℃ for further analysis.
RNA Extraction and Quantitative RT-PCR (qRT-PCR)Viral nucleic acids were extracted using the QIAGEN Viral DNA/RNA Kit (Valencia, CA) according to the manufacturer's protocol. Viral RNA or DNA loads were determined using the ABI 7500 real-time PCR system (Applied Biosystems). The RT-PCR reactions were prepared using the ABI Taqman EZ RT-PCR kit (Applied Biosystems, CA) according to the manufacturer's instruction. The primers for the detection of HRV-C were as follows: forward primer, 5x-AAAGATTGGACAGGGTGTGAAGA-3x; reverse primer, 5x-GAAACACGGACACCCAAAGTAGT-3x; and probe-linked with FAM/MGB, 5x-CCGGCCCCTGAAT-3x. The primers for the detection HBoV were as follows: forward primer, 5′-CCTATATAAGCTGCTGCACTTCCTG-3′; and reverse primer, 5′-AAGCCATAGTAGACTCACCACAAG-3′. The RT-PCR conditions were as follows: (ⅰ) reverse transcription at 60 ℃ for 30 min; (ⅱ) denaturation for 2 min at 95 ℃, and (ⅲ) 40 cycles of PCR amplification, with 30 s of denaturation (at 95 ℃) and 1 min of annealing and extension (at 60 ℃).
Enzyme-linked Immunosorbent Assay (ELISA)Tumor necrosis factor alpha (TNF-α), interferon-inducible protein-10 (IP-10), Interleukin-6 (IL-6), and interleukin-8 (IL-8) levels were measured using ELISA kits (R&D system, USA) according to the manufacturer's instructions. The intensity of the color was measured with a TECAN Microplate Reader at 450 nm. The levels of cytokines in samples were calculated based on a standard curve and were corrected for protein concentration.
StatisticsAll statistical analyses were performed using GraphPad Prism unless otherwise noted.
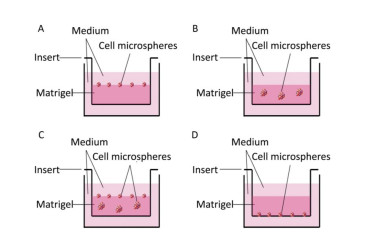
RESULTS Differences in Morphological Features between 2D-and 3D-cultured CellsFirst, we investigated the optimum location of cells and their number when seeding the Matrigel layer. Cells were seeded on (Figure 1A), within (Figure 1B), within and on (Figure 1C), and under (Figure 1D) the Matrigel layer. After being cultured for 7 days, we found that more cell microspheroids grew within and on Matrigel (Figure 1C). Comparatively, only a few microspheroids were found when the cells were seeded under the Matrigel layer (Figure 1D). We also found that 1 × 104 to 1 × 106 cells/per Transwell insert (12-well plate) were suitable for the formation of 3D microspheroids at day 7. The number and size of 3D cell microspheroids were ideal at a density of 1 × 105 cells/insert and the diameter of the microspheroids ranged from 100 to 200 μm (Figure 1B).
|
Download:
|
| Figure 1 Schematic of the 3D cultured cells grown on (A), within or in/on (B and C), and under (D) the Matrigel layer. | |
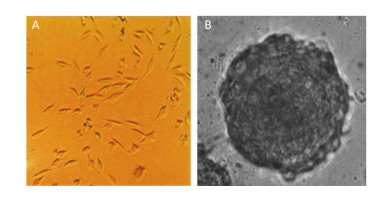
The morphological features of the 2D and 3D cultured HAE cells at day 7 were then investigated using a phase-contrast microscope. Figure 2 shows that 2D-grown cells were flat and spread as a monolayer on the tissue culture dish (Figure 2A), whereas 3D-grown cells cultured within the Matrigel microenvironment formed microspheres (Figure 2B).
|
Download:
|
| Figure 2 Different morphological features between 2D and 3D cultured cells. The morphology of 2D and 3D cultured HAE cells were captured under phase-contrast microscope (A & B). 3D cultures form microspheres (B), while 2D cells are flat and spread as a monolayer (A). | |
The cytokeratin family of proteins forms the intermediary filaments of keratin, which are present in the cytoskeleton of all epithelial cells[31]. They interact with desmosomes and hemidesmosomes, thereby assisting cell-cell adhesion and barrier functions[32-33]. Monoclonal anti-pan-cytokeratin is a broadly reactive reagent that recognizes epitopes present in most human epithelial tissues.
