2. Guizhou Provincial Center for Disease Control and Prevention, Guiyang 550004, Guizhou, China
Methods The PM was dissected from the midgut of the third instar larvae, and protein extracted from the PM was evaluated using SDS-PAGE. A 1D-PAGE lane containing all protein bands was cut from top to bottom, the proteins in-gel trypsinised and analysed via shotgun liquid chromatography-tandem mass spectrometry (LC-MS/MS).
Results In total, 374 proteins, with molecular weights varying from 8.225 kD to 996.065 kD and isoelectric points ranging from 3.83 to 11.24 were successfully identified, most identified proteins were mainly related to immunity, digestion, nutrient metabolism and PM structure. Furthermore, many of these proteins were functionally associated with pattern binding, polysaccharide binding, structural constituent of peritrophic membrane and chitin binding, according to Gene Ontology annotation.
Conclusion The PM protein composition, which provides a basis for further functional investigations of the identified proteins, will be useful for understanding the housefly larval gut immune system and may help to identify potential targets and exploit new bioinsecticides.
In many insects,the midgut epithelium is generally lined with an extracellular,semi-permeable structure referred to as the peritrophic matrix (PM),PM and mucous secretions are somewhat similar,but there are big differences too. The PM is essential for insect digestive physiology,as it protects the midgut epithelium from abrasion by food particles and toxins,serves as the first biophysical barrier that alters the temporal kinetics of host immune responses to pathogens ingested during feeding,and increases digestive efficiency by compartmentalizing of digestive processes[1, 2, 3, 4, 5, 6]. The insect PM is composed of chitin and glycoproteins,exhibiting characteristic chitin-binding activity[7]. PM proteins are classified into four categories on the basis of extractability under different conditions,and most of these proteins play significant roles in the functions of the PM[7]. Studies of the red flour beetle Tribolium castaneum have demonstrated that specific individual PM proteins may regulate PM permeability,and that a gradient of PM barrier function is essential for survival[8]. The PM not only plays important roles in facilitating food digestion and protecting the gut epithelium,but also may act as a significant structural target for insect control[9, 10].
The housefly (Musca domestica L.; Diptera: Muscidae) is a major domestic,medical,and veterinary pest that causes more than 100 human and animal diseases,including bacterial,protozoan,helminthic,viral,and rickettsial infections[11, 12]. Although insecticides have been widely used to control insect pests,the housefly has shown a remarkable and rapid ability to evolve resistance[13, 14]. Therefore,effective novel strategies of housefly control are vital for limiting the spread of disease,the evolution of resistance,and the economic losses associated with reduced production[15]. Because bacteria are both nutritional and developmental requirement of housefly larvae[16],adult flies associate with microbe-rich animal waste or septic substrates throughout their life cycle[17],but rarely show signs of disease,indicating the presence of efficient systems for gut defense in conjunction with biophysical barriers against microbes[18]. Bacteria do not pass through the PM and are sequestered therein by size exclusion,in addition,some bacterial species appear to be immobilized within the PM by an unknown mechanism[19].
Housefly larvae play a vital role in ecosystems as decomposers of organic waste[14]. Adaptation to different ecological environments may have led to the evolution of a housefly defense response. Therefore,a comparison of the PM proteome from Musca domestica with those from species exposed to different niches and microorganisms,such as Anopheles and Drosophila, may bevery significant,and will provide insights into housefly survival maintenance while in close contact with many pathogens[12, 20]. The housefly niches is unique relative to that of other insects (e.g.,Anopheles gambiae[21],Helicoverpa armigera[22] and Glossina morsitans morsitans[23],Bombyx mori[9, 24]),for which the PM composition have been reported. Increasing evidence suggests that the housefly gut is a primary site of pathogen replication after oral infection[25, 26]. Therefore,the importance of the PM as a crucial component of the local intestinal immune system merits further research. However,few studies have investigated the housefly’ PM,and therefore,the available information with which to understand its biological function is very limited. For a better understanding of how the PM performs these functions,a thorough revision of the molecular architecture of the housefly PM is required.
In this study,we attempted to achieve a comprehensive identification of proteins in the PM of the housefly,Musca domestica.
