A retrospective surveillance study on enterovirus D68 was performed in Beijing, China, following the largest and most widespread EV-D68 infection, which occurred in the USA. From January 2011 to July 2015, EV-D68 was identified in 12 individuals with respiratory infections in Beijing, China. The phylogenetic relationships based on the genomic sequence alignment showed that there were two lineages circulating in Beijing from 2011 to 2015. Eight EV-D68 strains belonged to group 1 and four belonged to group 3. All EV-D68 strains from Beijing in 2014 were separately clustered into subgroup Ⅱ of group 1. Based on these results, we concluded that the Beijing EV-D68 strains had little association with the EV-D68 strains circulating in the 2014 USA outbreak.
Enterovirus D68 (EV-D68) was first identified from cases of bronchiolitis and pneumonia in California in 1962[1]. It can cause a mild to severe respiratory infection. More recently, EV-D68 associated with respiratory infections has been reported worldwide[2-5]. The ongoing 2014 US outbreak is the largest and most widespread EV-D68 outbreak to date[6]. As of January 8, 2015, 1153 people in 46 states and the District of Columbia had come down with a respiratory illness caused by EV-D68, and 13 children died from EV-D68 infection. Several genomes of EV-D68 strains have been obtained during the current USA outbreak[7]. EV-D68 genomes from other geographic regions, including Japan, France, and New Zealand, have also been generated and deposited in the GenBank database maintained by NCBI. Currently, two EV-D68 genomes from China are available in GenBank, one (accession number: KF726085) from 2008 and the other (accession number: KP240936) from our previous study in 2014[8].
In Beijing, the Respiratory Virus Surveillance System (RVSS) was established by the Beijing Center for Disease Prevention and Control in 2011. The RVSS now covers 30 sentinel hospitals distributed in all 16 districts of Beijing. Clinical specimens were collected from respiratory infection cases for laboratory diagnostic testing. To better understand the EV-D68-associated respiratory infections in Beijing, we performed a retrospective surveillance study from January 5, 2011, to July 30, 2015, in the RVSS. A total of 7945 clinical specimens from the RVSS were chosen for EV-D68 detection with a commercial real-time RT-PCR kit purchased from Jiangsu Uninovo Biological Technology Company. Among the chosen specimens, 555 were enterovirus positive, and 12 were EV-D68 positive. Of the 12 EV-D68-positive cases, 11 were pediatric patients, and one was a 76-year-old man. The detection of EV-D68 in different age groups would provide useful epidemiologic data for monitoring the spread of EV-D68 in Beijing, China. The genomes of the 12 EV-D68 isolates were obtained by Sanger sequencing with seven pairs of overlapping primers and deposited in GenBank (accession numbers: KT285319, KT285320, KT280496 to KT280504, and KT306743). The lengths for the genomes, 5’-UTR, ORF, and 3’-UTR are shown in Table 1.
|
|
Table 1 The Sequence Characterizations of 12 EV-D68 Genomes |
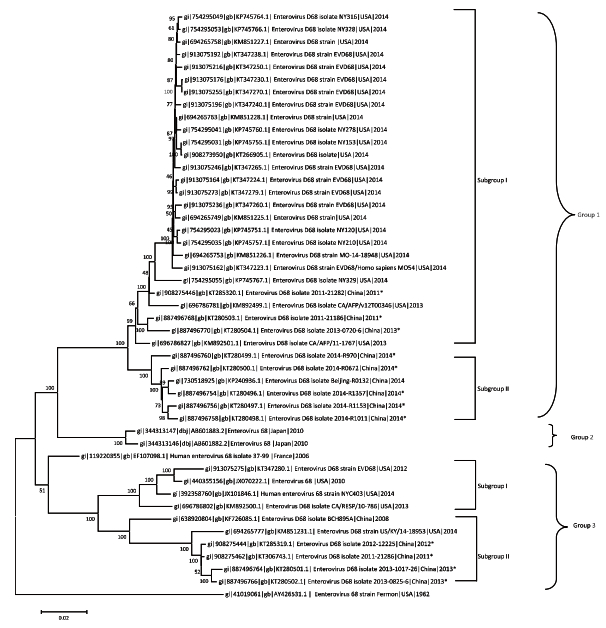
The phylogenetic relationships of the 12 EV-D68 isolates from China with 36 representative EV-D68 genomes (from the USA, China, Japan, France, and New Zealand) were estimated by the maximum-likelihood method using MEGA 6.0 software with the branch support estimated using 1000 bootstrap replicates. The results are shown in Figure 1. The EV-D68 strains segregated into three distinct clusters (groups 1-3), and eight of the 12 EV-D68 strains from Beijing belonged to group 1, whereas the remaining four strains belonged to group 3. Most of the representative EV-D68 strains circulating in the USA in 2014 were clustered into subgroup Ⅰ of group 1. The EV-D68 strains from 2011-2013 from Beijing, China, distributed to subgroup Ⅰ of group 1 or subgroup Ⅱ group 3. Of note, all EV-D68 strains from Beijing in 2014, including one EV-D68 strain from our previous study, were separately clustered into subgroup Ⅱ of group 1. Based on these results, we concluded that the Beijing strains had little association with the EV-D68 strains circulating in the 2014 USA outbreak.
