2. Department of Physiology and Neurobiology, Xinxiang Medical University, Xinxiang 453003, Henan, China
Long non-coding RNAs (lncRNAs) are a large and diverse class of transcribed RNA molecules measuring more than 200 bp in length. LncRNAs are non-protein-coding transcripts and comprise a family of non-protein-coding RNAs. Moreover, lncRNAs have been found to play important roles in several biological processes such as cell development, proliferation, invasion, and migration[1]. Aberrant lncRNA expression in humans can cause a variety of diseases[2]. Cleft palate is the most common congenital maxillofacial deformity among birth defects. A cleft palate may occur at any stage of palatal development. For example, defective palatal shelf growth can cause a cleft palate. A palate with a failed or delayed elevation also causes a cleft palate, and a palate with blocked fusion also results in a cleft palate. Several studies have indicated that the development of a cleft palate involves various types of genes and environmental factors[1, 3].
2, 3, 7, 8-tetrachlorodibenzo-p-dioxin (TCDD) is a wide-range and highly toxic environmental contaminant. It is also a strong teratogen for the development of a cleft palate. The incidence of cleft palate development was found to be 100% in each pregnant mouse treated once with 64 µg/kg body weight of TCDD[4]. It is believed that TCDD inhibits palatal fusion by suppressing the disappearance of the medial edge epithelial (MEE) cells on the palatal shelves[5]. Although the mechanism involved in TCDD-induced cleft palate has been described[6], there are limited data reporting the involvement of lncRNAs in TCDD-induced cleft palate.
LncRNAs provide new insight into the opportunities for studying the pathogenesis of cleft palate. The long non-coding RNA H19 (lncRNA H19) encodes a 2.3-kb non-coding mRNA, which is predominantly found in the cytoplasm with a minor fraction in the nucleus[7-8]. LncRNA H19 is expressed in various tissues and has several functions through activation both in the nucleus and in the cytoplasm. It has been closely associated with all types of diseases[9]. It was initially found and associated with cleft palate in transforming growth factor-β3 (TGF-β3)-knockout mice in 2013[10]. LncRNA H19 is located adjacent to the paternally expressed insulin-like growth factor 2 (IGF2) gene, and its target gene is IGF2. The IGF2 gene plays an important role in growth and development before birth. It has been hypothesized that the interaction between lncRNA H19 and IGF2 genes plays a role in the development of a cleft palate. However, the involvement of lncRNA H19 and IGF2 in TCDD-induced mouse cleft palate has been rarely reported. In this study, we explored the possible mechanism of involvement of lncRNA H19 and IGF2 genes in mouse cleft palate induced by TCDD.
Animals A total of 40 female and 10 male C57BL/6 mice, aged 8-12 weeks and weighing 25-30 g, were purchased from Beijing Weitong Lihua Experimental Animal Technology Co., Ltd. [Beijing, China; certificate No. SCXK (Jing) 2012-0001]. All animal experiments were carried out in accordance with the Animal Care Committee of Xinxiang Medical University, China, and the experiments were performed according to the ethical guidelines of the institution. Female mice were placed along with male mice (4 F: 1 M) for approximately 18 h and checked for the presence of vaginal plugs, which was considered as evidence of mating. The mice were housed at room temperature of 22 ℃ and relative humidity of 60% with a normal 12 h/12 h light/dark cycle. The animals were maintained on standard chow and water that were available ad libitum.
Construction of Cleft Palate Model and Sample Collection TCDD (Sigma, Saint Louis, MO, USA) dissolved in corn oil[11] was administered orally to each pregnant mouse (64 µg/kg body weight) in the treated group on embryo day 10 (E10)[12]. The same volume of corn oil (Jinglongyu, Qinghuangdao, China) was administered to the control mice. Three pregnant mice were sacrificed at each time point on E13, E14, E15, and E17, respectively. First, the fetuses were removed from the uterus, and then, five palates of the fetuses at each time point were harvested from both groups under a stereomicroscope (Olympus, Tokyo, Japan) and fixed with 10% formalin. Some palates were sectioned (5-mm thick), stained with hematoxylin and eosin (H & E), and evaluated under a microscope. Other palates were immediately preserved in RNAlater solution (Ambion, Austin, TX, USA) at 4 ℃ overnight and then stored at -80 ℃ for further RNA isolation.