MATERIALS AND METHODS Housefly Larval Rearing Conditions and Isolation of Larval PMHouse flies were reared at the Department of Parasitology,Guizhou Medical University (Guiyang,China)[27]. Larvae were raised in a climate-controlled room at 25 °C with a relative humidity of 75%-85% and were provided,medium comprising wheat bran (500 g),heat-inactivated yeast (30 g),and water (1500 mL) until pupation. After eclosion,adult flies were fed water,sugar,and milk powder. Flies were maintained at 25 °C under a 12 h light /12 h dark cycle (LD12:12).
The PM was isolated from the midgut of the third instar larvae. The larval midgut was dissected,and the PM allowed slid to out when the end of the midgut was cut transversely,the PM was subsequently washed with a 0.75% NaCl solution until no food debris remained. Approximately 200 PMs were pooled and stored at -80 °C until further use.
Sample Preparation and Gel ElectrophoresisThe PM was homogenized on ice for 5 min,and total proteins were extracted with 20 μL of lysis buffer (2.5% SDS,10% glycerin,5% β-mercaptoethanol,and 50 mmol/L Tris-HCl,pH 8.8) per mg of sample weight[24]. PMs were disrupted after incubation with the lysis buffer,mixed several times during a 1 h incubation at 4 °C,and centrifuged at 13,000 rpm and 4 °C for 20 min,after which the supernatant was recovered and recentrifuged. Protein concentrations were measured according to the Bradford method[28]. Each sample was subsequently boiled for 10 min and centrifuged at 13,000 rpm and 4 °C for 10 min,after which the proteins in the supernatant were separated by one-dimensional sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS- PAGE) with a 5% stacking gel and 12% resolving gel,according to the standard method. The gels were then stained with Coomassie Brilliant Blue R250 (Sigma-Aldrich,St. Louis,MO,USA).
In-gel DigestionA 1D-PAGE lane containing all protein bands was manually cut from top to bottom,excised gel slices were washed twice with ultrapure water and destained in 25 mmol/L ammonium bicarbonate/ 50% acetonitrile (pH 8.0) at room temperature for 30 min. Gel slices were subsequently dehydrated for 30 min in 50% acetonitrile and 100% acetonitrile,respectively. The acetonitrile was then removed and the gel slices were reduced for 1 h at 37 °C with 1 mmol/L dithiothreitol in 25 mmol/L ammonium bicarbonate,followed by alkylation with 50 mmol/L iodoacetamide in 25 mmol/L ammonium bicarbonate for 30 min at room temperature. Subsequently,the gel slices were washed for 10 min in 25 mmol/L ammonium bicarbonate and dehydrated again for 30 min in 50% and 100% acetonitrile,respectively. Thereafter,the 100% acetonitrile was removed,and the gels were rehydrated in 10 µL of digest solution [0.02 μg/μL trypsin in cover solution (25 mmol/L ammonium bicarbonate and 10% acetonitrile)] for 30 min,after which 20 µL of cover solution were subsequently added for a 16 h digestion step at 37 °C; the supernatants were then transferred into fresh tubes,and the gels were subjected to a single extraction step with 50 µL of extraction buffer (5% TFA and 67% acetonitrile) at 37 °C for 30 min. The peptide extracts and supernatants from the gel slices were combined and completely dried for liquid chromatography- tandem mass spectrometry (LC-MS/MS).
LC-MS/MS Analysis and Data SearchSamples were re-suspended in Nano-RPLC buffer A (0.1% formic acid,2% acetonitrile). Online nano-RPLC was conducted on an eksigent nano-LC-Ultra™ 2D system (AB SCIEX,Framingham,MA,USA). The samples were loaded on a C18 nano-LC trap column (100 µm × 3 cm,C18,3 µm,150 Å) and washed with Nano-RPLC buffer A at a rate of 2 μL/min for 10 min. An elution gradient of 5%-35% acetonitrile (0.1% formic acid) over a 90 min gradient period was used on an analytical ChromXP C18 column (75 µm × 15 cm,C18,3 µm,120 Å) with a spray tip. Data acquisition was performed with a triple time-of-flight (TOF) 5600 system (AB SCIEX) fitted with a nanospray III source (AB SCIEX),with a pulled quartz tip as the emitter (New Objective,Woburn,MA,USA). Data were acquired using an ion spray voltage of 2.5 kV,curtain gas of 30 PSI,nebulizer gas of 5 PSI,and an interface heater temperature of 150 °C. For information dependent acquisition (IDA),survey scans were acquired over a 250 ms period,and as many as 35 product ion scans were collected if they exceeded a threshold of 150 counts per second (counts/s) with a 2+ to 5+ charge-state. The total cycle time was fixed at 2.5 s. A rolling collision energy setting was applied to all precursor ions for collision-induced dissociation (CID). Dynamic exclusion was set for ½ half of the peak width (18 s),and the precursor was accordingly refreshed from the exclusion list.
