2. 中国农科院农业资源与农业区划研究所,北京 100086;
3. 作物遗传与种质创新国 家重点实验室南京农业大学,南京 210095
2. Institute of Agricultural Resources and Regional Planning, Chinese Academy of Agricultural Sciences, Bejjing 100086;
3. State Key Laboratory of Crop Genetics and Germplasm Enhancement, Nanjing Agricultural University, Nanjing 210095
我国农业生产上由于化肥的过量施用,造成了农田环境污染和农产品安全等问题,导致土壤质量的降低,影响土壤的持续利用。根际微生物被看做植物的第二基因组,对宿主植物的营养和健康发挥着重要的作用,促进化肥减施增效和农业绿色发展。根际微生物中的植物促生菌(Plant Growth-Promoting Rhizobacteria,PGPRs)由于其根际定殖能力强、对植物具有多种益生作用,是目前微生物肥料的主要生产菌种。
PGPRs对植物的促生机制分为直接促生和间接促生两种,直接促生机制主要包括活化养分,如固氮、溶磷、解钾、活化各种中微量营养元素等[1-2]、调节植物激素水平,如分泌有益激素及降解有害激素等[3]、产生挥发性促生物质[4];间接机制一般包括抑制植物土传病害[5]、诱导植物系统抗性和诱导植物产生对非生物胁迫的系统耐性等。
PGPRs在根际的定殖能调控植物根系构型(Root System Architecture,RSA),促进侧根和根毛的发育,增强植物的生长和根际养分吸收,提高肥料利用率。PGPRs通过自身分泌的各种激素和信号物质调控植物根系发育的信号通路和分子途径,会影响根系的主根、侧根和和根毛等的发育,最终影响植物的根系形态。目前已经鉴定了一些PGPRs影响RSA的激素和信号,也初步阐释了这些物质调控植物根系发育的分子机制。但是PGPRs种类众多,对根系的影响也复杂多样,为了利用PGPRs及其代谢物质调控形成不同作物的理想根系形态,促进根际养分的吸收利用,仍需系统发掘PGPRs调控根系形态的新型信号物质,深入揭示它们的作用机制,促进PGPRs在减施增效和农业绿色发展中的应用。本文综述了目前已知的PGPRs分泌的调控RSA的激素和信号,总结了调控机制的研究进展,并对今后的研究提出展望。
1 PGPRs影响主根、侧根和根毛的生长从而调控植物的根系构型植物根系自身具有高度的可塑性,PGPRs对植物RSA的影响是多方面的。大量研究证明PGPRs对主根伸长的影响有的表现为促进,有的表现为抑制;但侧根和根毛的数量都表现为显著促进,植株根系的生物量也得到提升[6]。
部分PGPRs会抑制植物主根的生长,如假单胞菌WCS417[7]和巨大芽孢杆菌UMCV1都通过影响拟南芥根部细胞在增殖和分化之间的转换来改变其根系结构,其中假单胞菌WCS417增加了分生区细胞的分裂、而巨大芽孢杆菌UMCV1减少了分生区细胞的分裂,但这两株菌直接接种于拟南芥根部时都抑制了主根的伸长[8-11]。
大多数PGPRs都能促进植物主根的伸长,如伯克氏菌PsJN能提升拟南芥主根长度、增加根毛数量以及根生物量[12]。Ryu等[4]测定了7株产生挥发性有机物(Volatile Organic Compounds,VOCs)的PGPRs对拟南芥生长的影响,结果发现其中枯草芽孢杆菌GB03和解淀粉芽孢杆菌IN937a可以显著促进拟南芥的地上部发育,其原因可能与PGPR对植物RSA的调控紧密相关。Gutierrez-Luna等[13]从柠檬根际土壤中分离得到3株PGPRs菌株,产生的VOCs都可以改变植物的RSA。
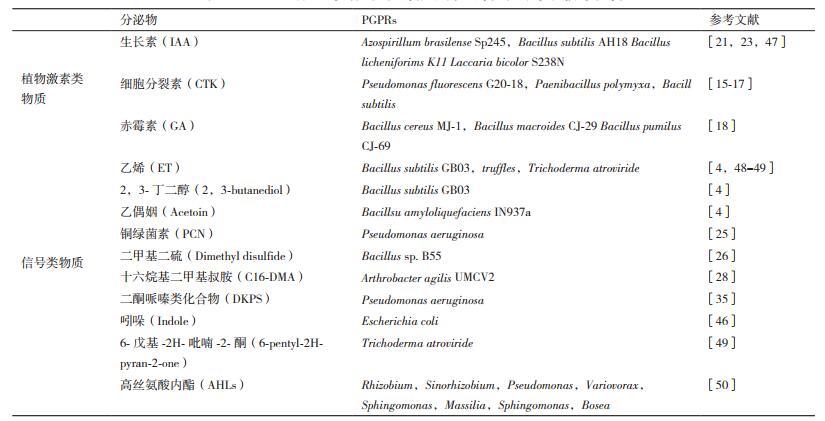
2 PGPRs通过分泌植物激素和信号物质调控植物的根系构型PGPRs主要通过分泌丰富的代谢物调控植物根系发育信号途径影响植物的RSA。这些代谢物可以分为两大类:一是植物激素类物质,包括生长素(IAA)、赤霉素(GA)、细胞分裂素(CTK)、脱落酸(ABA)、乙烯(ET)和油菜素甾醇(BR)等,这些激素可以通过激活其下游信号通路直接调控植物根系发育;第二大类是PGPRs分泌的激素之外的一些信号分子,它们通过影响植物内源激素的稳态和根系发育的过程来调控植物根系发育。
