2. State Key Laboratory of Infectious Diseases Prevention and Control, Collaborative Innovation Center for Diagnosis and Treatment of Infectious Diseases, National Institute for Communicable Disease Control and Prevention, Chinese Center for Disease Control and Prevention Beijing 102206, China;
3. Key Laboratory of Laboratory Medicine, Ministry of Education, College of Laboratory Medicine and Life Sciences, Wenzhou Medical University, Wenzhou 325035, Zhejiang, China;
4. Department of Clinical Laboratory, Shanghai Public Health Clinical Center, Affiliated to Fudan University, Shanghai 201508, China;
5. Department of Spine Surgery, The Xiangya Hospital of Central South University, Changsha 410008, Hunan, China
Objective In this study, milk from a cow with mastitis was analyzed to determine the presence of mycobacterial infection.Milk quality and security problems pertaining to the safe consumption of dairy products were also discussed in this study.
Methods Milk was preprocessed with 4% NaOH.Then, mycobacteria were isolated from the milk sample on L-J medium.The isolate was identified using multiple loci Polymerase Chain Reaction (PCR) and multi-locus sequence analysis with 16S rRNA, sodA, hsp65, and ITS genes.The drug sensitivity of the isolate to 27 antibiotics was tested through alamar blue assay.
Results Smooth, moist, pale yellow colonies appeared on the L-J medium within a week after inoculation.Based on the results of multiple loci PCR analysis, the isolate was preliminarily identified as non-tuberculous mycobacteria.The 16S rRNA, sodA, hsp65, and ITS gene sequences of the isolate exhibited 99%, 99%, 99%, and 100% similarities, respectively, with those of the published reference strains of Mycobacterium elephantis(M.elephantis).The drug sensitivity results showed that the strain is resistant to isoniazid, p-aminosalicylic acid, and trimesulf but is sensitive to ofloxacin, rifampicin, amikacin, capreomycin, moxifloxacin, kanamycin, levofloxacin, cycloserine, ethambutol, streptomycin, tobramycin, rifabutin, ciprofloxacin, linezolid, cefoxitin, clarithromycin, and minocycline.
Conclusion To the best of our knowledge, this study is initially to report the isolation of M.elephantis from the milk of a cow with mastitis in China.
The group of nontuberculous mycobacteria (NTM) comprises 169 recognized species and 13 subspecies that are widely distributed in various environments (http://www.bacterio.net/mycobacterium.html). In recent years, the reported number of NTM species and the incidence of their associated diseases have increased globally. NTM have been isolated from water, soil, air, food, protozoa, plants, animals, and humans[1-2]. Under certain conditions, opportunistic NTM may become pathogenic; by contrast, saprophytic NTM never or very rarely cause diseases[3]. Nevertheless, approximately one-third of NTM species are associated with human diseases[4-5]. Mycobacterium elephantis (M. elephantis), a fast-growing mycobacterial species, was first isolated from a lung abscess of a Sri Lankan elephant that had died of chronic respiratory disease[6]. M. elephantis has been isolated from patients in developed countries, including Canada, Italy, and Belgium; nevertheless, its clinical relevance remains unknown[7-10]. Thus far, the isolation and identification of M. elephantis from milk specimens has not been reported in China. In this paper, we report the first Chinese case of the isolation of M. elephantis from the milk a cow with mastitis. In addition, we discuss the identification and drug sensitivity of this bacterium in this paper.
MATERIALS AND METHODS Milk SampleFresh milk was collected from a cow with mastitis by a technician of a dairy cattle breeding company in Shaanxi Province, China. The sample was not sterilized.