Quantitative Real-time Polymerase Chain Reaction (qRT-PCR) The harvested palates from both the TCDD-treated and the control groups were lysed in Trizol lysis solution (Invitrogen, Carlsbad, CA, USA). Total RNA of five palates was isolated according to the manufacturer's instructions. First-strand cDNA was synthesized using PrimeScript™ Ⅱ 1st strand cDNA Synthesis Kit (TakaRa Biotechnology, Kyoto, Japan) and amplified by qRT-PCR using the SYBR Premix Ex Taq Kit (TaKaRa) by ABI 7900 PRISM system (Applied Biosystems, Carlsbad, CA, USA). 18 s rRNA was used as an internal control. The conditions for qRT-PCR for lncRNA H19 and IGF2 genes were as follows: polymerase activation for 15 min at 95 ℃, 40 cycles at 95 ℃ for 15 s, 56 ℃ for 20 s, and 72 ℃ for 30 s. All primers were synthesized by Dingguo Co. The primer sets used in qRT-PCR were as follows: 18 s rRNA-forward: 5'-CGGACATCTAAGGGCATCA-3', 18 s rRNA-reverse: 5'-AAGACGGACCAGAGCGAAA-3'; LncRNA H19-for ward: 5'-TACCCCGGGATGACTTCA TC-3', LncRNA H19-reverse: 5'-TATCTCCGGGACTCC AAACC-3', IGF2-forward: 5'-GTGTGTGTCAGCCAAG CATG-3', IGF2-reverse: 5'-CAAATGTGGGGACAC AGAGG-3'.
Statistical Analysis The data of tests were made automatically using statistical product and service solutions (SPSS) software, version 13.0 (SPSS, Chicago, IL, USA). Double-sided Student's t-test was used for comparison between the two groups. Multiple group comparisons were performed using one-way analysis of variance, and Tukey's post hoc test was used to determine the differences between the groups. All data were presented as mean ± standard deviation (SD). Differences were considered to be statistically significant when P < 0.05, or P < 0.01.
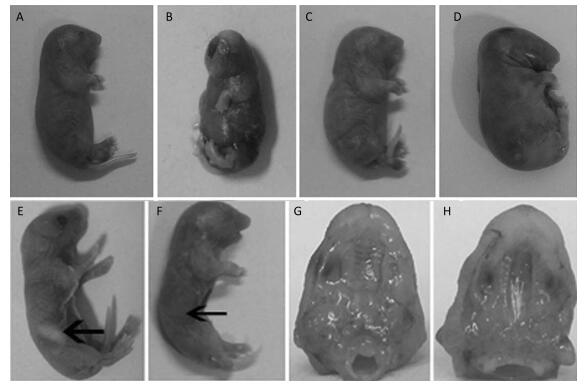
The Effects of TCDD on Fetal Mice After the pregnant C57BL/6 mice delivered, we found that some fetuses had short upper limb (Figure 1A), cerebral fissure (Figure 1B), webbed neck (Figure 1C), and short neck (Figure 1D), and the rate of cleft palate occurrence was 100% in the TCDD-treated group, a finding consistent with that reported by Pu Y[13]. In the control group mice, the fetuses had milk in their stomachs (Figure 1E), whereas those in the TCDD-treated group had no milk in their stomachs (Figure 1F), and they soon died of starvation. The fetuses in the control group had normal and palatal wrinkles obviously (Figure 1G), while the fetuses occurred cleft palates and no palatal wrinkles (Figure 1H). These results were in agreement with another study using TCDD-induced mice[14] and were similar to the birth defects observed in children due to TCDD contamination after the war in Vietnam[15].