Based on the combined MS and MS/MS spectra,proteins were successfully identified at a 95% or higher confidence interval,using their scores in the MASCOT V2.3 search engine (Matrix Science Ltd.,London,UK),and the following search parameters: Musca domestica database,trypsin as the digestion enzyme,two missed cleavage sites,fixed modifications of Carbamidomethyl (C),partial modifications of Acetyl (Protein N-term),Deamidated (NQ),Dioxidation (W),Oxidation (M) Phospho (ST),and Phospho (Y),±15 ppm precursor ion tolerance,and ±0.15 Da fragment ion tolerance.
Bioinformatics AnalysisThe gene ontology (GO) analysis used different mapping steps to link all blast hits to the functional information stored in the GO database with the Database for Annotation,Visualization and Integrated Discovery (DAVID 6.7) toolkit[29]. Public resources such as the NCBI,PIR,and databases used to create links between protein IDs and corresponding GO information. All annotations were associated with evidence code that provided information about the quality of this functional assignment.
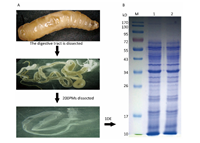
RESULTS Proteomic Analysis of the PMTo understand the protein composition of the Musca domestica PM and comprehensively analyze its potential function,proteins extracted from the PM of actively feeding third instar larvae were separated by SDS-PAGE as shown in Figure 1. A slice cut from the gel was analyzed using Shotgun LC-MS/MS after tryptic in-gel digestion. A total of 374 proteins were successfully identified in the housefly PM dataset,and detailed information about these proteins were listed in the supplementary Table S1(www.besjournal.com for details).
|
Download:
|
| Figure 1. Experimental flow diagram. (A) Dissected peritrophic matrix (PM) from Musca domestica larvae. (B) Proteins from the PMs were separated by one-dimensional electrophoresis SDS-PAGE. M: marker. 1-2: proteins extracted from housefly PM. | |
|
|
supplementary Table S1. The Proteins Identified from the Housefly Larval PM |
The molecular weight (MW) and isoelectric point (pI) distributions of the identified proteins were shown in Figure 2. The MWs mostly ranged from 8.225 kD to 996.065 kD,with an outlier of 2,265.385 kD (XP_005182933.1). Among the identified proteins,63.37% (237/374) were smaller than 100 kD. The acidic and basic proteins identified by LC-MS/MS had pIs ranging from 3.83 to 11.24,and 56.95% (213/374) of proteins were within the range of pI 4.00-7.00. The results can be further summarized as follows: 18 proteins were hypothetical proteins of unknown function,and 71 proteins had predicted signal peptides. There are 8 predicted proteins with chitin-binding domains (CBDs),belonging to the carbohydrate-binding module,CBM14 family (ChtBD2 family or peritrophin A-type),which contain 6 characteristically spaced cysteine residues[7]. Not surprisingly,extracellular matrix associated proteins,serine proteases,trypsin,carboxypeptidase,and proteins of uncharacterized or unknown function were relatively more abundant than proteins involved in digestion and energy metabolism.

|
Download:
|
| Figure 2.Theoretical two dimensional distribution of the identified proteins. (A) MW distribution. (B) pI distribution. | |
The GO analysis was performed with different mapping steps to link all blast hits to the functional information stored in the GO database with the DAVID toolkit. Public resources such as the NCBI,PIR and GO databases were used to create links with proteins ID and corresponding gene ontology information. All annotations were associated with evidence codes that provided information about the quality of the functional assignments. Out of the 374 queried proteins,318 homologous with D. melanogaster were identified. Of these,257 were mapped into the DAVID database,the rest have not yet been annotated in the DAVID database. These proteins with GO annotations were classified into the following three functional categories: biological process,cell component and molecular function.