大部分PGPRs可以产生CTK[14-16],例如枯草芽孢杆菌会分泌CTK从而增加植物内源CTK的含量影响生菜根系的发育[17]。有些PGPRs能分泌GA,如蜡样芽孢杆菌MJ-1产生GA来增加红辣椒内源GA的含量,改变其根系结构[18]。PGPRs分泌的生长素吲哚乙酸(IAA)同样能诱导植物根系生长[19-22]。例如,枯草芽孢杆菌AH18和地衣芽孢杆菌K11都有分泌植物激素IAA的能力,当接种不同浓度的枯草芽孢杆菌AH18和地衣芽孢杆菌K11于黄瓜根部时,其根系生长状态存在梯度效应,黄瓜主根的>长随着接种浓度的升高而增大,当接种量过高时对主根存在抑制效果[23],证明PGPRs分泌的不同水平IAA对植物根系的影响有显著差异性,IAA的促根效用与剂量有关。微生物分泌的植物激素类物质,多数被植物根系直接吸收利用,但极少数PGPRs代谢产生的挥发气体乙烯(ET)在调控植物根系发育过程中扮演信号类物质的角色[13]。
PGPRs还能分泌一些非激素的信号物质影响植物的根系发育。PGPRs的群感信号分子高丝氨酸内酯(N-Acyl-homoserine lactones,AHLs)能够在高浓度下通过改变植物体内CTK的应答来诱导侧根的生长及抑制主根的伸长,在低浓度下通过作用于植物G蛋白信号和钙调蛋白的机制,增加分生区细胞的分裂和伸长来增加主根的延伸[24];某些PGPRs产生的另一种群感信号二酮哌嗪类化合物(DKPs)能够激活植物IAA应答基因的表达,和植物内IAA受体蛋白结合,诱导植物侧根生长[25]。PGPRs产生的挥发性有机物(VOCs)也具有调控植物根系发育的功能,如芽孢杆菌B55可以通过产生的挥发性的二甲基二硫,能够恢复乙烯不敏感突变株的根系结构[26]。枯草芽孢杆菌GB03和解淀粉芽孢杆菌IN937a分泌的VOCs类物质2,3-丁二醇和乙偶姻可能通过影响根系发育来促进植物地上部的生长[4, 27]。运动节杆菌UMCV2产生的挥发性有机物十六烷基二甲基叔胺(N,N-dimethyl hexadecylamine,C16-DMA)及有机胺类通过茉莉酸信号途径介导根系的发育[28]。这些研究证明PGPRs通过分泌多种的植物激素和信号物质调控植物根系的发育和结构(表 1)。然而,PGPRs种类众多,分泌的代谢物也复杂多样,许多调控植物根际发育的新型信号物质仍待挖掘。
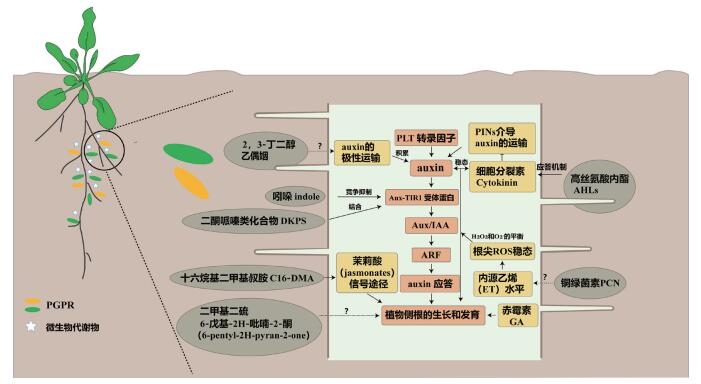
PGPRs所分泌的信号类物质对植物根系结构的影响,主要通过对内源植物激素通路的调控。植物主根的发育受胚胎时期所形成根尖干细胞的控制[27],在根系发育过程中,IAA途径下游的PLT转录因子控制IAA的浓度梯度,来维持干细胞的稳态,从而控制主根的伸长[29-31]。o在主根的“过渡区(Transition Zone)”(位于分生组织的尖端,该区域的细胞不分裂,细胞延长缓慢)[32], CTK可以抑制IAA极性运输蛋白(PINs)的表达,导致主根局部IAA浓度梯度的改变,诱导细胞的分化而影响主根的伸长[33-34]。一些PGPRs的代谢物可以直接介导植物内源IAA的转运,影响植物根系的构型。研究发现植物根际促生菌枯草芽孢杆菌GB03分泌的VOCs可以促进植物根系发育,当施用于带有IAA信号报告基因DR5:GUS标记的拟南芥时,可观察到IAA信号显著增强,但施用于eir1突变株时,其促生效果受到抑制,证明枯草芽孢杆菌GB03分泌的VOCs通过调控拟南芥内源IAA的极性运输以诱导细胞的伸长,从而影响拟南芥主根的发育[35]。铜绿假单胞菌分泌的铜绿菌素(PCN),在群感信号分子的调控下,不依赖植物IAA和CTK信号通路诱导根系构型变化,其中PCN可能协调内源ET水平,改变主根根尖部分活性氧(ROS)稳态(H2O2和O2-的平衡)而抑制植物主根生长[36]。
植物的侧根由主根成熟区的中柱鞘细胞脱分化形成,IAA在中柱鞘细胞中会诱导侧根原基的产生[37-38]。研究表明,拟南芥侧根在主根上的周期性发生受到主根靠近根尖的信号震荡区(Oscillation zone,包括过渡区和伸长区)震荡基因(Oscillating genes)的调控,震荡基因的表达上调会增强根尖信号震荡区的IAA信号,进而诱导中柱鞘细胞的分裂和侧根形成点(Pre-branch site)的形成,生长素受体TIR1和根冠的生长素合成途径可以通过维持信号震荡区的生长素信号强度来诱导侧根形成点的发生[39-40]。
在侧根形成点发育成为侧根的过程中,生长素信号途径也起着决定性的作用。在中柱鞘细胞第一次细胞分裂前,细胞中的IAA应答途径被活化,从而选择性的激活特异的中柱鞘细胞分裂形成侧根原基[41],这一过程受到AUX/IAA转录抑制因子IAA14和IAA28、IAA转录调控因子ARF7和ARF19及其下游信号基因GATA23等的严密调控[42]。此外,IAA途径下游靶标基因PLT3,PLT5和PLT7在侧根原基形成后,参与对侧根原基发育的调控,对侧根持续的萌发产生重要影响[43-44]。因此,IAA信号不仅在信号震荡区诱导侧根形成点的形成,也调控侧根原基的发生和发育,是侧根发生的最重要的调控因子[45]。