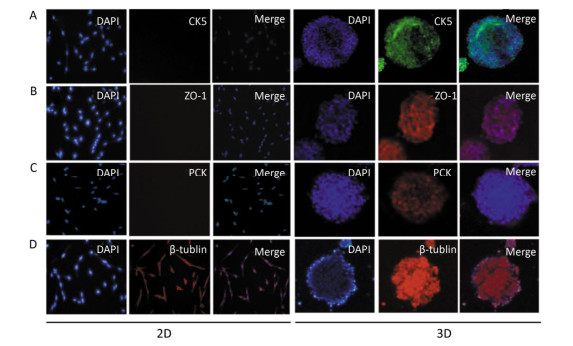
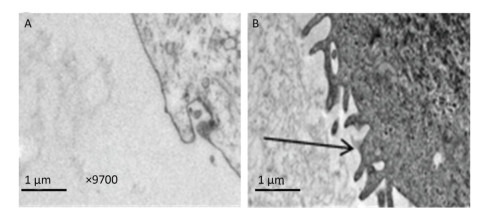
In this experiment, we initially identified the expression of pan-cytokeratin (PCK) in 3D-cultured HAE cells, and the results showed that PCK was expressed in 3D-cultured HAE cells; however, 2D-cultured HAE cells did not express this marker (Figure 3A). Subsequently, we examined the expression of CK5 and ZO-1; the results also showed that CK5 and ZO-1 were only expressed in 3D-cultured HAE cells, but not in 2D-cultured HAE cells (Figure 3B-C). β-tubulin was expressed in 3D-cultured and 2D-cultured HAE cells (Figure 3D). Finally, we identified the surface characteristics of 3D-cultured HAE cells using TEM, results showed that cilium-like protuberances appeared in the microspheroids of 3D-cultured HAE cells (Figure 4A), but not in 2D-cultured cells (Figure 4B).
|
Download:
|
| Figure 3 Signature protein expressions of organotypic tissue in 3D cultured HAE cells. The 2D and 3D cultured cells were immunostained with anti-cytokeratin 5 (A), anti-ZO-1 (B), anti-pan-cytokeratin (C) and anti-β-tublin (D), and the nuclei were counterstained with DAPI. The representative confocal images are shown (×10). | |
|
Download:
|
| Figure 4 More cilium-like protuberances of human respiration tract appeared in 3D cultured cells. The 2D (A) and 3D (B) cultured HAE cells were made the thin section for TEM detection (×9700). Many ciliary in 3D cultured HAE cells were observed (B). Arrows indicates the ciliary on cell. | |
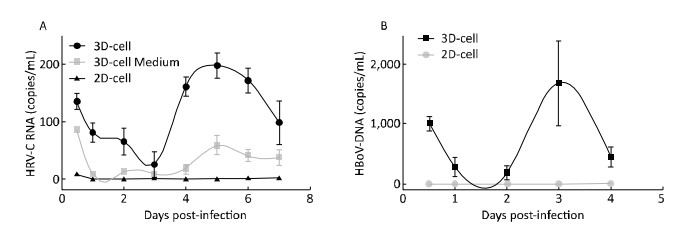
As we observed some organoid features in 3D-cultured HAE cells (primary cell line), we sought to investigate whether HRV-C could propagate in this 3D system. We incubated 3D and 2D culture with HRV-C viruses for 2 h, and subsequently washed out the remaining viruses that did not enter the cells. Subsequently, cells and media were harvested at 6 h and 1, 2, 3, 4, 5, 6, 7, 8, and 9 days post-inoculation. We next sought to evaluate whether these 3D-cultured HAE cells could support HRV-C and HBoV replication, after identifying the biological characteristics of these cells. Results showed that HRV-C RNA content in 3D cultures decreased 3 days post-inoculation, as compared to that detected 6 h post-inoculation; it then dramatically increased at the 5th day reaching its peak level. At this time, HRV-C RNA content began to decline. The level of HRV-C released into the supernatant followed a similar trend for HRV-C infection of 3D-cultured HAE cells. However, viral RNA was not detected using qRT-PCR in the 2D cell culture system (Figure 5A).
