2. National Institute for Communicable Disease Control and Prevention, State Key Laboratory for Infectious Disease Prevention and Control, Chinese Center for Disease Control and Prevention, Beijing 102206, China;
3. Department of Respiratory Medicine, Zhejiang Provincial People's Hospital, Hangzhou 310014, Zhejiang, China
Objective Lower respiratory tract infections continue to pose a significant threat to human health. It is important to accurately and rapidly detect respiratory bacteria. To compensate for the limits of current respiratory bacteria detection methods, we developed a combination of multiplex polymerase chain reaction (PCR) and capillary electrophoresis (MPCE) assay to detect thirteen bacterial pathogens responsible for lower respiratory tract infections, including Streptococcus pneumoniae, Haemophilus influenzae, Moraxella catarrhalis, Pseudomonas aeruginosa, Klebsiella pneumoniae, Escherichia coli, Staphylococcus aureus, Mycoplasma pneumoniae, Legionella spp., Bordetella pertussis, Mycobacterium tuberculosis complex, Corynebacterium diphtheriae, and Streptococcus pyogenes.
Methods Three multiplex PCR reactions were built, and the products were analyzed by capillary electrophoresis using the high-throughput DNA analyzer. The specificity of the MPCE assay was examined and the detection limit was evaluated using DNA samples from each bacterial strain and the simulative samples of each strain. This assay was further evaluated using 152 clinical specimens and compared with real-time PCR reactions. For this assay, three nested-multiplex-PCRs were used to detect these clinical specimens.
Results The detection limits of the MPCE assay for the 13 pathogens were very low and ranged from 10-7 to 10-2 ng/μL. Furthermore, analysis of the 152 clinical specimens yielded a specificity ranging from 96.5%-100.0%, and a sensitivity of 100.0% for the 13 pathogens.
Conclusion This study revealed that the MPCE assay is a rapid, reliable, and high-throughput method with high specificity and sensitivity. This assay has great potential in the molecular epidemiological survey of respiratory pathogens.
A lower respiratory tract infection (LRTI) is an acute illness usually presenting with cough as the main symptom, and with at least one other LRT symptom such as fever, sputum production, dyspnea, wheezing, or chest discomfort/pain and no alternative explanation[1]. LRTIs have a high mortality and morbidity worldwide, and The Global Burden of Disease Study reported that LRTIs are the second leading cause of deaths in 2013[2]. The age-standardized mortality rate of LRTIs has been reported to be 41.7 in 100, 000 (95% CI 37.1-44.1)[2].
A large and growing number of microbial pathogens, including bacteria, atypical agents, viruses, and fungi, have been reported to be responsible for LRTI[3-5]. The clinical presentations of LRTI caused by different pathogens are very similar, making differentiation by clinical symptoms and pathology alone difficult.
Traditional methods used to identify respiratory pathogens include Gram stain of the sputum, culture of respiratory secretions and blood serological tests[6]. Although these traditional methods have their advantages in identifying respiratory pathogens to some extent, the disadvantages cannot be ignored. Such as time-consuming, labor-intensive and poor sensitivity. Molecular methods such as polymerase chain reaction (PCR) and real-time PCR have been developed for the rapid detection and identification for respiratory pathogens[7-10]. These methods have high sensitivity and specificity, and can be used to analyze clinical specimens, including blood, sputum, and urine. However, most molecular methods cannot detect more than five pathogens per reaction[7-10]. Therefore, designing a rapid, labor-saving, and high-throughput method to detect and differentiate respiratory pathogens in a single reaction would be greatly useful in enhancing rapid response for the prompt treatment and control for LRTI.
The Applied Biosystems 3730-XL 96-capillary DNA analyzer, which has been designed by Applied Biosystems (ABI, USA) Company, is based on the technology of capillary electrophoresis separation. It allows for multiplexed detection of molecules, and various molecular targets can be detected by using four types of fluorescent tags in 96 samples within a single 96-well plate. Additionally, fourteen 96-well plates could be placed in this analyzer at the same time, and the analyzer can automatically detect the different plates one by one. Compared with other methods, this is a high-throughput, rapid detection method with high specificity and sensitivity, and products differing by as little as 7-10 bp in size can be separated by this system[11].
In this study, a ABI 3730-XL DNA analyzer-based multiplex PCR and capillary electrophoresis (MPCE) assay was developed and applied for the simultaneous detection of 13 respiratory bacteria, including Streptococcus pneumoniae, Haemophilus influenzae, Moraxella catarrhalis, Pseudomonas aeruginosa, Klebsiella pneumoniae, Escherichia coli, Staphylococcus aureus, Mycoplasma pneumoniae, Legionella spp., Bordetella pertussis, Mycobacterium tuberculosis complex, Corynebacterium diphtheriae, and Streptococcus pyogenes. The sensitivity and specificity of the MPCE assay were tested, and 152 clinical samples collected from patients with LRTI were assayed for bacterial detection using this MPCE assay. The results using this method were then compared with results using the single real-time PCR assays mentioned below.
METHODS Bacterial Strains, Simulative Samples, Clinical Specimens, and ControlsThe 13 respiratory bacteria detected in this study are listed in Table S1 available in www.besjournal.com.
|
|
Table S1 List of Target Bacteria, Source, Genes, Primers, and Their Amplicon Sizes |
Simulative samples were composed by mixing sputum from individuals who didn't have respiratory infection symptoms and 11 target bacteria (from the 13 except for M. tuberculosis and M. pneumoniae). The sputum from these individuals were confirmed negative for the 13 target bacteria using 13 single real-time PCR assays. Serial tenfold dilutions (108-100CFU/mL) of 11 bacteria (from the 13 target bacteria, except M. tuberculosis and M. pneumoniae) were mixed with the sputum samples (100 μL) separately. And the plate count method was used to measure bacteria in solution mentioned above (CFU/mL).