值得注意的是,铜绿假单胞菌产生的群感信号DKPs和已知的IAA有相似的二维杂环结构,同样能和植物内的IAA受体蛋白TIR1结合,激活下游IAA应答基因的表达,诱导植物侧根的形成[25]。大肠杆菌产生的吲哚会影响植物内源的IAA信号,当施用于植物根部时会促进侧根原基的发育[46]。推测是吲哚在低浓度时会扮演和IAA相似的功能促进AUX/IAA转录抑制因子的泛素化降解,增加IAA转录调控因子(ARFs)对下游IAA应答基因的转录激活,从而促进侧根的生长;而在高浓度时吲哚则会扮演IAA的竞争抑制物抑制AUX/IAA的降解,降低ARF对下游基因的调控能力,说明吲哚可以转化为IAA的拮抗物质干预植物内源IAA信号通路。综上所述,PGPRs对植物RSA的改变,通常伴随着植物内源激素响应的变化。PGPRs代谢物对植物根系发育内源激素途径的影响机制总结如图 1所示。
|
| 图 1 PGPRs产生信号分子调控植物根系构型分子机制 |
根系是植物吸收水分和养分的主要门户,调控植物RSA是促进根际养分吸收利用、提高养分利用效率的重要途径。植物的根系发育既受到复杂的内源信号途径的控制,又受根际微生物、养分含量、含水量、有害元素胁迫等生物和非生物因素的影响。培育根系构型合理、养分利用高效的作物新品种是目前的主要育种目标之一,但是这个周期较长;利用根际微生物对根系构型的调控和影响促进作物的养分吸收利用是简便有效的可行途径之一,具有重要的应用价值。
根际微生物作为植物的第二基因组,其研究对我国农业绿色发展战略意义重大,也是目前国际前沿研究热点。根际微生物已经连续两年被中科院科技战略咨询研究院的报告列为国际重点研究前沿,并被美国科学家评为未来农业的五大发展方向之一,显示其未来的巨大应用潜力。但是根际微生物在农业绿色发展中的潜力还未被充分利用,对PGPRs与植物互作的信号、功能、机制和应用缺乏深入的研究和系统的认识是限制其广泛应用的瓶颈。
未来在PGPRs调控植物RSA以促进植物生长和养分利用方面,应重点围绕以下几个方向从基础研究到实际应用系统开展工作,促进PGPRs在农业减肥增效和绿色发展中的应用。第一,深入挖掘PGPRs调控植物RSA的新型信号分子;第二,系统阐释PGPRs新型信号分子调控RSA的分子途径;第三,根据各种作物高效吸收不同养分的根系形态,探索综合利用不同PGPRs菌株和信号组合调控形成理想RSA的技术体系;第四,实现PGPRs和新型信号分子产品化(微生物肥料和植物生长调节剂)和产业化的研发,服务于我国化肥减施增效和农业绿色发展战略的实施。
| [1] |
Das AJ, Kumar M, Kumar R. Plant growth promoting PGPR :an alternative of chemical fertilizer for sustainable environment friendly agriculture[J]. Turkish Journal of Agriculture and Forestry, 2013, 1: 21-23. |
| [2] |
Adesemoye AO, Torbert HA, Kloepper JW. Plant growth-promoting rhizobacteria allow reduced application rates of chemical fertilizers[J]. Microbial Ecology, 2009, 58: 921-929. DOI:10.1007/s00248-009-9531-y |
| [3] |
Huang XF, Zhou D, Guo J, et al. Bacillus spp. from rainforest soil promote plant growth under limited nitrogen conditions[J]. Journal of Applied Microbiology, 2014, 118: 672-684. |
| [4] |
Ryu CM, Farag MA, Hu CH, et al. Bacterial volatiles promote growth in Arabidopsis[J]. Proceedings of the National Academy of Sciences of the United States of America, 2003, 100: 4927-4932. DOI:10.1073/pnas.0730845100 |
| [5] |