|
Download:
|
| Figure 5 The viruses propagation in 2D and 3D cultures measured by qRT-PCR. (A) HAE cultures were infected by HRV-C; (B) HAE cultures were infected by HBoV. Each data point represents the mean of three independent experiments at least. | |
We also examined the infectivity of the tested DNA virus (HBoV) in 2D-and 3D-cultured HAE cells. HBoV DNA content decreased in 3D cultures from 6 h to 2 days post-inoculation, and then increased on the 3rd day, compared to that at the 2nd day, reaching its peak. The content of HBoV DNA declined to undetectable levels by 5 days post-infection. We could not detect the propagation of HBoV in the 2D cell culture system by real-time RT-PCR (Figure 5B).
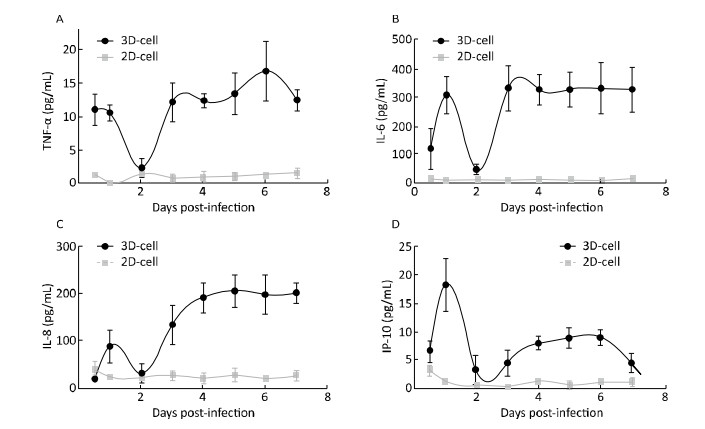
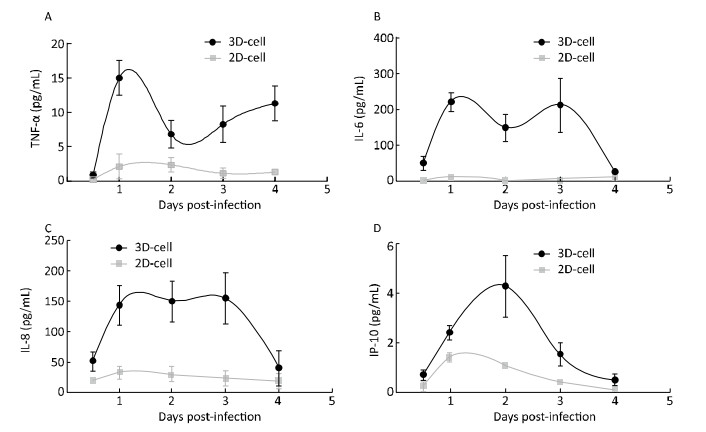
Cytokine were Inducd in HRV-C-and HBoV-infected-3D Cultured HAE CellsWhen cells are infected by viruses, they secrete cytokines such as TNF-α, IP-10, IL-6, and IL-8 to activate an immune response[12, 25]. To further confirm whether 3D cultures were infected by the virus, cytokines in the media of 3D HAE cultures were examined by ELISA at the indicated time points after infection with HRV-C and HBoV.