Then, 215 adult patients who were hospitalized in the Department of Respiratory Medicine, Shengjing Hospital, China Medical University, Shenyang, China, from May 2015 to January 2016, and were diagnosed with LRTI, including 180 cases of pneumonia, 14 cases of acute bronchitis, 17 cases of acute exacerbation of chronic obstructive pulmonary disease, and 4 cases of pulmonary abscess, were recruited to this study. A total of 215 clinical specimens were gathered from these 215 patients. The following criteria were used to diagnose LRTI: fever and/or an increased blood leukocyte count (≥ 11 × 109/L), increased focal symptoms from the lower airways, with at least one of the following three newly developed symptoms: increased dyspnea, increased coughing, and/or increased purulent sputum[12]. The patients were required to gargle with water in the morning before they spat. The second mouth sputum samples from deep lower respiratory tract were collected in sterile containers and immediately used for Gram staining and microscopic analysis. Samples with fewer than 10 epithelial cells and more than 25 leukocytes in each field at 100 × magnification were included in this study as available sputum samples[13]. And finally, 152 sputum samples from 152 patients were eligible and tested in this study.
For the testing of clinical samples, the targeting 13 respiratory bacteria were used as positive controls, and the sputum from individuals which were confirmed negative for the 13 target bacteria using 13 single real-time PCR assays were used as negative controls. All these samples mentioned above were stored at -80 ℃ until tested.
DNA ExtractionThe target DNA of 11 bacteria were extracted using a QIAamp DNA Mini Kit (Qiagen, Germany), according to the manufacturer's instructions. The 11 bacterial specimens were serially diluted tenfold from 108 to 100 CFU/mL. The clinical specimens were diluted 1:1 with an equal volume of phosphate-buffered saline and homogenized by vortexing for 1 min. Homogenized sputum was transferred into a microcentrifuge tube, and centrifuged at 8, 000 ×g for 5 min. Finally, 200 µL of homogenized sputum on the bottom including the pellet was used for DNA extraction. DNA extraction was carried out using a QIAmp DNA Mini Kit (Qiagen) according to the recommended protocol (Blood and Body Fluids Spin Protocol). The DNA was eluted in a final volume of 100 μL. The DNA of the simulative samples was extracted in the same manner as that described for the clinical specimens.
All the purified DNA, and the DNAs of M. tuberculosis and M. pneumoniae, which had been supplied by the National Institute for Communicable Disease Control and Prevention, Chinese Center for Disease Control and Prevention were stored at -20 ℃ in 1.5 mL microcentrifuge tubes until analysis. And DNA concentrations were measured by NanoDrop 2000 (Thermo Fisher Scientific, Shanghai, China)
Primer DesignGene sequences for S. aureus were obtained through the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/). The S. aureus primers were designed in a highly conserved region for increased specificity and evaluated by the NCBI Primer-Blast and Primer Premier 5.0 software (Table S1). The primer sequences, their target genes, and the size of the resulting amplicons are shown in Table S1. All the forward primers were fluorescently labeled at the 5'-end with carboxyfluorescein (FAM).
Multiplex PCR for Bacterial Strains and Simulative SamplesFirst, primers for the target bacterial strains were confirmed by simplex PCR reactions. The mPCR was affected by many factors such as annealing time, annealing temperature, and primer concentrations. These conditions were optimized for the mPCR to ensure specific amplification of the selected targets. Finally, three mPCR reactions were built. The three mPCR systems are listed in Table S2 available in www.besjournal.com, and the three mPCR reactions were performed in a Thermo Cycler (SensoQuest LabCycler, Senso, Germany). For the first mPCR, cycling parameters were as follows: denaturation at 95 ℃ for 10 min; followed by 35 cycles of 95 ℃ for 30 s, 54 ℃ for 45 s, and 72 ℃ for 1 min; and a final extension at 72 ℃ for 10 min. For the second and the third mPCR reactions, cycling parameters were as follows: denaturation at 95 ℃ for 10 min; followed by 35 cycles of 95 ℃ for 30 s, 60 ℃ for 45 s, and 72 ℃ for 1 min; and a final extension at 72 ℃ for 10 min. The reaction products were resolved on a 2.0% agarose gel stained with GoldView, visualized under UV light, and analyzed using a Gel Doc system (BioRad, CA, USA).
|
|
Table S2 Three mPCR Systems |
Nested mPCR was used to obtain the target DNA data from clinical specimens. After optimization, three nested mPCR reactions were built. All the primers used for the first round and the second round of these nested mPCR were just the same ones listed in Table S1. The three nested mPCR systems are listed in Table S4 available in www.besjournal.com, and the cycling parameters can be seen in Table S3 available in www.besjournal.com. And then the nested mPCR reactions were performed and analyzed as that described above.
|
|
Table S4 Three Nested mPCR Systems |
|
|
Table S3 Thermal Cycler Parameters for MPCR and Nested mPCR |
After amplification, 3 µL of each amplified product was mixed and homogenized by vortexing for 10 min. Then, 1 µL of the mixture was mixed with 8.5 µL of Hi-Di™ Formamide (Applied Biosystems) and 0.5 µL of GeneScan™ 1200 LIZ® Size Standard (Applied Biosystems). This mixture was denatured for 5 min at 95 ℃ and cooled for 4 min at 4 ℃. The denatured DNA fragments were then separated by capillary electrophoresis using an ABI 3730-XL DNA analyzer (Applied Biosystems) and the results were examined using Genemapper software v. 4.0 (Applied Biosystems). The peak height of each gene was determined from the electropherogram.