Antibiotics and ChemicalsCation-adjusted Mueller-Hinton broth (CAMHB) was purchased from Difco (Detroit, MI, USA). Alamar blue was purchased from AbD Serotec (Oxford, UK). Twenty-seven types of antibiotics were purchased from Sigma-Aldrich Company (St. Louis, MO, USA). Gatifloxacin and rifabutin were procured from Toronto Research Chemicals Inc. (Toronto, Canada). The details of the 27 antibiotics are shown in Table 1.
|
|
Table 1 Drug Sensitivity Test Results of the Isolate from Milk Samples |
The reference strain NTM95080 (originally purchased from National Institutes for Food and Drug Control) and H37Rv were provided by and stored at the Tuberculosis Laboratory, National Institute for Communicable Disease Control and Prevention, Chinese Center for Disease Control and Prevention.
Specimen Processing and Mycobacterial CultureA 30-mL milk sample was placed in a 50-mL centrifuge tube and centrifuged at 12, 000 rpm for 15 min to obtain a precipitate. The precipitate was preprocessed with an equal volume of 4% NaOH (1:1) for 15 min to eliminate contaminants. Then, 45 mL of PBS (0.01 mol/L pH 7.2) was added to the sample to dilute NaOH. The sample was centrifuged again at 12, 000 rpm for 15 min to obtain a precipitate. The precipitate was resuspended in 1.0 mL PBS. To culture mycobacteria, L-J medium was inoculated with 0.5 mL of the suspension.
Multiple Loci Polymerase Chain Reaction (PCR)After three generations of culture on L-J culture, individual colonies were selected and placed in 1.5-mL screw-capped centrifuge tubes with 0.5 mL water. Bacteria were inactivated by incubating the tubes in a water bath at 80 ℃ for 30 min. Bacterial cells were collected by centrifuging the incubated tubes at 12, 000 rpm for 5 min. The precipitate was resuspended in 300 μL water. To release genomic DNA from the bacterial suspension, the tubes were placed in a metal bath at 100 ℃ for 20 min, then centrifuged at 12, 000 rpm for 5 min. The supernatant was then collected for PCR amplification. Multiple loci PCR with seven target genes, including 16S rRNA, Rv0577, IS1561, Rv1510, Rv1970, Rv3877, and Rv3120, was used to identify the strain as Mycobacterium tuberculosis complex or NTM. Refer to reference[11] for the identification criteria and the PCR primers used to amplify the target gene loci from the chromosomal DNA of the isolate.
Multi-locus Sequence AnalysisMulti-locus sequence analysis (MLSA) was used to identify the Mycobacterium species of the isolate based on the sequences of the 16S rRNA, SodA, hsp65, and ITS genes. hsp65 was amplified using the hsp65F and hsp65R primers reported by Huard Richard C, et al.[11]. SodA was amplified using the SodlgF and SodlgF primers reported by Adékambi Toïdi, et al.[12]. 16s-23s ITS was amplified using the 16sF and 23sR primers reported by Roth A, et al.[13]. A total of 2 μL of DNA, 1 μL of primers, and 8.5 μL distilled water were added to 12.5 μL 2 × Taq PCR MasterMix to a final volume of 25 μL. PCR products were sent to Tsingke Company (Beijing, China) for two-way sequencing and sequence splicing to obtain the complete sequence of the PCR products. The sequencing results of 16SrRNA/SodA/hsp65/ITS were submitted to the National Center for Biotechnology Information (NCBI) website for homology comparison. Those that displayed a similarity of over 98% were accepted as species[14]. MEGA 6.01 software was used for cluster analysis. Alignment and evolutionary tree construction were conducted using MUSCLE with Maximum Likelihood method.
Drug Sensitivity TestingWe performed the alamar blue assay to test the sensitivity of the isolate ShaanX15001 and the reference strain NTM95080 to 27 antibacterial drugs. Minimum inhibitory concentration (MIC) testing was performed in accordance with the guidelines established and approved by Clinical and Laboratory Standards Institute[15]. We combined the existing MIC thresholds of the tested drugs to estimate the susceptibility of the isolate to different antibacterial drugs.