Cytokine levels in the media significantly increased in 3D-cultured HAE cells after infection with HRV-C (Figure 6) and HBoV (Figure 7), as compared to levels in 2D-cultured cells. The cytokine increases appeared as two peaks in 3D cultures after infection with HRV-C. The first peak appeared in the early stage after HRV-C infection (at 2 days post-infection), whereas the second peak appeared at the 5th (for IL-8 and IP-10) or 6th (for TNF-α) day after infection, which might represent the stage of HRV-C amplification. However, the levels of cytokines in the media 2D cultures only increased slightly at 6 h after virus inoculation, and then decreased to background levels.
|
Download:
|
| Figure 6 Cytokine secretion measured in the 2D and 3D cultured HAE cells after infection with HRV-C. TNF-α (A), IL-6 (B), IL-8 (C), and IP-10 (D) levels in the media of HAE cultures at the indicated day after infection were measured by ELISA assay. Each data point represents the mean of three independent experiments at least. | |
|
Download:
|
| Figure 7 Cytokine secretion measured in the 2D and 3D cultured HAE cells after infection with HBoV. TNF-α (A), IL-6 (B), IL-8 (C), and IP-10 (D) levels in the media of HAE cultures at the indicated day after infection were measured by ELISA assay. Each data point represents the mean of three independent experiments at least. | |
Emerging data indicate a significant role for HRV-C and HBoV in respiratory tract disease, particularly in children[34-36]. HRV-C and HBoV could not propagate using the traditional cell culture model; thus, studies on virus isolation, virus-host cell interaction, and vaccine development for these types of viruses are limited. In addition, the cellular receptors for these viruses remain unknown[37]. HRV-C viruses are clearly rhinoviruses, but unlike HRV-A and HRV-B, they do not readily propagate in traditional cell culture systems, including WI-38, WisL, BEAS-2B, A549, and HeLa lines[38]. Thus, we can only construct infectious clone plasmids to generate progeny virus in vitro.
Although HRV-C can propagate in HAE cells cultured with the ALI technique[39-40], the amount of virus is not sufficient for extensive biological studies; meanwhile, cells must be cultured for more than 15 days before virus inoculation. In this study, we sought to develop a time-saving organoid culture system comprising respiratory epithelial cells to mimic the host microenvironment, and to propagate HRV-C and HBoV, which cannot propagate in traditional 2D cell culture systems.
In this study, we constructed a human organotypic tissue model using HAE cells in vitro for the propagation of HRV-C and HBoV. Cells were cultured in Matrigel for 7 more days, and then the 3D microspheres were observed by light microscopy. TEM and immunostaining assays indicated that 3D cultures had the properties of organotypic tissue (respiratory epithelium). They had more brush borders with cilia and expressed the signature protein of the respiratory tract, properties that are not observed with 2D cells. Next, these 3D cultures were inoculated with HRV-C and HBoV, both of which cannot propagate in traditional 2D cells. Fortunately, the viral titer increased and cytokines were secreted in 3D cultures after infection, as detected by qRT-PCR and ELISA, respectively. This suggests that HRV-C and HBoV can propagate in our 3D cell culture system using Matrigel as a scaffold.
To determine the optimal conditions for virus propagation, different cell densities and Matrigel concentrations were tested. A primary cell density of 105 cells/insert (12-well Transwell plate) and a ratio of cell suspension/Matrigel of 1:(6-8) (v/v) were optimal for viral replication (data not shown). We cultured different cell lines (A549, BEAS-2B, MCF-7, HNEPC, HAE, and HBEC-3KT) in Matrigel, and then attempted to infect them with HRV-C and HBoV. Finally, we identified two types of human normal airway epithelial cells, HAE (primary cell line) and BEAS-2B cell (immortal cell line), that were suitable for 3D growth and for inoculation and isolation of the viruses. It is likely that the other cell lines (A549, MCF-7, HNEPC, and HBEC-3KT) no longer express the receptor or specific structures (such as cilia) required for respiratory virus invasion. The results of this study showed that both HRV-C and HBoV can propagate in 3D-cultured HAE cells (primary), but not well in BEAS-2B (immortal) cultures. This might be due to decreased differentiation ability and absence of specific features of organotypic tissue in the immortal cell lines, compared to those features in primary cell lines.