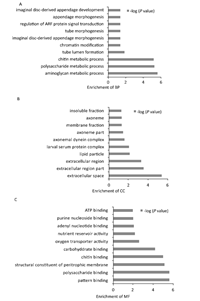
In this dataset,44 biological processes were enriched. Eighteen terms had P<0.05,the top 10 enriched processes are shown in Figure 3A,this demonstrates that most queried proteins were involved in biological processes related to metabolic processes,such as aminoglycan,polysaccharide and chitin metabolism. Fifteen different cellular component terms are enriched. Of these,9 had P<0.05,the top 10 terms are shown in Figure 3B. Most queried proteins were located in the extracellular space and membrane. Additionally,30 different molecular functional groups were enriched. Of these,22 had P<0.05. The top 10 enriched molecular functions are shown in Figure 3C,the main function groups represented among the queried proteins were binding proteins involved in various metabolic processes. GO enrichment revealed that the queried proteins were predominately binding proteins,located in the membrane and extracellular space and involved in biological processes related to various metabolic processes.
|
Download:
|
| Figure 3. GO analysis of the functional categories. (A) Distribution of enriched biological processes (BP). (B) Distribution of cell component (CC) enrichment. (C) Distribution of molecular function (MF) enrichment. | |
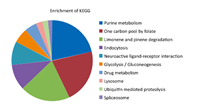
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment was performed by comparing annotated enzymes from the query dataset against the KEGG database. As shown in Figure 4,the pathways were classified into the following ten categories: purine metabolism,one carbon pool by folate,limonene and pinene degradation,endocytosis,neuroactive ligand- receptor interaction,glycolysis/gluconeogenesis,drug metabolism,lysosome,ubiquitin mediated proteolysis,spliceosome. Pathway revealed that the queried proteins were mainly involved in purine metabolic pathways. However,only 15.2% (39/257) of the queried data were enriched in the KEGG pathway database,indicating the presence of huge knowledge gaps.
|
Download:
|
| Figure 4. KEGG pathway enrichment. | |
This study provides the advanced comprehensive analysis of housefly larval PM proteins using LC-MS/MS. Herein,we successfully identified 374 proteins,a higher number than that previously reported in other insect studies. Given the relatively simple composition of the PM (estimated approximately 305 proteins[9]),this large number of identified proteins was initially surprising. We attribute the large number to two reasons: 1) the houseflies are polyphagous insects that consume large amounts of septic substrates and have a high growth rate,and 2) the sensitivity of the mass spectrometry techniques. In this study,we focused on these identified proteins were involved in PM structure,immunity,and nutrients metabolism,which could imply the different functions of the PM in the gut.
PM Proteins with Chitin-binding DomainsPeritrophins are structural PM proteins with one or more characteristic chitin-binding domains (CBDs) defined by the consensus CX15-17CX5-6CX9CX12 CX6-7C where X is any amino acid other than cysteine[7]. In this study,a total of eight peritrophins were identified via mass spectrometry analysis (Table 1). Peritrophin-44 and peritrophin-48 are known to be the most abundant integral PM proteins and are secreted during PM formation. Among them peritrophin-44 is the first characterization of a PM protein from Lucilia cuprina. Peritrophin-44 could be binding to chitin fibrils in the PM,consequently cross-linking and locking the chitin fibrils together while concurrently dictating the PM permeability characteristics[30],this feature could have vital implications for the nature of the digestive process in the housefly gut by partitioning the ingested proteins and digestive enzymes between the ecto- and endo-PM spaces. Chondroitin proteoglycan-2-like protein (gi|557772194|) is a 748 amino acid long peritrophin with a conserved insect intestinal mucin flanked by eight Peritrophin A Domains (PAD) and a predicted signal peptide sequ- ence between residues 21/22. The PADs are typically 43-53 residues in length and contain the consensus CX11-21CX5CX9-19CX10-14CX4-16C. PADs are ubiquitous among insects and are the most common chitin-binding domains in dipteran larvae[23]. This type of domain is abundant in invertebrate mucins and can also be found in other PM-associated proteins such as cuticular proteins,and chitinases[31],as these proteins are also associated with chitin,although so far no