Canbolat MY, Bilen S, Akmak R, et al. Effect of plant growth-promoting bacteria and soil compaction on barley seedling growth, nutrient uptake, soil properties and rhizosphere microflora[J]. Biology and Fertility of Soils, 2006, 42: 350-357. DOI:10.1007/s00374-005-0034-9 |
| [6] |
Shahzad SM, Khalid A, Arshad M, et al. Improving nodulation, growth and yield of Cicer arietinum L. through bacterial ACC-deaminase induced changes in root architecture[J]. European Journal of Soil Biology, 2010, 46: 342-347. DOI:10.1016/j.ejsobi.2010.05.007 |
| [7] |
Berendsen RL, van Verk MC, Stringlis IA, et al. Unearthing the genomes of plant beneficial Pseudomonas model strains WCS358, WCS374 and WCS417[J]. BMC Genomics, 2015, 16: 539. DOI:10.1186/s12864-015-1632-z |
| [8] |
Zamioudis C, Mastranesti P, Dhonukshe P, et al. Unraveling root developmental programs initiated by beneficial Pseudomonas spp. bacteria[J]. Plant Physiology, 2013, 162(304): 318. |
| [9] |
López-Bucio J, Camposcuevas JC, HernándezCalderón E, et al. Bacillus megaterium rhizobacteria promote growth and alter root-system architecture through an auxin- and ethylene-independent signaling mechanism in Arabidopsis thaliana[J]. Molecular Plant-Microbe Interactions, 2007, 20: 207-217. DOI:10.1094/MPMI-20-2-0207 |
| [10] |
Patten CL, Glick BR. Role of Pseudomonas putida indole acetic acid in development of the host plant root system[J]. Applied and Environmental Microbiology, 2002, 68: 3795-3801. DOI:10.1128/AEM.68.8.3795-3801.2002 |
| [11] |
Zou C, Li U, Yu D, et al. Bacillus megaterium strain XTBG34 promotes plant growth by producing 2-pentylfuran[J]. Journal of Microbiology, 2010, 48: 460-466. DOI:10.1007/s12275-010-0068-z |
| [12] |
Zú iga A, Poupin MJ, Raúl AD, et al. Quorum sensing and indole-3-acetic acid degradation play a role in colonization and plant growth promotion of Arabidopsis thaliana by Burkholderia phytofirmans PsJN[J]. Molecular Plant-Microbe Interactions, 2013, 26: 546-553. DOI:10.1094/MPMI-10-12-0241-R |
| [13] |
Gutierrez-Luna FM, López-Bucio J, Altamirano-Hernández J, et al. Plant growth-promoting rhizobacteria modulate root-system architecture in Arabidopsis thaliana through volatile organic compound emission[J]. Symbiosis, 2010, 51: 75-83. DOI:10.1007/s13199-010-0066-2 |