|
Download:
|
| Figure 1 The effects on morphology in the control and TCDD-treated groups. Some fetuses in the TCDD-treated group had short upper limb (A), cerebral fissure (B), webbed neck (C), and short neck (D), and the rate of cleft palate occurrence was 100%. Fetuses in the control group showed stomach with milk (E), but fetuses in the TCDD-treated group had no milk in their stomachs (F). The palatal wrinkles were obvious in the TCDD-treated group (G), whereas no palatal wrinkles were observed in the control group (H). The arrowhead indicates the site of a fetus' stomach | |
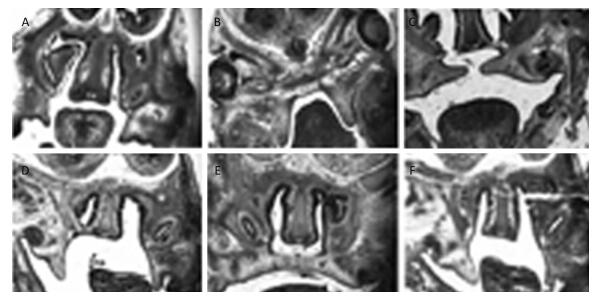
Morphological and Histological Observations of Palatal Development The morphological and histological alterations were examined and are shown in Figure 2. In detail, the budding of the palatal shelves began to form on E13 in the fetuses of the control group (Figure 2A). In contrast, the formation of the palatal shelves in the fetuses of the TCDD-treated group was arrested on E13 (Figure 2B). In the control group, the palatal shelves of the fetuses elevated and reached a horizontal position above the tongue on E14 (Figure 2C), whereas the palate began to elevate on E14 in the fetuses of the TCDD-treated group (Figure 2D). On E15, the palatal shelves of the fetuses of untreated mice started to fuse (Figure 2E); however, the palatal shelves of the fetuses elevated unsuccessfully in the TCDD-treated group and led to the formation of a cleft palate (Figure 2F). These results were also reported in another study with TCDD-induced mice[14].
|
Download:
|
| Figure 2 Histological analysis of the palatal shelves in the control and TCDD-treated groups from E13 to E15. The palate shelves of the fetuses elevated on E13 in the control group (A). The palatal shelves of the fetuses failed to elevate on E13 in the TCDD-treated group (B). The palatal shelves of the fetuses elevated and reached a horizontal position above the dorsum of the tongue on E14 in the control group (C). The palatal shelves of the fetuses delayed to elevate on E14 in the TCDD-treated group (D). The palatal shelves of the fetuses contacted each other in the end on E15 in the control group (E). The palatal shelves of the fetuses failed to contact each other and resulted in a cleft palate in the TCDD-treated group (F). H & E staining × 40 in both groups | |
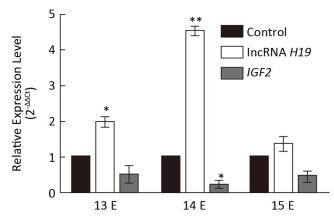
The Expression Levels of lncRNA H19 and IGF2 at Different Developmental Stages of Cleft Palate Very important palatal developments occur during E13 to E15, such as both sides of the palatal shelves are above the tongue on E13, there is rapid growth of the palate on E14, and they beg into touch each other on E15. However, the involvement of lncRNA H19 and the target gene IGF2 in this important time period has not yet been reported. Therefore, to study the expression patterns of lncRNA H19 and IGF2 parallel to cleft palate formation, the palatal tissues were analyzed on E13, E14, and E15. It was observed that the expression levels of lncRNA H19 and IGF2 varied with the development of the palate (Figure 3). In detail, the expression levels of lncRNA H19 significantly increased by 1.99 ± 0.23-fold, 4.52 ± 0.21-fold, and 1.36 ± 0.33-fold compared with those of the corresponding control palatal tissues on E13, E14, and E15, respectively. This