Sensitivity of the MPCE AssayTo determine the relative sensitivities of the MPCE assay, serial tenfold dilutions (102-10-6ng/μL) of the 13 target DNA samples were extracted. Besides, simulative samples were mixed with different concentrations of bacteria (108-100 CFU/mL), respectively, and later total DNA was extracted, respectively. Moreover, different concentrations of 13 target DNA and simulative samples for 11 bacteria of the same order of magnitude were prepared separately and tested by the MPCE assay to ascertain the ability of this assay to co-amplify multiple gene targets and gene targets present at different concentrations in simulative samples.
Specificity of the MPCE AssayThe species used to assess the specificity of the assay are listed in Table S1. For each pathogen, monoplex PCR assays were performed with its target DNA, and negative controls (other DNA except the target template) were simultaneously tested for each organism. To determine the specificity of the MPCE assay, DNA of the 13 target bacteria were analyzed in the 3 mPCR systems, and after amplification, the 13 mPCR products (5 μL each) were mixed and analyzed again in the capillary electrophoresis system.
Detection in Clinical SamplesA total of 152 sputum samples were collected from Shengjing Hospital, China Medical University in Shenyang, China. DNA was extracted from the samples and tested by both the MPCE (nested mPCR) assay and single real-time PCR assays.
Real-time PCR for Bacterial DetectionThe 13 pairs of specific primers and probes and amplification conditions used in 13 single real-time PCR reactions are shown in Table S5 available in www.besjournal.com.
|
|
Table S5 Real-time PCR Assays for Detection of Thirteen Respiratory Pathogens |
We used a Stratagene Mx3000P (Stratagene, La Jolla, CA) for the real-time PCR assay. The reaction system consisted of 12.5 μL of Premix Ex Taq (Takara) and 1.25 μL of each primer and fluorescent probe. The final volume of the system was adjusted to 25 μL with 0.5 μL of ROX Reference Dye II (Takara) and distilled water. The specificities of the 13 real-time PCR reactions have been proven in previous studies[25-33], and the relevant references have been listed in Table S5 available in www.besjournal.com. The analytical sensitivities for these 13 pathogens were measured by 10-fold serial dilutions of each target DNA from 108-100 CFU/mL. The threshold cycle (CT) values of all the dilutions for these 13 pathogens were confirmed in triplicate.
Statistical AnalysesConsistency between the results of the MPCE assay and real-time PCR assays was verified using Cohen's kappa test in SPSS 17.0 software (IBM SPSS, United States). The kappa value was graded as follows: < 0, no agreement; 0-0.20, slight; 0.21-0.40, fair; 0.41-0.60, moderate; 0.61-0.80, substantial; and 0.81-1, almost perfect agreement.
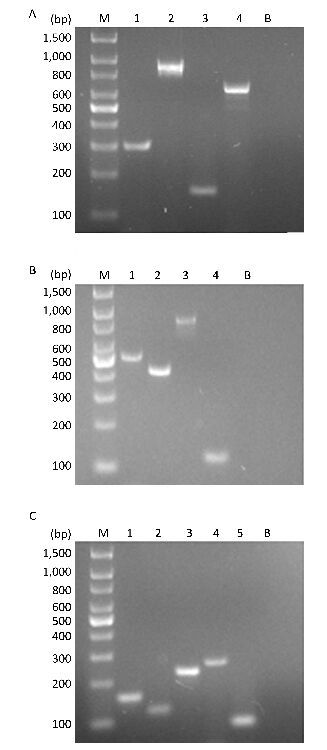
RESULTS Establishment of the Three mPCR AssaysThe three mPCR reactions were optimized by evaluating the primer concentrations, annealing time, and temperature, and no cross-amplification was observed (Figure 1).
|
Download:
|
| Figure 1 (A) Multiplex PCR products from species-specific primers for H. influenzae 296 bp (lane 1), E. coli 903 bp (lane 2), M. pneumoniae 153 bp (lane 3), and Legionella spp. 650 bp (lane 4). (B) Multiplex PCR products from species-specific primers for P. aeruginosa 504 bp (lane 1), K. pneumoniae 423 bp (lane 2), B. pertussis 872 bp (lane 3), M. tuberculosis 123 bp (lane 4), respectively. (C) Multiplex PCR products from species-specific primers for S. pneumoniae 160 bp (lane 1), M. catarrhalis 140 bp (lane 2), S. aureus 233 bp (lane 3), and C. diphtheriae 258 bp (lane 4), S. pyogenes 93 bp (lane 5), respectively. M, 100-base pair (bp) DNA ladder. B, blank. Numbers on the left indicate the bps of the DNA ladder marker. | |
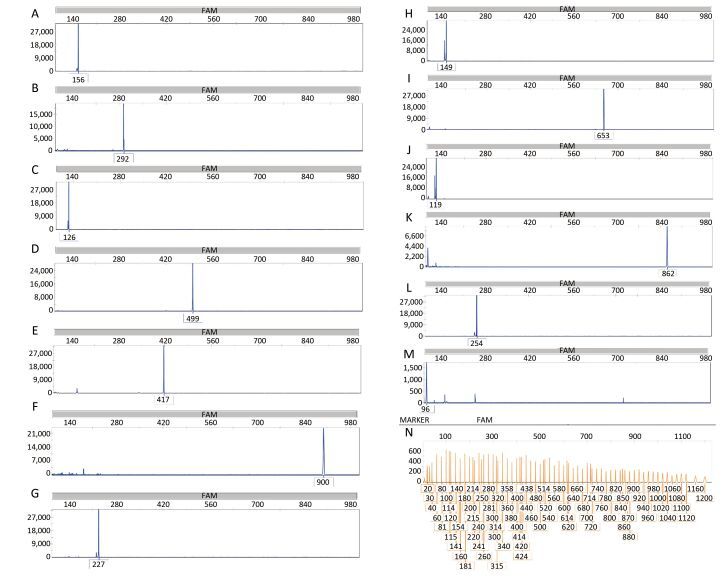
The target DNA samples of all the 13 pathogens were used to evaluate the specificity of the primers. The results revealed that each primer pair could detect the targeted pathogen without cross-amplification. First, all bacterial targets were individually tested in the MPCE assay, and the results are presented in Figure 2.