Preparation of Microbial DilutionColonies that were growing well in L-J medium were scraped and placed in normal saline, suspended to 0.5 McIntosh concentration (approximately 0.5 mg/mL), and diluted with liquid medium at a ratio of 1:200 to a final concentration of approximately 105 CFU/mL of inoculant suspension.
Establishment of a Drug Concentration GradientA total of 100 μL liquid CAMHB was added to each well of a 96-well plate except to border wells. Two-fold diluted drugs were added to the plate to establish the corresponding drug test range, as shown in Table 1. Finally, 100 μL of bacterial suspension was added to each well. Three negative controls were set. The drug-free control well (CAMHB + inoculum) was used as a reference for the addition of alamar blue. The well containing CAMHB without inoculum was used as reference to decide the interference of CAMHB with alamar blue. A series of control wells containing different concentration gradients of each drug and drug-CAMHB mixture were used as a reference for the interference of CAMHB mixture with alamar blue. The plates were sealed in individual Ziploc bags and then incubated at 37 ℃.
Interpretation of Experimental ResultsAfter in-cubation at 37 ℃ for 24 h, the first drug-free control wells were examined using an indicator (20 μL alamar blue and 50 μL sterile 5% Tween-80). The plates were then re-incubated for 24 h. If the control well turned pink, all of the other wells containing drugs received the indicator. After an additional 24 h of incubation, the colors of all wells were recorded. If the color of the first drug-free growth control well did not change to pink, the second drug-free control well received the indicator and the above steps were repeated. MIC was based on the range between the non-discolored hole that corresponded to the lowest drug concentration and discolored hole that corresponded to the largest drug concentration[16]. The MIC value reflected the bacteriostatic activity of the drug. A low value indicated high bacteriostatic activity. The final MIC of each drug was calculated as the mean of two or three tests. The MIC thresholds indicating sensitivity, moderate susceptibility, and resistance were interpreted in references[15, 17-19].
Ethics StatementExperimental animals were scientifically and rationally treated in accordance with the '3R' principle: 'Reduction, Replacement, and Refinement.' This study was performed in strict accordance with the recommendations provided in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. This study was approved by the Ethics Committee of the National Institute for Communicable Disease Control and Prevention, Chinese Center for Disease Control and Prevention. The company that provided the milk for this study also provided written informed consent.
RESULTS Culture ResultsSeveral smooth and moist pale yellow colonies appeared on the L-J medium within a week after inoculation.
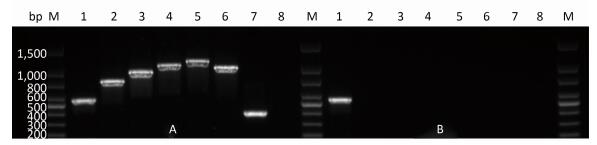
Multiple Loci PCRThe results of multiple loci PCR analysis were consistent with those for mycobacteria other than tuberculosis, e.g., NTM. In the results, only 16S rRNA corresponded with the target band. The results are shown in Figure 1.