result was similar to that of a previous study, which reported a high expression level of lncRNA H19 in the critical period of embryo development[16]. This finding suggested that lncRNA H19 might have an important function in the development of the palate induced by TCDD. LncRNAs predominantly participate in diverse physiological processes in organisms, exert important biological functions, and can regulate the expression of target genes at transcriptional, post-transcriptional, and epigenetic levels. The target gene of lncRNA H19 is IGF2. Therefore, we examined the expression levels of IGF2 during the important stages on E13, E14, and E15. The expression levels of IGF2 gene significantly reduced to 0.52 ± 0.42, 0.24 ± 0.16, and 0.49 ± 0.22 compared with those of the corresponding control palatal tissues on E13, E14, and E15, respectively. The expression level of IGF2 gene was always lower than that of the corresponding control, which was opposite to that of lncRNA H19. This result was consistent with the expression levels of lncRNA H19 and IGF2 in two growth disorders with opposite phenotypes of fetoplacental development[17-18], which suggested that lncRNA H19 might exert its function through the interaction with IGF2 in the cleft palate induced by TCDD. There may be methylation of the lncRNA H19 and IGF2 differentially methylated domain[19]. Obviously, the precise mechanisms involved in the lncRNA H19-and IGF2-regulated TCDD-induced cleft palate need further exploration. Although the mechanism and function of lncRNA H19/IGF2 in cleft palate biology remain unclear, the present study findings demonstrated that lncRNA H19 could be a potential indicator for TCDD-induced cleft palate.
|
Download:
|
| Figure 3 Relative expression of lncRNA H19 and IGF2 genes in different developmental stages of palatogenesis. Expression levels of lncRNA H19 and IGF2 genes were compared with those of the control group on E13, E14, and E15, respectively, which were determined by qRT-PCR. The data are mean ± standard deviation of six replicate experiments. *, P < 0.05 or **, P < 0.01 compared with the corresponding control values | |
In summary, this study revealed that lncRNA H19 and IGF2 mediated the development of cleft palate induced by TCDD, which provides a new insight into the role of lncRNA in TCDD-induced cleft palate.
AcknowledgmentsConceived and designed the manuscript: GAO Li Yun; Searched and read papers: ZHANG Feng Quan, Zhao Wei Hui, YAO Yong Cheng, HAN Guang Liang; Wrote the manuscript: Gao Li Yun; Revised the manuscript critically: WANG Xiao, LI Qiang, GAO Shan Shan, WU Wei Dong; Made the final approval of the manuscript to be published: GAO Li Yun.
Declaration of Conflict of InterestThe authors declared no potential conflicts of interest with respect to the research, authorship, and publication of this article.
| 1. | Martinelli M, Girardi A, Cura F, et al. Non-syndromic cleft lip with or without cleft palate in Asian populations:Association analysis on three gene polymorphisms of the folate pathway. Arch Oral Biol, 2015, 61: 79. |
| 2. | Wu T, Du Y. LncRNAs:From Basic Research to Medical Application. Int J Biol Sci, 2017, 13: 295. doi:10.7150/ijbs.16968 |
| 3. | Sabbagh HJ, Alamoudi NM, Abdulhameed FD, et al. Environmental Risk Factors in the Etiology of Nonsyndromic Orofacial Clefts in the Western Region of Saudi Arabia. Cleft Palate Craniofac J, 2016, 53: 435–43. doi:10.1597/14-136 |