|
Download:
|
| Figure 2 Specificity results of the MPCE assay for thirteen individual pathogen templates. The MPCE assay was performed with individual templates for S. pneumoniae 156 bp (A), H. influenzae 292 bp (B), M. catarrhalis 126 bp (C), P. aeruginosa 499 bp (D), K. pneumoniae 417 bp (E), E. coli 900 bp (F), S. aureus 227 bp (G), M. pneumoniae 149 bp (H), L. pneumophila 653 bp (I), M. tuberculosis 119 bp (J), B. pertussis 862 bp (K), C. diphtheriae 254 bp (L), and S. pyogenes 92 bp (M). Distilled water was used as the negative control (N). Blue peaks denote specific amplification peaks, and orange peaks denote the marker peaks. | |
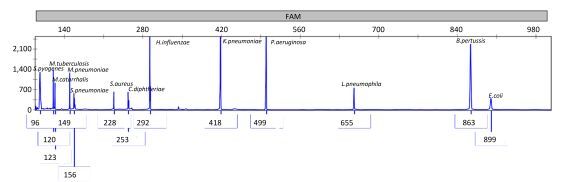
Second, to further determine the specificity of the MPCE assay, DNA of the 13 target bacteria were analyzed in three mPCR systems, and the resultant amplified mPCR products were mixed and analyzed in the capillary electrophoresis system, and the results are presented in Figure 3. As shown in Figure 3, the resultant amplicon sizes for the 13 pathogens in the mixture were similar to their corresponding target sequences (Table S1), verifying the specificity of this method.
|
Download:
|
| Figure 3 Results of the specificity of the MPCE assay for mixed templates of 13 pathogen strains. The concentrations of the target DNA templates for S. pyogenes, M. tuberculosis, M. catarrhalis, M. pneumoniae, S. pneumoniae, S. aureus, C. diphtheriae, H. influenzae, K. pneumoniae, P. aeruginosa, L. pneumophila, B. pertussis, and E. coli were 208.9, 20.1, 148.8, 15.5, 80.1, 22.7, 130.2, 105.1, 103.5, 204.9, 45.4, 13.3, and 192.3 ng/μL, respectively. | |
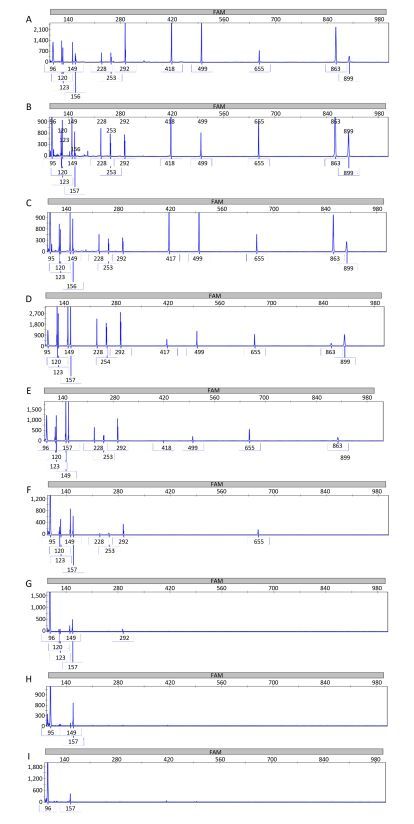
The sensitivity of the MPCE assay was examined. The mPCR reaction products obtained from three mPCR assays were mixed with the same order of magnitude of each sample and tested in an ABI 3730-XL DNA analyzer. The detection limits in the MPCE assay can be seen in Table 1 and Figure 4. The detection limits for these pathogens ranged from 10-7 to 10-2 ng/μL. The lowest detection limit was 10-7 ng/μL for S. pneumoniae.