|
Download:
|
| Figure 1 Phylogenetic relationships of 12 EV-D68 strains from Beijing with other strains sampled from different geographic locations. The phylogenetic relationships of the representative EV-D68 genomes in GenBank were estimated using the maximum-likelihood method with 1000 bootstrap replicates using MEGA 6.0 software (http://www.megasoftware.net). Bootstrap support values are indicated at the node of each cluster. The scale bar represents nucleotide substitutions per site. The 12 EV-D68 strains from this study are indicated with an asterisk. The GenBank accession number and the country of origin are shown for each EV-D68 strain. | |
The 5’-UTR sequences of the Beijing EV-D68 strains were aligned with the prototype and the USA strain circulating in 2014. In contrast to the prototype, all of the EV-D68 strains had the ‘CCTCAAAACCTCCAGTACATAAC’ sequence deletion corresponding to nt 682 to 704 of the prototype genome. Eight of the Beijing EV-D68 strains clustered in group 1 had the ‘TTATTTATAACA’ sequence deletion corresponding to nt 721 to 732 of the prototype genome. The others in group 3 did not have this sequence deletion. ‘TTATTTATAACA’ sequence deletions were also present in most of the USA strains circulating in 2014. The 5’-UTR of enterovirus contains an internal ribosome entry site that is associated with the translational efficiency and virulence of the enterovirus[9-10]. The sequence deletions in the Beijing strains in 2014 and most of the USA strains circulating in 2014 may have played an important role in the spread of these strains worldwide. Moreover, a 3-nt deletion (AAT) in the polyprotein gene of the Beijing EV-D68 strains in group 3 was identified corresponding to nt 2794-2796 of the prototype. The same sequence deletion was not present in the Beijing EV-D68 strains in group 1.
The results above indicated that there were sporadic respiratory infection cases caused by the EV-D68 infections during the last 4 years in Beijing, China. The Beijing EV-D68 strains had little association with the EV-D68 strains circulating in the 2014 USA outbreak according to the results of phylogenetic analysis.
We thank sentinel hospitals of RVSS in Beijing for collecting samples and investigating cases. We acknowledge Dr. Jean Lutamyo Mbisa and Kevin E. Brown of Public Health England for their reading throughout the original manuscript.
| 1. | Schieble JH, Fox VL, Lennette EH. A probable new human picornavirus associated with respiratory diseases[J]. Am J Epidemiol , 1967, 85 :297–310. |
| 2. | Kaida A, Kubo H, Sekiguchi J, et al. Enterovirus 68 in children with acute respiratory tract infections, Osaka, Japan[J]. Emerg Infect Dis , 2011, 17 :1494–7. |
| 3. | Rahamat-Langendoen J, Riezebos-Brilman A, Borger R, et al. Upsurge of human enterovirus 68 infections in patients with severe respiratory tract infections[J]. J Clin Virol , 2011, 52 :103–6. doi:10.1016/j.jcv.2011.06.019 |
| 4. | Meijer A, Benschop KS, Donker GA, et al. Continued seasonal circulation of enterovirus D68 in the Netherlands, 2011-2014[J]. Euro Surveill , 2014, 19 :1–6. |
| 5. | Piralla A, Girello A, Grignani M, et al. Phylogenetic characterization of enterovirus 68 strains in patients with respiratory syndromes in Italy[J]. J Med Virol , 2014, 86 :1590–3. doi:10.1002/jmv.v86.9 |
| 6. | Midgley CM, Jackson MA, Selvarangan R, et al. Severe respiratory illness associated with enterovirus d68-Missouri and Illinois, 2014. MMWR Morb[J]. Mortal. Wkly Rep , 2014, 63 :798–9. |
| 7. | Brown BA, Nix WA, Sheth M, et al. Seven Strains of Enterovirus D68 Detected in the United States during the 2014 Severe Respiratory Disease Outbreak[J]. Genome Announc , 2014, 2 :e01201–14. |
| 8. | Zhang T, Ren L, Luo M, et al. Enterovirus D68-associated severe pneumonia, China, 2014[J]. Emerg Infect Dis , 2015, 21 :916–8. doi:10.3201/eid2105.150036 |
| 9. | Martínez-Salas E, Pacheco A, Serrano P, et al. New insights into internal ribosome entry site elements relevant for viral gene expression[J]. J Gen Virol , 2008, 89 :611–26. doi:10.1099/vir.0.83426-0 |
| 10. | De Jesus N, Franco D, Paul A, et al. Mutation of a single conserved nucleotide between the cloverleaf and internal ribosome entry site attenuates poliovirus neurovirulence[J]. J Virol , 2005, 79 :14235–43. doi:10.1128/JVI.79.22.14235-14243.2005 |
 2016, Vol. 29
2016, Vol. 29