|
Download:
|
| Figure 1 Multiple loci PCR results for H37Rv and ShaanX15001. A: H37Rv, B: ShannX15001, M: DNA marker. Molecular weights from the top to the bottom of the lane followed the order of 1, 500 bp, 1, 000 bp, 800 bp, 600 bp, 500 bp, 400 bp, 300 bp, and 200 bp. Lanes 1, 2, 3, 4, 5, 6, 7, and 8 correspond to 16S rRNA, Rv0577, IS1561, Rv1510, Rv1970, Rv3877, Rv3120, and the negative control, respectively. | |
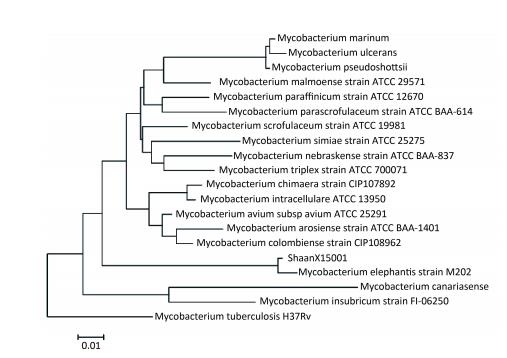
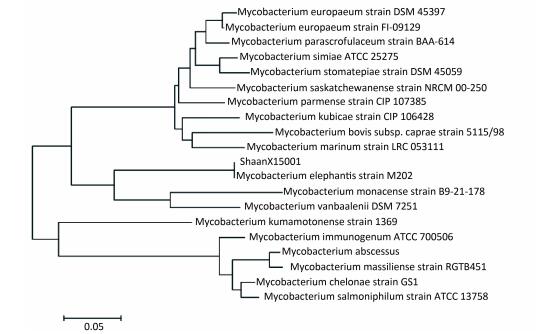
The sequences of 16S rRNA, SodA, hsp65, and ITS of ShaanX15001 revealed 99%, 99%, 99%, and 100% similarities, respectively, with those of the validated M. elephantis reference strains (NCBI). The clustering results for Hsp65 and ITS are shown in Figures 2 and 3, respectively. These results provide further evidence that ShaanX15001 is M. elephantis.
|
Download:
|
| Figure 2 Sequence analysis of hsp65 genes of partial nontuberculous mycobacterium strains and ShaanX15001 isolated from milk. | |
|
Download:
|
| Figure 3 Sequence analysis of the ITS genes of partial nontuberculous mycobacterium strains and ShannX15001 isolated from milk. | |
The drug sensitivity results showed that the strain was resistant to isoniazid, p-aminosalicylic acid, and trimesulf. The strain was sensitive to ofloxacin, rifampicin, amikacin, capreomycin, moxifloxacin, kanamycin, levofloxacin, cycloserine, ethambutol, streptomycin, tobramycin, rifabutin, ciprofloxacin, linezolid, cefoxitin sodium, clarithromycin, and minocycline (Table 1).
DISCUSSIONIdentifying NTM on the species level is difficult given the lack of a rapid and easy method for NTM identification. The hsp65 gene has been used to identify mycobacteria since 1993[20] given its high conservation among mycobacterial species and interspecific differences. However, in this study, we found that the ITS gene is better at distinguishing between species than hsp65 gene. Therefore, multiple genes should be used for species identification when the use of one gene could not provide definitive identification.
NTM can cause serious infections by direct transmission from the environment or after colonization. Bold et al. suggested that slowly growing mycobacteria are usually isolated from the lower respiratory tract of patients with NTM infections, whereas rapidly growing mycobacteria are more frequently isolated from other tissues[21]. Rapidly growing NTM species are more prevalent in Eastern Asia[22]. Tortoli stated that the environment is the most probable reservoir of M. elephantis for either human or animal infections[8]. These reports are all consistent with our results, which showed that M. elephantis is a rapidly growing mycobacteria that can isolated from the milk of a cow with mastitis.
M. elephantis, a rapidly growing mycobacterial species, was first isolated in 2, 000 from the lung abscess of an elephant. Since then, this mycobacterium has been occasionally isolated from the clinical samples of human patients and is most commonly isolated from sputum[6-8]. M. elephantis has also been isolated from a cervical lymph node sample, although its relationship with the clinical disease is unclear[7]. This study is the first to report a Chinese case of the isolation of M. elephantis from milk taken from a cow with mastitis.
Milk is pasteurized to ensure the safety of milk and dairy products for human consumption. However, to retain nutrients and active substances in milk, milk companies may apply temperatures lower than 60 ℃ during pasteurization. During pasteurization, milk is subjected to low temperatures for a long period or to high temperatures for a short period. The optimal growth temperature of M. elephantis is 42 ℃; this species does not grow at 52 ℃[7]. Nevertheless, we posit that M. elephantis may remain in milk due to incomplete pasteurization. Patients who drink milk as a nutritional supplement may acquire M. elephantis infections from milk, thus aggravating their illness. If the milk is used to produce milk powder for babies, babies who drink the milk powder could become seriously ill.