|
|
Table 1 Detection Limits of the MPCE Assay |
|
Download:
|
| Figure 4 Sensitivity results of the MPCE assay for the thirteen pathogen templates. The concentration of the target DNA templates for S. pyogenes (96 bp), M. tuberculosis (120 bp), M. catarrhalis (123 bp), M. pneumoniae (149 bp), S. pneumoniae (156 bp), S. aureus (228 bp), C. diphtheriae (253 bp), H. influenzae (292 bp), K. pneumoniae (418 bp), P. aeruginosa (499 bp), L. pneumophila (655 bp), B. pertussis (863 bp), and E. coli (899 bp) were 208.9, 20.1, 148.8, 15.5, 80.1, 22.7, 130.2, 105.1, 103.5, 204.9, 45.4, 13.3, and 192.3 ng/μL, respectively. The MPCE assay was performed using a mixture of target DNA templates of different concentrations ranging from 108(A), 107(B), 106 (C), 105(D), 104(E), 103(F), 102(G), 101(H) to 100 (I) CFU/mL. All the 11 target genes (13 target genes except M. tuberculosis and M. pneumoniae) were detected simultaneously at 105 to 108 CFU/mL. P. aeruginosa, K. pneumoniae, and E. coli could be detected at 104CFU/mL. S. aureus, C. diphtheriae, and L. pneumophila could be detected at 103CFU/mL. M. catarrhalis, and H. influenzae could be detected at 102CFU/mL. S. pneumoniae and S. pyogenes could be detected at 100CFU/mL. M. pneumoniae and M. tuberculosis could be detected at 10-6 and 10-5 ng/μL. Blue peaks denote specific amplification peaks. | |
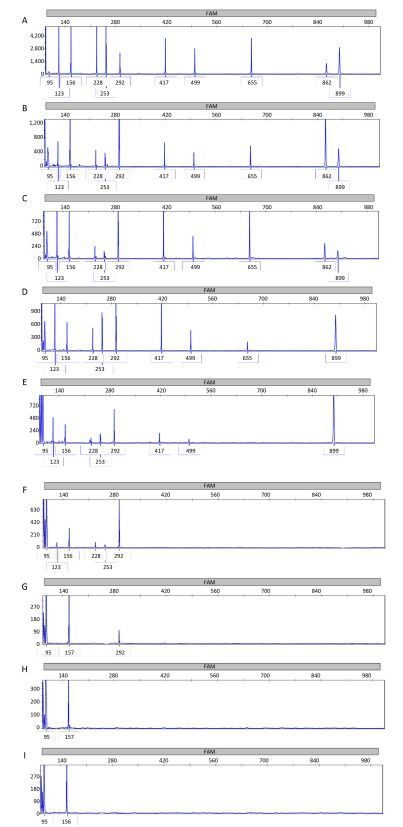
Simulative DNA samples for 11 bacteria were extracted and analyzed in three mPCR assays, respectively. And the mPCR reaction products obtained from three mPCR assays were mixed with the same order of magnitude of each sample and tested in an ABI 3730-XL DNA analyzer. As shown in Table 1 and Figure 5, the detection limits for these simulative samples ranged from 10-7 to 10-2 ng/μL. The lowest detection limits was 10-7 ng/μL for S. pneumoniae.
|
Download:
|
| Figure 5 Results of the sensitivity of the MPCE assay for simulative samples. The concentrations of the target DNA templates for S. pyogenes (95 bp), M. catarrhalis (123 bp), S. pneumoniae (156 bp), S. aureus (228 bp), C. diphtheriae (253 bp), H. influenzae (292 bp), K. pneumoniae (417 bp), P. aeruginosa (499 bp), L. pneumophila (655 bp), B. pertussis (862 bp), and E. coli (899 bp) were 152.4, 95.9, 51.3, 18.9, 98.2, 89.3, 95.0, 171.1, 40.3, 11.0, and 153.3 ng/μL, respectively. The MPCE assay was performed using different concentrations of simulative samples containing 11 bacterial species ranging from 108(A), 107(B), 106 (C), 105(D), 104(E), 103(F), 102(G), 101(H), to 100 (I) CFU/mL. All 11 target simulative samples were simultaneously detected at 106 to 108 CFU/mL. L. pneumophila could be detected at 105CFU/mL. P. aeruginosa, K. pneumoniae, and E. coli could be detected at 104CFU/mL. S. aureus, C. diphtheriae, and M. catarrhalis could be detected at 103CFU/mL. H. influenzae could be detected at 102CFU/mL. S. pneumoniae and S. pyogenes could be detected at 100CFU/mL. Blue peaks denote specific amplification peaks. | |
A total of 152 clinical samples were assayed using the three nested mPCR assays, and tested in an ABI 3730-XL DNA analyzer. And the results were confirmed by the simplex real-time PCR for all the 13 species listed in Table S5. The MPCE (nested mPCR) assay successfully detected and differentiated all 13 target respiratory pathogens. Compared with the results of the simplex real-time PCR, the consistency of MPCE assay exceeded 96.1% for all the 13 target pathogens (Table 2).
|
|
Table 2 Performance of the MPCE Assay Compared to the Real-time PCR Assay |
Of the 152 clinical samples, the MPCE assay dete-cted S. pneumoniae, M. pneumoniae, M. tuberculosis, C. diphtheriae, and S. pyogenes in more samples than the real-time PCR assay. Furthermore, the MPCE assay detected B. pertussis in fewer samples than the real-time PCR assay. The MPCE assay and the simplex real-time PCR assay both identified the same number of positive samples for H. influenzae, M. catarrhalis, P. aeruginosa, K. pneumoniae, E. coli, Legionella spp. and S. aureus. In this study, some samples were detected positive by the MPCE assay but negative by the real-time PCR assay. These samples were then confirmed as true positives by combination of simplex PCR and capillary electrophoresis with the primers designed for the target genes. Besides, these samples were also confirmed as true positives by 16SrRNA sequence analysis. A total of 152 clinical samples were assayed using the MPCE assay, and coinfections were identified in 17 samples.
The obtained sensitivity and specificity of this MPCE assay relative to those of real-time PCR were as follows: 100.0% and 96.5% for S. pneumoniae, 100.0% and 99.3% for M. pneumoniae, 100.0% and 99.3% for M. tuberculosis, 100.0% and 96.6% for C. diphtheriae, 100.0% and 96.9% for S. pyogenes, 100.0% and 100.0% for H. influenzae, M. catarrhalis, P. aeruginosa, K. pneumoniae, E. coli, S. aureus, Legionella spp., and B. pertussis, respectively.