The results of this study showed that HRV-C and HBoV virus contents deceased at 2 or 3 days post-inoculation, and subsequently increased for several days. The period with diminished viral detection is called the 'eclipse period'. This is the time following infection but before productive virus growth, and it is often associated with a decrease in the amount of viral DNA or RNA.
It has been observed that many infants generate HRV-specific cytokine responses during HRV infection, such as increased levels of TNF-α, IP-10, IL-6, and IL-8[41-42]. The immune response to virus includes the vigorous production of cytokines and chemokines including IL-8 and IL-6[43-45]. Cytokines/chemokines play a critical role in regulating local inflammatory processes in the lung and subsequent tissue damage. Concentrations of these cytokines/chemokines secreted by respiratory epithelial cells might correlate with disease severity[46-47]. The results of this study showed that two peaks of cytokine secretion occur in 3D cultures after infection. We think that the inoculated virus initially enters the cell, stimulating the secretion of cytokines/chemokines, which results in the first peak. The cytokines activate the cellular self-protection mechanism, which results in the subsequent decrease. Upon amplification of new virus, cells secrete more cytokines/chemokines, resulting in the appearance of the second peak. Cytokine secretion by 3D-cultured HAE cells, after infection with HRV-C or HBoV, indicates that 3D cells exhibit an activated innate immune response against HRV-C and HBoV; however, further studies are required to elucidate the exact mechanisms of antiviral innate immunity and virus replication.
In summary, our 3D culture system using Matrigel as a scaffold in vitro is a simple, time-saving, and low-cost system that does not need specialized equipment, unlike 3D culture systems with rotating instruments. The morphology and function of the 3D-cultured human cells are similar to those in human tissues. Although the efficiency of propagation of HRV-C or HBoV virus is very low and culture conditions need to be optimized, the results of this study suggest that this in vitro 3D culture model is a good platform for the propagation of viruses that are difficult to culture using traditional monolayer cell and animal models.
CONFLICT OF INTERESTNo conflict of interest to declare.
AUTHOR CONTRIBUTIONSCHEN YX designed and performed the experiments, data analysis, and wrote the manuscript. XIE GC and PAN D helped with experiments, data interpretation, and manuscript preparation. DU YR and PANG LL provided materials and helped with data analysis. SONG JD helped with the TEM experiment and data analysis. DUAN ZJ and HU BR were involved in the experimental design, data analysis, and manuscript revision. All authors read and approved the final manuscript.