The control of cow tuberculosis must adhere to the policy of 'Prevention First' combined with the comprehensive preventative and control measures of monitoring, quarantine, culling, and disinfection. Cows should be regularly monitored, and tuberculosis-positive cattle should be immediately removed from the herd and subjected to clinical examination, and, if necessary, to bacteriological examination. Sick cows should receive treatment in accordance with the relevant provisions of the country. The pool, barn, stadium, and relevant vehicles and personnel should be disinfected.
Finally, more attention should be paid to the screening of milk products released into the market to ensure the safety of dairy products and avoid potential risks.
DISCLOSURE OF CONFLICT OF INTERESTNone.
AUTHOR CONTRIBUTIONSWAN Kang Lin, LYU Jian Xin, JI Ling Yun, and XU Dong Lei conceived and designed the experiments in this study. JI Ling Yun, XU Dong Lei, LI Gui Lian, YIN Shu Peng, and WEI Jian Hao performed the experiments. JI Ling Yun, XU Dong Lei, LIU Hai Can, JIANG Yi, ZENG Hao, and LOU Yong Liang analyzed the data. WAN Kang Lin and LYU Jian Xin contributed reagents, materials, and analytical tools. JI Ling Yun and WAN Kang Lin wrote the article.
| 1. | Falkinham JO 3rd. Surrounded by mycobacteria:nontuberculous mycobacteria in the human environment. J Appl Microbiol, 2009, 107: 356–67. doi:10.1111/jam.2009.107.issue-2 |
| 2. | Castillo-Rodal AI, Mazari-Hiriart M, Lloret-Sanchez LT, et al. Potentially pathogenic nontuberculous mycobacteria found in aquatic systems. Analysis from a reclaimed water and water distribution system in Mexico City. Eur J Clin Microbiol Infect Dis, 2012, 31: 693–94. |
| 3. | Somoskovi A, Salfinger M. Nontuberculous mycobacteria in respiratory infections:advances in diagnosis and identification. Clin Lab Med, 2014, 34: 271–95. doi:10.1016/j.cll.2014.03.001 |
| 4. | Thomson RM. Changing epidemiology of pulmonary nontuberculous mycobacteria infections. Emerg Infect Dis, 2010, 16: 1576–83. doi:10.3201/eid1610.091201 |
| 5. | Cassidy PM, Hedberg K, Saulson A, et al. Nontuberculous mycobacterial disease prevalence and risk factors:a changing epidemiology. Clin Infect Dis, 2009, 49: e124–9. doi:10.1086/599195 |
| 6. | Shojaei H, Magee JG, Freeman R, et al. Mycobacterium elephantis sp. nov., a rapidly growing non-chromogenic Mycobacterium isolated from an elephant. Int J Syst Evol Micr, 2000, 50(Pt 5): 1817–20. |
| 7. | Christine Turenne, Pamela Chedore, Joyce Wolfe, et al. Phenotypic and molecular characterization of clinical isolates of Mycobacterium elephantis from human specimens. J Clin Microbiol, 2002, 40: 1230–6. doi:10.1128/JCM.40.4.1230-1236.2002 |
| 8. | Tortoli E, Rindi L, Bartoloni A, et al. Mycobacterium elephantis:not an exceptional finding in clinical specimens. Eur J Clin Microbiol Infect Dis, 2003, 22: 427–30. doi:10.1007/s10096-003-0950-2 |
| 9. | Springer B, Stockman LTeschner K, Roberts GD, et al. Two-laboratory collaborative study on identification of mycobacteria:molecular versus phenotypic methods. J Clin Microbiol, 1996, 34: 296–303. |