experimental evidence has proved this. Another protein identified and involved in immunity was peritrophin-1-like protein (gi|557773941|),homologous with peritrophin-like of Eriocheir sinensis,which might indicate that this protein has a similar anti-bacterial innate immune function as that observed in the Chinese mitten crab[32]. The mammalian mucins were likely to have a number of functions,i.e.,lubrication of the passage of food through the gut,protection of epithelial cells from digestive proteases,and protection from invasion by bacteria[7]. Invertebrate mucins had many of the molecular features of mammalian mucins. Peritrophin-55 (gi|755880471|),a mucin-like glycoprotein with one CBD domain was identified. A predicted signal peptide between residues 19/20 suggests that peritrophin-55 is secreted into the PM after synthesis. An ortholog of Lucilia cuprina peritrophin-55 which had been reported to function as probably lubricates the surface of the PM and protects the midgut from invasion by bacteria[33]. The current analysis also identified a novel 158-amino-acid protein (gi|557772100|) with a predicted molecular weight of 17.3 kD,including an 18 amino acid signal peptide,and 2 conserved chitin binding type 2 domains (ChtBD2) characterized by having a six cysteine motif. A chitin-binding protein,which had ChtBD2 from Penaeus monodon,and which were homologous to insect peritrophin,was demonstrated to be involved in white spot syndrome virus (WSSV) infection[34]. These results suggested that gi|557772100| proteins may play a role in immune defense in housefly. We hypothesized that these peritrophin proteins might main participate in formation of the peritrophic matrix. Further characterization of these novel peritrophin proteins should provide additional insights into the molecular function of the housefly larval PM.
|
|
Table 1. Putative Peritrophic Matrix Proteins Identified by Mass Spectrometry with Predicted Chitin-binding Domains |
During the third instar larval stage,the housefly digests large amounts of decaying organic substrates to obtain nutrition and energy for the pupal and adult stages. At this stage,the epithelial cells synthesize and secrete many enzymes for food digestion through the larval PM into the midgut or bounding to the PM. In the present study,the majority of the identified proteins were hydrolytic enzymes,including glycosyl hydrolase,alpha-amylase,laccase-1,and digestive enzymes such as trypsin,serine proteases,cysteine proteases,and carboxypeptidase. PMs may accelerate digestion in housefly larvae via PM-bound digestive enzymes. In Spodoptera frugiperda,trypsin was found to be partly processed into a soluble form and partly bound to vesicle membranes incorporated into the PM[35]. Glycosyl hydrolase was reported to collaboratively digest host-symbiont lignocellulose in the lower gut of the termite Reticulitermes flavipes[36],and may have a similar function in housefly larvae. Alpha amylase is widely distributed among plant,animal,and microbial species and significantly catalyzes the hydrolysis of starch (amylose and amylopectin),resulting in the release of maltose[37]. Laccase,a multicopper oxidase,is present in bacteria,fungi,plants,and insects[38]. Previous results have suggested that the laccase-1 may play an important role in the detoxification of phenolic compounds or the metabolism of copper or iron in the larval diet[39].
Serine proteases (SPs) are considered important proteolytic enzymes[40]. In the housefly larval gut,SPs comprise one of the most important groups of digestive enzymes. In this study,10 putative serine proteases were identified,9 of which had been described according to MEROPS database (MER536669,MER536852,MER536854,MER537154,MER536853,MER537153,MER536615,MER536616,MER536866). These proteins not only facilitate digestion,but also play important roles in the insect innate immune systems[41]. Recently,Bactrocera dorsalis SPs were reported to play an indirect role in relieving insecticide toxicity stress[42]. Cysteine proteases have been characterized as major digestive enzymes in many coleopteran species such as the cowpea bruchid (Callosobruchus maculatus Fabricius) and the western corn rootworm (Diabrotica virgifera virgifera LeConte)[43, 44]. Carboxypeptidase is typically expressed in the midgut[45] and is often associated with the lepidopteran PM[46]. In future work,we will investigate the functions of these digestion related proteins in the housefly larval gut.