| 1. | Bhattacharya S, Srinivasan K, Abdisalaam S, et al. RAD51 interconnects between DNA replication, DNA repair and immunity. Nucleic Acids Res, 2017, 45: 4590–605. doi:10.1093/nar/gkx126 |
| 2. | Nakada Y, Canseco DC, Thet S, et al. Hypoxia induces heart regeneration in adult mice. Nature, 2017, 541: 222–7. |
| 3. | Yu L, Shang ZF, Abdisalaam S, et al. Tumor suppressor protein DAB2IP participates in chromosomal stability maintenance through activating spindle assembly checkpoint and stabilizing kinetochore-microtubule attachments. Nucleic Acids Res, 2016, 44: 8842–54. doi:10.1093/nar/gkw746 |
| 4. | Bhattacharya S, Asaithamby A. Ionizing radiation and heart risks. Semin Cell Dev Biol, 2016, 58: 14–25. doi:10.1016/j.semcdb.2016.01.045 |
| 5. | McFadden CH, Hallacy TM, Flint DB, et al. Time-Lapse Monitoring of DNA Damage Colocalized With Particle Tracks in Single Living Cells. Int J Radiat Oncol, 2016, 96: 221–7. doi:10.1016/j.ijrobp.2016.04.007 |
| 6. | McFadden C, Flint D, Sadetaporn D, et al. A Portable Confocal Microscope to Image Live Cell Damage Response Induced by Therapeutic Radiation. Med Phys, 2016, 43: 3824. |
| 7. | Sadetaporn D, Flint D, McFadden C, et al. Confocal Microscopy Imaging of Cell Cycle Distribution in Cells Treated with Pegylated Gold Nanoshells. Med Phys, 2016, 43: 3617. |
| 8. | Hao W, Bernard K, Patel N, et al. Infection and propagation of human rhinovirus C in human airway epithelial cells. Journal of virology, 2012, 86: 13524–32. doi:10.1128/JVI.02094-12 |
| 9. | Kimura W, Xiao F, Canseco DC, et al. Hypoxia fate mapping identifies cycling cardiomyocytes in the adult heart (vol 523, pg 226, 2015). Nature, 2016: 532. |
| 10. | Su FT, Bhattacharya S, Abdisalaam S, et al. Replication stress induced site-specific phosphorylation targets WRN to the ubiquitin-proteasome pathway. Oncotarget, 2016, 7: 46–65. |
| 11. | Zhu J, Su F, Mukherjee S, et al. FANCD2 influences replication fork processes and genome stability in response to clustered DSBs (vol 14, pg 1809, 2015). Cell Cycle, 2016, 15: 2377. doi:10.1080/15384101.2016.1222785 |
| 12. | Bunch H, Lawney BP, Lin YF, et al. Transcriptional elongation requires DNA break-induced signalling. Nat Commun, 2015, 6: 10191. doi:10.1038/ncomms10191 |
| 13. | Kimura W, Xiao F, Canseco DC, et al. Hypoxia fate mapping identifies cycling cardiomyocytes in the adult heart. Nature, 2015, 523: 226–43. doi:10.1038/nature14582 |
| 14. | Xue G, Ren Z, Grabham PW, et al. Reprogramming mediated radio-resistance of 3D-grown cancer cells. J Radiat Res, 2015, 56: 656–62. doi:10.1093/jrr/rrv018 |
| 15. | Roskelley CD, Desprez PY, Bissell MJ. Extracellular matrix-dependent tissue-specific gene expression in mammary epithelial cells requires both physical and biochemical signal transduction. Proc Natl Acad Sci U S A, 1994, 91: 12378–82. doi:10.1073/pnas.91.26.12378 |
| 16. | Le Beyec J, Xu R, Lee SY, et al. Cell shape regulates global histone acetylation in human mammary epithelial cells. Exp Cell Res, 2007, 313: 3066–75. doi:10.1016/j.yexcr.2007.04.022 |
| 17. | Lelievre SA. Contributions of extracellular matrix signaling and tissue architecture to nuclear mechanisms and spatial organization of gene expression control. Biochim Biophys Acta, 2009, 1790: 925–35. doi:10.1016/j.bbagen.2009.03.013 |