The detection results of our MPCE assay were also compared with conventional diagnostic methods used in our hospital, such as microscopy, blood serological tests, and urinary antigen tests. For M. tuberculosis, nine of the 152 clinical samples detected positive by MPCE assay were also confirmed positive by Ziehl-Neelsen stain. For S. pneumoniae, 41 clinical samples detected positive by MPCE assay were also confirmed positive by urinary antigen test (Binax). For Legionella spp., three clinical samples detected positive by MPCE assay were also confirmed positive by urinary antigen test (Binax). For M. pneumoniae, seven clinical samples detected positive by MPCE assay were also confirmed IgM positive by paired blood serological tests (SERODIA-MYCOII, Tokyo, Japan). These results demonstrated that the MPCE assay was comparable to the conventional diagnostic methods used in the hospital.
DISCUSSIONS. pyogenes, M. tuberculosis, M. catarrhalis, M. pneumoniae, S. pneumoniae, S. aureus, C. diphtheriae, H. influenzae, K. pneumoniae, P. aeruginosa, Legionella spp., B. pertussis, and E. coli are the most commonly reported causative pathogens in LRTI worldwide[1-2, 4-5]. All these bacteria have similar clinical manifestations, and may present as multiple infections, further complicating the LRTI and increasing its morbidity. Conventional diagnostic methods such as isolation and serological investigations for identifying these causative bacteria are laborious and time-consuming. Furthermore, in the case of multiple infections, conventional diagnostic methods need to be carried out separately for each pathogen. Many molecular tests, including single PCR, multiplex PCR, single real-time PCR, and multiplex real-time PCR, for the detection of several of the above mentioned LRTI agents have been developed[10, 16, 27, 29-30]. However, these methods can detect only a limited number of bacterial species at a time[7-10]. Therefore, there is an urgent need for a high-throughput and rapid method that can detect multiple causative agents in LRTI for effective disease surveillance and disease management.
The MPCE assay based on the technology of capillary electrophoresis separation is a high-throughput, rapid detection method with high specificity and sensitivity, and products differing by as little as 7-10 bp in size can be separated by this system[11]. Additionally, this assay is very useful because it enables the simultaneous detection of a broad range of bacterial agents related to respiratory infections, which is an optional feature, meaning that it can be used as a single-target assay for each pathogen or organized into different multiplex assays for different diagnostic purposes, as required. Moreover, this method can also be expanded to include more pathogens, further increasing its abilities. For example, one way to achieve this is to use different fluorescent dyes as well, so the different pathogens could be separated by both size and fluorescence, similar to the strategy that was described before[34]. Previously, capillary electrophoresis was successfully applied to investigate ethnic variations of polymorphism and to analyze the differences in genetic polymor-phism[35-36]. Recently, capillary electrophoresis has been used for the rapid identification of Candida fungal infections and for the multiplex detection of nine foodborne pathogens[37-38]. To our knowledge, this is the first study to investigate the microbial pathogens responsible for LRTI using this method.
In this study, 13 specific primer pairs were used to develop a MPCE assay based on the ABI 3730-XL analyzer for the simultaneous detection of 13 common LRTI pathogens, namely, S. pyogenes, M. tuberculosis, M. catarrhalis, M. pneumoniae, S. pneumoniae, S. aureus, C. diphtheriae, H. influenzae, K. pneumoniae, P. aeruginosa, L. pneumophila, B. pertussis, and E. coli. Compared with the results of the real-time PCR reactions (Table 2), the consistency of the MPCE assay exceeded 96.1% for all 13 target pathogens, which demonstrated that the MPCE assay had comparable detection to real-time PCR. In addition, the detection limits for most of the pathogens in our assay are very low. For example, 10-7 ng/μL for S. pneumoniae; 10-6 ng/μL for M. pneumoniae and S. pyogenes; 10-5 ng/μL for M. tuberculosis; 10-4 ng/μL for H. influenzae, M. catarrhalis, S. aureus, and Legionella spp.
Another advantage of the MPCE assay is that it is a rapid and high-throughput method. The entire reaction can be completed within 4.5 h. And the analyzer can detect fourteen 96-well plates with 96 samples one by one automatically.
However, some of the samples that were found to be positive by the single real-time PCR assay tested negative by the MPCE assay. The reason for the low detection rate of B. pertussis in our assay may be related to the PCR primers used, which targeted the B. pertussis S1 subunit, which is important for the detection of toxin-producing B. pertussis, while the primers in the real-time PCR assay targeted IS481, which is the pertussis toxin promotor region specific to B. pertussis. To improve the specificity of our assay, new primers with high specificity and sensitivity should be designed in the future. In this study, we used sputum samples to verify the validity of the MPCE assay, and we tried our best to avoid contamination in the process of collecting the samples. Although this method has its limitation in etiological diagnosis using sputum samples, the MPCE assay is still a newly built method with high-throughput and is easy to operate with high reliability. Based on the findings of the present study, this is a promising new method with potential for identifying respiratory pathogens in LRTI.
CONCLUSIONSIn conclusion, this study illustrates that the MPCE assay is a rapid, reliable, labor-saving, and high-throughput method with high specificity and sensitivity for the identification of 13 common respiratory pathogens responsible for LRTI. This assay has great potential in the molecular epidemiological survey of respiratory pathogens.