| 10. | Potters D, Seghers M, Muyldermans G, et al. Recovery of Mycobacterium elephantis from sputum of a patient in Belgium. J Clin Microbiol, 2003, 41: 1344. doi:10.1128/JCM.41.3.1344.2003 |
| 11. | Huard Richard C, Butler W Ray, Dick Van Soolingen, et al. PCR-based method to differentiate the subspecies of the Mycobacterium tuberculosis complex on the basis of genomic deletions. J Clin Microbiol, 2003, 41: 1637–50. doi:10.1128/JCM.41.4.1637-1650.2003 |
| 12. | Adékambi Toïdi, Drancourt Michel. Dissection of phylogenetic relationships among 19 rapidly growing Mycobacterium species by 16S rRNA, hsp65, sodA, recA and rpoB gene sequencing. Int J Syst Evol Micr, 2004, 54: 2095–105. doi:10.1099/ijs.0.63094-0 |
| 13. | Roth A, Reischl U, Streubel A, et al. Novel diagnostic algorithm for identification of mycobacteria using genus-specific amplification of the 16S-23S rRNA gene spacer and restriction endonucleases. J Clin Microbiol, 2000, 38: 1094–104. |
| 14. | Turenne CY, Tschetter L, Wolfe J, et al. Necessity of quality-controlled 16S rRNA gene sequence databases:identifying nontuberculous Mycobacterium species. J Clin Microbiol, 2001, 39: 3637–48. doi:10.1128/JCM.39.10.3638-3648.2001 |
| 15. | Institue Clinical Laboratory Standards. Susceptibility testing of mycobacteria, Nocardiae, and other aerobic actinomycetes. approved standard-Second Edition. Document M24-A2, 31, 2011. https://www.researchgate.net/publication/285850333_Susceptibility_testing_of_mycobacteria_nocardiae_and_other_aerobic_actinomycetes |
| 16. | Franzblau SG, Witzig RS, McLaughlin JC, et al. Rapid, low-technology MIC determination with clinical Mycobacterium tuberculosis isolates by using the microplate Alamar Blue assay. J Clin Microbiol, 1998, 36: 362–6. |
| 17. | Li G, Lian LL, Wan L, et al. Antimicrobial Susceptibility of Standard Strains of Nontuberculous Mycobacteria by Microplate Alamar Blue Assay. Plos One, 2013, 8: e84065. doi:10.1371/journal.pone.0084065 |
| 18. | Agnieszka Broda, Heather Jebbari, Kate Beaton, et al. Comparative Drug Resistance of Mycobacterium abscessus and M. chelonae Isolates from Patients with and without Cystic Fibrosis in the United Kingdom. J Clin Microbiol, 2012, 51: 217–23. |
| 19. | Van Ingen Jakko, Tridia Van Der Laan, Dekhuijzen Richard, et al. In vitro drug susceptibility of 2275 clinical non-tuberculous Mycobacterium isolates of 49 species in The Netherlands. Int J Antimicrob Ag, 2009, 35: 169–73. |
| 20. | Telenti A, Marchesi F, Balz M, et al. Rapid identification of mycobacteria to the species level by polymerase chain reaction and restriction enzyme analysis. J Clin Microbiol, 1993, 31: 175–8. |
| 21. | Bodle Ethan E, Cunningham Jennifer A, Phyllis Della Latta, et al. Epidemiology of Nontuberculous Mycobacteria in Patients without HIV Infection, New York City. Emerg Infect Dis, 2008, 14: 390–6. doi:10.3201/eid1403.061143 |
| 22. | Simons Sami, Van Ingen Jakko, Hsueh Po-Ren, et al. Nontuberculous mycobacteria in respiratory tract infections, eastern Asia. Emerg Infect Dis, 2011, 17: 343–9. doi:10.3201/eid170310060 |
 2017, Vol. 30
2017, Vol. 30