| 18. | Hackethal J, Muhleder S, Hofer A, et al. An Effective Method of Atelocollagen Type 1/3 Isolation from Human Placenta and Its In Vitro Characterization in Two-Dimesional and Three-Dimensional Cell Culture Applications. Tissue Engineering Part C, Methods, 2017, 23: 274–85. doi:10.1089/ten.tec.2017.0016 |
| 19. | Sieh S, Taubenberger AV, Rizzi SC, et al. Phenotypic Characterization of Prostate Cancer LNCaP Cells Cultured within a Bioengineered Microenvironment. Plos One, 2012, 7: e40217. doi:10.1371/journal.pone.0040217 |
| 20. | Sempere LF, Gunn JR, Korc M. A novel 3-dimensional culture system uncovers growth stimulatory actions by TGF beta in pancreatic cancer cells. Cancer Biol Ther, 2011, 12: 198–207. doi:10.4161/cbt.12.3.15979 |
| 21. | Rangarajan A, Hong SJ, Gifford A, et al. Species-and Cell Type-Specific Requirements for Cellular Transformation (vol 6, pg 171, 2004). Cancer Cell, 2013, 24: 394–8. doi:10.1016/j.ccr.2013.08.028 |
| 22. | Rangarajan A, Hong SJ, Gifford A, et al. Species-and cell type-specific requirements for cellular transformation. Cancer Cell, 2004, 6: 171–83. doi:10.1016/j.ccr.2004.07.009 |
| 23. | Yamada KM, Cukierman E. Modeling tissue morphogenesis and cancer in 3D. Cell, 2007, 130: 601–10. doi:10.1016/j.cell.2007.08.006 |
| 24. | Stetten AZ, Moraca G, Corcoran TE, et al. Enabling Marangoni flow at air-liquid interfaces through deposition of aerosolized lipid dispersions. J Colloid Interf Sci, 2016, 484: 270–8. doi:10.1016/j.jcis.2016.08.076 |
| 25. | Aufderheide M, Forster C, Beschay M, et al. A new computer-controlled air-liquid interface cultivation system for the generation of differentiated cell cultures of the airway epithelium. Exp Toxicol Pathol, 2016, 68: 77–87. doi:10.1016/j.etp.2015.10.001 |
| 26. | Okubo T, Hosaka M, Nakae D. In vitro effects induced by diesel exhaust at an air-liquid interface in a human lung alveolar carcinoma cell line A549. Exp Toxicol Pathol, 2015, 67: 383–8. doi:10.1016/j.etp.2015.03.004 |
| 27. | Jing XF, Park JH, Peters TM, et al. Toxicity of copper oxide nanoparticles in lung epithelial cells exposed at the air-liquid interface compared with in vivo assessment. Toxicol in Vitro, 2015, 29: 502–11. doi:10.1016/j.tiv.2014.12.023 |
| 28. | Zhu JY, Su FT, Mukherjee S, et al. FANCD2 influences replication fork processes and genome stability in response to clustered DSBs. Cell Cycle, 2015, 14: 1809–22. doi:10.1080/15384101.2015.1036210 |
| 29. | Bochkov YA, Palmenberg AC, Lee WM, et al. Molecular modeling, organ culture and reverse genetics for a newly identified human rhinovirus C. Nat Med, 2011, 17: 627. doi:10.1038/nm.2358 |
| 30. | Canseco DC, Kimura W, Garg S, et al. Human Ventricular Unloading Induces Cardiomyocyte Proliferation. J Am Coll Cardiol, 2015, 65: 892–900. doi:10.1016/j.jacc.2014.12.027 |
| 31. | Vasca V, Vasca E, Freiman P, et al. Keratin 5 expression in squamocellular carcinoma of the head and neck. Oncol Lett, 2014, 8: 2501–4. doi:10.3892/ol.2014.2591 |
| 32. | Kouklis PD, Hutton E, Fuchs E. Making a Connection-Direct Binding between Keratin Intermediate Filaments and Desmosomal Proteins. J Cell Biol, 1994, 127: 1049–60. doi:10.1083/jcb.127.4.1049 |
| 33. | Osmani N, Labouesse M. Remodeling of keratin-coupled cell adhesion complexes. Current opinion in cell biology, 2015, 32: 30–8. doi:10.1016/j.ceb.2014.10.004 |