ETHICS APPROVALThe research protocol for the current study has been approved by The Ethics Committee of the Shengjing Hospital of China Medical University, Shenyang, Liaoning, China (2016PS238K).
CONFLICT OF INTERESTAll the authors declare that there are no conflicts of interests.
AUTHORS' CONTRIBUTIONSJLX carried out the experiments of this study, except those documented in Figure 1 which were performed by RHY, and took part in writing the manuscript. ZHJ participated in data analysis and took part in writing the manuscript. CY and QT participated in conceiving and coordinating experimental work, and took part in writing the manuscript. And they also provided materials for this study. ZSH and HBY collected all the sputum samples for the study, and wrote the manuscript. All authors read and approved the final manuscript.
| 1. | Woodhead M, Blasi F, Ewig S, et al. Guidelines for the management of adult lower respiratory tract infections-full version. Clin Microbiol Infect, 2011, 17: E1–59. |
| 2. | Prina E, Ranzani OT, Torres A. Community-acquired pneumonia. Lancet, 2015, 386: 1097–108. doi:10.1016/S0140-6736(15)60733-4 |
| 3. | Johansson N, Kalin M, Tiveljung-Lindell A, et al. Etiology of community-acquired pneumonia:increased microbiological yield with new diagnostic methods. Clin Infect Dis, 2010, 50: 202–9. doi:10.1086/648977 |
| 4. | Lim WS, Baudouin SV, George RC, et al. BTS guidelines for the management of community acquired pneumonia in adults:update 2009. Thorax, 2009, 64: iii1–55. |
| 5. | Mandell LA, Wunderink RG, Anzueto A, et al. Infectious Diseases Society of America/American Thoracic Societyconsensus guidelines on the management of community-acquired pneumonia in adults. Clin Infect Dis, 2007, 44: S27–72. doi:10.1086/511159 |
| 6. | Mandell LA. Community-acquired pneumonia:An overview. Postgrad Med, 2015, 127: 607–15. doi:10.1080/00325481.2015.1074030 |
| 7. | Pai R, Gertz RE, Beall B. Sequential multiplex PCR approach for determining capsular serotypes of Streptococcus pneumoniae isolates. J Clin Microbiol, 2006, 44: 124–31. doi:10.1128/JCM.44.1.124-131.2006 |
| 8. | Tian GZ, Zhang LJ, Wang XL, et al. Rapid detection of Haemophilus influenzae and Haemophilus parainfluenzae in nasopharyngeal swabs by multiplex PCR. Biomed Environ Sci, 2012, 25: 367–71. |
| 9. | Gok U, Bulut Y, Keles E, et al. Bacteriological and PCR analysis of clinical material aspirated from otitis media with effusions. Int J Pediatr Otorhinolaryngol, 2001, 60: 49–54. doi:10.1016/S0165-5876(01)00510-9 |
| 10. | Thong KL, Lai MY, Teh C SJ, et al. Simultaneous detection of methicillin-resistant Staphylococcus aureus, Acinetobacter baumannii, Escherichiacoli, Klebsiella pneumoniae and Pseudomonas aeruginosa by multiplex PCR. Trop Biomed, 2011, 28: 21–3. |
| 11. | Yang MJ, Luo L, Nie K, et al. Genotyping of 11 human papillomaviruses by multiplex PCR with a GeXP analyzer. J Med Virol, 2012, 84: 957–63. doi:10.1002/jmv.23275 |
| 12. | Abdeldaim GM, Strålin K, Korsgaard J, et al. Multiplexquantitative PCR for detection of lower respiratory tract infection and meningitis caused by Streptococcus pneumoniae, Haemophilus influenzae and Neisseria meningitidis. BMC Microbiol, 2010, 10: 310. doi:10.1186/1471-2180-10-310 |
| 13. | Aydemir O, Aydemir Y, Ozdemir M. The role of multiplex PCR test in identification of bacterial pathogens in lower respiratory tract infections. Pak J Med Sci, 2014, 30: 1011–6. |
| 14. | Mousavi SF, Nobari S, Rahmati Ghezelgeh F, et al. Serotyping of Streptococcus pneumoniae isolated from Tehran by Multiplex PCR: Are serotypes of clinical and carrier isolates identical?Iran J Microbiol, 2013;5, 220-6. |
| 15. | Pai R, Gertz RE, Beall B. Sequential multiplex PCR approach for determining capsular serotypes of Streptococcus pneumoniae isolates. J Clin Microbiol, 2006, 44: 124–31. doi:10.1128/JCM.44.1.124-131.2006 |
| 16. | Strålin K, Bäckman A, Holmberg H, et al. Design of a multiplex PCR for Streptococcus pneumoniae, Haemophilus influenzae, Mycoplasma pneumoniae and Chlamydophila pneumoniae to be used on sputum samples. APMIS, 2005, 113: 99–111. doi:10.1111/apm.2005.113.issue-2 |
| 17. | Hall-Stoodley L, Hu FZ, Gieseke A, et al. Direct detection of bacterial biofilms on the middle-ear mucosa of children with chronic otitis media. JAMA, 2006, 296: 202–11. doi:10.1001/jama.296.2.202 |
| 18. | Chen L, Chavda KD, Findlay J, et al. Multiplex PCR for identification of two capsular types in epidemic KPC-producing Klebsiella pneumoniae sequence type 258 strains. Antimicrob Agents Chemother, 2014, 58: 4196–9. doi:10.1128/AAC.02673-14 |