Proteins Related to Housefly Larval Innate ImmunitySeveral identified proteins were related to the immune response. Musca domestica antifungal peptide-1 was first isolated from the hemolymph of housefly larvae and has been confirmed as a novel insect antifungal peptide[27]. SPs comprise a large group of digestive enzymes in the larval gut that participate in the innate immune response in insects[41]. This study identified lysozyme,which can hydrolyze bacteria,and it plays an important role in insect digestion and innate immunity[47]. Croquemort-like protein is a member of the CD36 superfamily,one major function of this protein is scavenger receptor activity,in which molecular patterns presented by bacteria,viruses,and pathogen-infected cells are recognized[48]. As a PM component,croquemort-like protein may have multifunctional roles ranging from homoeostasis maintenance to immune system mediation and is potentially involved in primary pathogen clearance[23]. Peptidoglycan recognition proteins (PGRPs) are key regulators of the insect innate antibacterial response. In this study,PGRP-SC2 and PGRP-LA were observed in the PM. In Drosophila, an ortholog of PGRP-SC2 is responsible for regulating the larval gut to prevent immune deficiency (IMD) signaling pathway activation following bacterial ingestion[49]. Recently,Drosophila PGRP-LA has been reported to positively regulate the IMD signaling pathway in barrier epithelial tissues[50]. The identification of these proteins in the present study suggested that PM may play a vital role in housefly larval gut immunity via signal transduction in response to bacterial activation.
Additionally,we obtained a rather large number of proteins with unknown functions,which should not be neglected because they might make important contributions to the molecular architecture of the housefly PM. In conclusion,the PM proteins provide a foundation for further investigations of biochemical functions in the PM of Musca domestica. The determination of protein expression profiles in the housefly PM may accelerate the identification of molecular targets that could be used to explore novel and environmentally benign control strategies. Regarding further work,the physiological function of these identified proteins should be studied,as this will facilitate an understanding of the housefly PM formation mechanism and its role in gut local immune responses.
Author ContributionsConceived and designed the experiments: WANG Yu,XIU Jiang Fan,and WU Jian Wei. Performed the experiments: WANG Yu,LUO Man,SHANG Xiao Li,and WANG Tao. Analyzed the data: CHENG Jin Zhi and ZHAO Peng. Wrote the paper: WANG Yu.
COMPETING INTERESTSThe authors declare that they have no competing interests.
| 1. | Lehane MJ. Peritrophic matrix structure and function. AnnuRev Entomol,1997; 42,525-50. |
| 2. | Terra WR. The origin and functions of the insect peritrophic membrane and peritrophic gel. Arch Insect Biochem Physiol,2001; 47,47-61. |
| 3. | Hegedus D,Erlandson M,Gillott C,et al. New insights into peritrophic matrix synthesis,architecture,and function. AnnuRev Entomol,2009; 54,285-302. |
| 4. | Kuraishi T,Binggeli O,Opota O,et al. Genetic evidence for a protective role of the peritrophic matrix against intestinal bacterial infection in Drosophila melanogaster. Proc Natl Acad Sci USA,2011; 108,15966-71. |
| 5. | Weiss BL,Savage AF,Griffith BC,et al. The peritrophic matrix mediates differential infection outcomes in the tsetse fly gut following challenge with commensal,pathogenic,and parasitic microbes. J Immunol,2014; 193,773-82. |
| 6. | Yin J,Yang S,Li KB,et al. Identification and molecular characterization of a chitin-binding protein from the Beet Webworm,Loxostege sticticalis L. Int J Mol Sci,2014; 15,19147-61. |
| 7. | Tellam RL,Wijffels G,Willadsen P. Peritrophic matrix proteins. Insect Biochem Mol Biol,1999; 29,87-101. |