| 34. | Han TH, Chung JY, Hwang ES, et al. Detection of human rhinovirus C in children with acute lower respiratory tract infections in South Korea. Arch Virol, 2009, 154: 987–91. doi:10.1007/s00705-009-0383-z |
| 35. | Lau SKP, Yip CCY, Lin AWC, et al. Clinical and Molecular Epidemiology of Human Rhinovirus C in Children and Adults in Hong Kong Reveals a Possible Distinct Human Rhinovirus C Subgroup. J Infect Dis, 2009, 200: 1096–103. doi:10.1086/599179 |
| 36. | Linsuwanon P, Payungporn S, Samransamruajkit R, et al. High prevalence of human rhinovirus C infection in Thai children with acute lower respiratory tract disease. J Infection, 2009, 59: 115–21. doi:10.1016/j.jinf.2009.05.009 |
| 37. | Bochkov YA, Watters K, Ashraf S, et al. Cadherin-related family member 3, a childhood asthma susceptibility gene product, mediates rhinovirus C binding and replication. P Natl Acad Sci USA, 2015, 112: 5485–90. doi:10.1073/pnas.1421178112 |
| 38. | Bochkov YA, Palmenberg AC, Lee WM, et al. Molecular modeling, organ culture and reverse genetics for a newly identified human rhinovirus C. Nat Med, 2011, 17: 627–32. doi:10.1038/nm.2358 |
| 39. | Ashraf S, Brockman-Schneider R, Gern JE. Propagation of rhinovirus-C strains in human airway epithelial cells differentiated at air-liquid interface. Methods Mol Biol, 2015, 1221: 63–70. doi:10.1007/978-1-4939-1571-2 |
| 40. | Mello C, Aguayo E, Rodriguez M, et al. Multiple classes of antiviral agents exhibit in vitro activity against human rhinovirus type C. Antimicrob Agents Chemother, 2014, 58: 1546–55. doi:10.1128/AAC.01746-13 |
| 41. | Lee FE, Walsh EE, Falsey AR, et al. Human infant respiratory syncytial virus (RSV)-specific type 1 and 2 cytokine responses ex vivo during primary RSV infection. J Infect Dis, 2007, 195: 1779–88. doi:10.1086/522480 |
| 42. | Fraenkel DJ, Bardin PG, Sanderson G, et al. Lower airways inflammation during rhinovirus colds in normal and in asthmatic subjects. Am J Respir Crit Care Med, 1995, 151: 879–86. |
| 43. | Garofalo RP, Hintz KH, Hill V, et al. A comparison of epidemiologic and immunologic features of bronchiolitis caused by influenza virus and respiratory syncytial virus. J Med Virol, 2005, 75: 282–9. doi:10.1002/(ISSN)1096-9071 |
| 44. | Jamaluddin M, Choudhary S, Wang S, et al. Respiratory syncytial virus-inducible BCL-3 expression antagonizes the STAT/IRF and NF-kappaB signaling pathways by inducing histone deacetylase 1 recruitment to the interleukin-8 promoter. J Virol, 2005, 79: 15302–13. doi:10.1128/JVI.79.24.15302-15313.2005 |
| 45. | Legg JP, Hussain IR, Warner JA, et al. Type 1 and type 2 cytokine imbalance in acute respiratory syncytial virus bronchiolitis. Am J Respir Crit Care Med, 2003, 168: 633–9. doi:10.1164/rccm.200210-1148OC |
| 46. | Garofalo RP, Patti J, Hintz KA, et al. Macrophage inflammatory protein-1alpha (not T helper type 2 cytokines) is associated with severe forms of respiratory syncytial virus bronchiolitis. J Infect Dis, 2001, 184: 393–9. doi:10.1086/jid.2001.184.issue-4 |
| 47. | Welliver RC, Garofalo RP, Ogra PL. Beta-chemokines, but neither T helper type 1 nor T helper type 2 cytokines, correlate with severity of illness during respiratory syncytial virus infection. Pediatr Infect Dis J, 2002, 21: 457–61. doi:10.1097/00006454-200205000-00033 |
 2018, Vol. 31
2018, Vol. 31