| 19. | Qasem JA, Al-Khalaf BN, Qasem AA, et al. Application of three uniplex polymerase chain reaction assays for the detection of atypical bacteria in asthmatic patients in Kuwait. J Infect Public Health, 2013, 6: 134–41. doi:10.1016/j.jiph.2012.12.002 |
| 20. | Rafiee M, Jahangiri-Rad M, Hajjaran H, et al. Detection and identification of Legionella species in hospital water supplies through Polymerase Chain Reaction (16S rRNA). J Environ Health Sci Eng, 2014, 12: 83. doi:10.1186/2052-336X-12-83 |
| 21. | Xu YH, Wang YY, Zhang SM, et al. Sequencing of the S1 and Prn genes of three pertussis strains for the preparation of vaccine in China. Chin J Microbiol Immunol, 2006, 26: 137–42. |
| 22. | Makeshkumar V, Madhavan R, Narayanan S. Polymerase chain reaction targeting insertion sequence for the diagnosis of extrapulmonary tuberculosis. Indian J Med Res, 2014, 139: 161–6. |
| 23. | Pimenta FP, Matias GA, Pereira GA, et al. A PCR for dtxR gene:application to diagnosis of non-toxigenic and toxigenic Corynebacterium diphtheriae. Mol Cell Probes, 2008, 22: 189–92. doi:10.1016/j.mcp.2008.01.001 |
| 24. | Slinger R, Goldfarb D, Rajakumar D, et al. Rapid PCR detection of group A Streptococcus from flocked throat swabs:a retrospective clinical study. Ann Clin Microbiol Antimicrob, 2011, 10: 33. doi:10.1186/1476-0711-10-33 |
| 25. | Carvalho Mda G, Tondella ML, McCaustland K, et al. Evaluation and improvement of real-time PCR assays targeting lytA, ply, and psaA genes for detection of pneumococcal DNA. J Clin Microbiol, 2007, 45: 2460–6. doi:10.1128/JCM.02498-06 |
| 26. | Hirama T, Yamaguchi T, Miyazawa H, et al. Prediction of the pathogens that are the cause of pneumonia by the battlefield hypothesis. PLoS One, 2011, 6: e24474. doi:10.1371/journal.pone.0024474 |
| 27. | Smati M, Clermont O, Le Gal F, et al. Real-time PCR for quantitative analysis of human commensal Escherichia coli populations reveals a high frequency of subdominant phylogroups. Appl Environ Microbiol, 2013, 79: 5005–12. doi:10.1128/AEM.01423-13 |
| 28. | Gosiewski T, Szała L, Pietrzyk A, et al. Comparison of methods for isolation of bacterial and fungal DNA from human blood. Curr Microbiol, 2014, 68: 149–55. doi:10.1007/s00284-013-0451-1 |
| 29. | Chiba N, Murayama SY, Morozumi M, et al. Rapid detection of eight causative pathogens for the diagnosis of bacterial meningitis by real-time PCR. J Infect Chemother, 2009, 15: 92–8. doi:10.1007/s10156-009-0670-3 |
| 30. | Welti M, Jaton K, Altwegg M, et al. Development of a multiplex real-time quantitative PCR assay to detect Chlamydia pneumoniae, Legionella pneumophila and Mycoplasma pneumoniae in respiratory tract secretions. Diagn Microbiol Infect Dis, 2003, 45: 85–95. doi:10.1016/S0732-8893(02)00484-4 |
| 31. | Kösters K, Riffelmann M, Wirsing von König CH. Evaluation of a real-time PCR assay for detection of Bordetella pertussis and B.parapertussis in clinical samples. J Med Microbiol, 2001, 50: 436–40. doi:10.1099/0022-1317-50-5-436 |
| 32. | Schuhegger R, Lindermayer M, Kugler R, et al. Detection of toxigenic Corynebacterium diphtheriae and Corynebacterium ulcerans strains by a novel real-time PCR. J Clin Microbiol, 2008, 46: 2822–3. doi:10.1128/JCM.01010-08 |
| 33. | Morozumi M, Nakayama E, Iwata S, et al. Simultaneous detection of pathogens in clinical samples from patients with community-acquiredpneumonia by real-time PCR with pathogen-specific molecular beacon probes. J Clin Microbiol, 2006, 44: 1440–6. doi:10.1128/JCM.44.4.1440-1446.2006 |
| 34. | Cheng Pettersson A, Viskari A, Odén U, et al. Improved MPL mutation screening with multiplex PCR and capillary electrophoresis. Br J Haematol, 2016;19. doi: 10.1111/bjh.14253.[Epubaheadofprint] |
| 35. | Stewart S, Wickramasinghe D, Dorrance AE, et al. Comparison of three microsatellite analysis methods for detecting genetic diversity in Phytophthora sojae (Stramenopila:Oomycete). Biotechnol Lett, 2011, 33: 2217–23. doi:10.1007/s10529-011-0682-9 |
| 36. | Piras I, Falchi A, Moral P, et al. Frequencies of promoter pentanucleotide (TTTTA) n of CYP11A gene in European and North African populations. Genet Test, 2008, 12: 93–6. doi:10.1089/gte.2007.0060 |
| 37. | ObručováH, TihelkováR, KotáskováI, et al. Evaluation of fluorescent capillary electrophoresis for rapid identification of Candida fungal infections. J Clin Microbiol, 2016, 54: 1295–303. doi:10.1128/JCM.00118-16 |
| 38. | Villamizar-Rodríguez G, Fernández J, Marín L, et al. Multiplex detection of nine food-borne pathogens by mPCR and capillary electrophoresis after using a universalpre-enrichment medium. Front Microbiol, 2015, 3: 1194. |
 2017, Vol. 30
2017, Vol. 30