| 8. | Agrawal S,Kelkenberg M,Begum K,et al. Two essential peritrophic matrix proteins mediate matrix barrier functions in the insect midgut. Insect Biochem Mol Biol,2014; 49,24-34. |
| 9. | Zhong XW,Zhang LP,Zou Y,et al. Shotgun analysis on the peritrophic membrane of the silkworm Bombyx mori. BMB Rep,2012; 45,665-70. |
| 10. | Wang P,Granados RR. Molecular Structure of the Peritrophic Membrane (PM): Identification of potential PM target sites for insect control. Arch Insect Biochem Physiol,2001; 47,110-8. |
| 11. | Keiding J. The Housefly: Biology and Control; [Training and Information Guide]. Geneva: World Health Organization,Vector Biology and Control Division,1986. |
| 12. | Scott JG,Liu N,Kristensen M,et al. A case for sequencing the genome of Musca domestica (Diptera: Muscidae). J Med Entomol,2009; 46,175-82. |
| 13. | Li M,Reid WR,Zhang L,et al. A whole transcriptomal linkage analysis of gene co-regulation in insecticide resistant house flies,Musca domestica. BMC Genomics,2013; 14,803. |
| 14. | Scott JG,Warren WC,Beukeboom LW,et al. Genome of the housefly,Musca domestica L.,a global vector of diseases with adaptations to a septic environment. Genome Biol,2014; 15,466. |
| 15. | Højland DH,Jensen KMV,Kristensen M. Adaptation of Musca domestica L. field population to laboratory breeding causes transcriptional alterations. PLoS ONE,2014; 9,e85965. |
| 16. | Zurek L,Schal C,Watson DW. Diversity and contribution of the intestinal bacterial community to the development of Musca domestica (Diptera: Muscidae) larvae. J Med Entomol,2000; 37,924-8. |
| 17. | Nayduch D,Cho H,Joyner C. Staphylococcus aureus in the housefly: temporospatial fate of bacteria and expression of the antimicrobial peptide defensin. J Med Entomol,2013; 50,171-8. |
| 18. | Fleming A,Kumar HV,Joyner C,et al. Temporospatial fate of bacteria and immune effector expression in house flies fed GFP-Escherichia coli O157:H7. Med Vet Entomol,2014; 28,364-71. |
| 19. | McGaughey J,Nayduch D. Temporal and spatial fate of GFP-expressing motile and nonmotile aeromonas hydrophila in the housefly digestive tract. J Med Entomol,2009; 46,123-30. |
| 20. | Tang T,Li X,Yang X,et al. Transcriptional response of Musca domestica larvae to bacterial infection. PLoS ONE,2014; 9,e104867. |
| 21. | Dinglasan RR,Devenport M,Florens L,et al. The Anopheles gambiae adult midgut peritrophic matrix proteome. Insect Biochem Mol Biol,2009; 39,125-34. |
| 22. | Campbell PM,Cao AT,Hines ER,et al. Proteomic analysis of the peritrophic matrix from the gut of the caterpillar,Helicoverpa armigera. Insect Biochem Mol Biol,2008; 38,950-8. |
| 23. | Rose C,Belmonte R,Armstrong SD,et al. An investigation into the protein composition of the Teneral Glossina morsitans morsitans peritrophic matrix. PLoS Negl Trop Dis,2014; 8,e2691. |
| 24. | Hu XL,Chen L,Xiang XW,et al. Proteomic analysis of peritrophic membrane (PM) from the midgut of fifth-instar larvae,Bombyx mori. Mol Biol Rep,2012; 39,3427-34. |
| 25. | Wasala L,Talley JL,Desilva U,et al. Transfer of Escherichia coli O157:H7 to spinach by house flies,Musca domestica (Diptera: Muscidae). Phytopathology,2013; 103,373-80. |
| 26. | Joyner C,Mills MK,Nayduch D. Pseudomonas aeruginosa in Musca domestica L.: temporospatial examination of bacteria population dynamics and housefly antimicrobial responses. PLoS ONE,2013; 8,e79224. |
| 27. | Fu P,Wu JW,Guo G. Purification and molecular identification of an antifungal peptide from the hemolymph of Musca domestica (housefly). Cell Mol Immunol,2009; 6,245-51. |
| 28. | Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem,1976; 72,248-54. |
| 29. | Huang DW,Sherman BT,Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res,2009; 37,1-13. |
| 30. | Elvin CM,Vuocolo T,Pearson RD,et al. Characterization of a major peritrophic membrane protein,peritrophin-44,from the larvae of Lucilia cuprina: cDNA and deduced amino acid sequences. J Biol Chem,1996; 271,8925-35. |