| 4. | Yuan X, Lin Q, Pu Y, et al. Histone acetylation is involved in TCDD-induced cleft palate formation in fetal mice. Mol Med Rep, 2016, 14: 1139–45. doi:10.3892/mmr.2016.5348 |
| 5. | Takagi TN, Matsui KA, Yamashita K, et al. Pathogenesis of cleft palate in mouse embryos exposed to 2, 3, 7, 8-tetrachlorodibenzo-p-dioxin (TCDD). Teratog Carcinog Mutagen, 2000, 20: 73–86. doi:10.1002/(ISSN)1520-6866 |
| 6. | Gao Z, Bu Y, Liu X, et al. TCDD promoted EMT of hFPECs via AhR, which involved the activation of EGFR/ERK signaling. Toxicol Appl Pharmacol, 2016, 298: 48–55. doi:10.1016/j.taap.2016.03.005 |
| 7. | Gabory A, Jammes H, Dandolo L. The H19 locus:role of an imprinted non-coding RNA in growth and development. Bioessays, 2010, 32: 473–80. doi:10.1002/bies.200900170 |
| 8. | Monnier P, Martinet C, Pontis J, et al. H19 lncRNA controls gene expression of the Imprinted Gene Network by recruiting MBD1. Proc Natl Acad Sci USA, 2013, 110: 20693–8. doi:10.1073/pnas.1310201110 |
| 9. | He P, Zhang Z, Huang G, et al. miR-141 modulates osteoblastic cell proliferation by regulating the target gene of lncRNA H19 and lncRNA H19-derived miR-675. Am J Transl Res, 2016, 8: 1780–8. |
| 10. | Ozturk F, Li Y, Zhu X, et al. Systematic analysis of palatal transcriptome to identify cleft palate genes within TGFbeta3-knockout mice alleles:RNA-Seq analysis of TGFbeta3 Mice. BMC Genomics, 2013, 14: 113. doi:10.1186/1471-2164-14-113 |
| 11. | Prokopec SD, Houlahan KE, Sun RX, et al. Compendium of TCDD-mediated transcriptomic response datasets in mammalian model systems. BMC genomics, 2017, 18: 78. doi:10.1186/s12864-016-3446-z |
| 12. | Hu X, Gao JH, Liao YJ, et al. 2, 3, 7, 8-Tetrachlorodibenzo-p-dioxin Delays Palatal Shelf Elevation and Suppresses Wnt5a and Lymphoid Enhancing-Binding Factor 1 Signaling in Developing Palate. Cleft Palate Craniofac J, 2015, 52: 54–61. doi:10.1597/13-018 |
| 13. | Pu YL, Liu LL, Gan LQ, et al. Mechanism of cleft palate in mice induced by 2, 3, 7, 8-tetrachlorodibenzo-p-dioxin. Chinese Journal of Plastic Surgery, 2011, 27: 448–53. |
| 14. | Yuan X, He X, Zhang X, et al. Comparative Study of Folic Acid and alpha-Naphthoflavone on Reducing TCDD-Induced Cleft Palate in Fetal Mice. Cleft Palate Craniofac J, 2017, 54: 216–22. doi:10.1597/15-211 |
| 15. | Sycheva LP, Umnova NV, Kovalenko MA, et al. Dioxins and cytogenetic status of villagers after 40 years of agent Orange application in Vietnam. Chemosphere, 2016, 144: 1415–20. doi:10.1016/j.chemosphere.2015.10.009 |
| 16. | Liang S, Zhao MH, Ock SA, et al. Fluoride impairs oocyte maturation and subsequent embryonic development in mice. Environ Toxicol, 2016, 31: 1486. doi:10.1002/tox.v31.11 |
| 17. | Gicquel C, Rossignol S, Cabrol S, et al. Epimutation of the telomeric imprinting center region on chromosome 11p15 in Silver-Russell syndrome. Nat Genet, 2005, 37: 1003–7. doi:10.1038/ng1629 |
| 18. | Kappil MA, Li Q, Li A, et al. In utero exposures to environmental organic pollutants disrupt epigenetic marks linked to fetoplacental development. Environ Epigenet, 2016, 2. |
| 19. | Kanduri C. Long noncoding RNAs:Lessons from genomic imprinting. Biochim Biophys Acta, 2016, 1859: 102–11. doi:10.1016/j.bbagrm.2015.05.006 |
 2017, Vol. 30
2017, Vol. 30