| 31. | Shen Z,Jacobs LM. A type I peritrophic matrix protein from the malaria vector Anopheles gambiae binds to chitin. cloning,expression,and characterization. J Biol Chem,1998; 273,17665-70. |
| 32. | Huang Y,Ma FT,Wang W,et al. Identification and molecular characterization of a peritrophin-like gene,involved in the antibacterial response in Chinese mitten crab,Eriocheir sinensis. Dev Comp Immunol,2015; 50,129-38. |
| 33. | Tellam RL,Vuocolo T,Eisemann C,et al. Identification of an immune-protective mucin-like protein,peritrophin-55,from the peritrophic matrix of Lucilia cuprina larvae. Insect Biochem Mol Biol,2003; 33,239-52. |
| 34. | Chen KY,Hsu TC,Huang PY,et al. Penaeus monodon chitin-binding protein (PmCBP) is involved in white spot syndrome virus (WSSV) infection. Fish Shellfish Immunol,2009; 27,460-5. |
| 35. | Jordao BP,Capella AN,Terra WR,et al. Nature of the anchors of membrane bound aminopeptidase,amylase,and trypsin and secretory mechanisms in Spodoptera frugiperda (Lepidoptera) midgut cells. J Insect Physiol,1999; 45,29-37. |
| 36. | Scharf ME,Kovaleva ES,Jadhao S,et al. Functional and translational analyses of a beta-glucosidase gene (glycosyl hydrolase family 1) isolated from the gut of the lower termite Reticulitermes flavipes. Insect Biochem Mol Biol,2010; 40,611-20. |
| 37. | Ewelina C,Wojciech B,Monika B,et al. Cloning,expression,and purification of insect (Sitophilus oryzae) alpha-amylase,able to digest granular starch,in Yarrowia lipolytica host. Appl Microbiol Biotechnol,2015; 99,2727-39. |
| 38. | Mayer AM,Staples RC. Laccase: new functions for an old enzyme. Phytochemistry,2002; 60,551-65. |
| 39. | Dittmer N,Suderman R,Jiang H,et al. Characterization of cDNAs encoding putative laccase-like multicopper oxidases and developmental expression in the tobacco hornworm,Manduca sexta,and the malaria mosquito,Anopheles gambiae. Insect Biochem Mol Biol,2004; 34,29-41. |
| 40. | Wolfson JL,Murdock LL. Diversity in digestive proteinase activity among insects. J Chem Ecol,1990; 16,1089-102. |
| 41. | Gorman MJ,Paskewitz SM. Serine proteases as mediators of mosquito immune responses. Insect Biochem Mol Biol,2001; 31,257-62. |
| 42. | Hou MZ,Shen GM,Wei D,et al. Characterization of bactrocera dorsalis serine proteases and evidence for their indirect role in insecticide tolerance. Int J Mol Sci,2014; 15,3272-86. |
| 43. | Koiwa H,Shade RE,Zhu-Salzman K,et al. A plant defensive cystatin (soyacystatin) targets cathepsin L-like digestive cysteine proteinases (DvCALs) in the larval midgut of western corn rootworm (Diabrotica virgifera virgifera). FEBS Lett,2000; 471,67-70. |
| 44. | Zhu-Salzman K,Koiwa H,Salzman RA,et al. Cowpea bruchid Callosobruchus maculatus uses a three-component strategy to overcome a plant defensive cysteine protease inhibitor. Insect Mol. Biol,2003; 12,135-45. |
| 45. | Ferreira C,Rebola KG,Cardoso C,et al. Insect midgut carboxypeptidases with emphasis on S10 hemipteran and M14 lepidopteran carboxypeptidases. Insect Mol Biol,2015; 24,222-39. |
| 46. | Ferreira C,Capella AN,Sitnik R,et al. Digestive enzymes in midgut cells,endo- and ectoperitrophic contents,and peritrophic membranes of Spodoptera frugiperda (Lepidoptera) larvae. Arch Insect Biochem Physiol,1994; 26,299-313. |
| 47. | Ren Q,Zhao XF,Wang JX. Molecular characterization and expression analysis of a chicken-type lysozyme gene from housefly (Musca domestica). J Genet Genomics,2009; 36,7-16. |
| 48. | Silverstein RL,Febbraio M. CD36,a scavenger receptor involved in immunity,metabolism,angiogenesis,and behavior. Sci Signal,2009; 2,re3. |
| 49. | Bischoff V,Vignal C,Duvic B,et al. Downregulation of the Drosophila immune response by peptidoglycan recognition proteins SC1and SC2. PLoS Pathog,2006; 2,e14. |
| 50. | Gendrin M,Zaidman-Rexmy A,Broderick NA,et al. Functional analysis of PGRP-LA in Drosophila immunity. PLoS ONE,2013; 8,e69742. |
 2016, Vol.29
2016, Vol.29 