| [14] |
李晓倩. 茶树根际促生菌研究展望[J]. 山东农业大学学报:自然科学版, 2009, 40(2): 301-303. Li XQ. Studies and prospects on plant growth-promoting rhizobacteria of tea rhizosphere(Camelia sinensis)[J]. Journal of Shandong Agricultural University :Natural Science Edition, 2009, 40(2): 301-303. |
| [15] |
De Salamone IEG, Hynes RK, Nelson LM, et al. Cytokinin production by plant growth promoting rhizobacteria and selected mutants[J]. Canadian Journal of Microbiology, 2001, 47: 404-411. DOI:10.1139/w01-029 |
| [16] |
Timmusk S, Nicander B, Granhall U, et al. Cytokinin production by Paenibacillus polymyxa[J]. Soil Biology Biochemistry, 1999, 31: 1847-1852. DOI:10.1016/S0038-0717(99)00113-3 |
| [17] |
Arkhipova TN, Veselov SU, Melentiev AI, et al. Ability of bacterium Bacillus subtilis to produce cytokinins and to influence the growth and endogenous hormone content of lettuce plants[J]. Plant Soil, 2005, 272: 201-209. DOI:10.1007/s11104-004-5047-x |
| [18] |
Joo GJ, Kim YM, Lee IJ, et al. Growth promotion of red pepper plug seedlings and the production of gibberellins by Bacillus cereus, Bacillus macroides and Bacillus pumilus[J]. Biotechnology Letters, 2004, 26: 487. DOI:10.1023/B:BILE.0000019555.87121.34 |
| [19] |
Khalid A, Arshad M, Zahir ZA. Screening plant growth-promoting rhizobacteria for improving growth and yield of wheat[J]. Journal of Applied Microbiology, 2004, 96: 73-480. |
| [20] |
Asghar H, Zahir Z, Arshad M, et al. Relationship between in vitro production of auxins by rhizobacteria and their growth-promoting activities in Brassica juncea L[J]. Biology and Fertility of Soils, 2002, 35: 231-237. DOI:10.1007/s00374-002-0462-8 |
| [21] |
Remans R, Beebe S, Blair M, et al. Physiological and genetic analysis of root responsiveness to auxin-producing plant growth-promoting bacteria in common bean(Phaseolus vulgaris L.)[J]. Plant Soil, 2007, 302: 149-161. |
| [22] |
Spaepen S, Bossuyt S, Engelen K, et al. Phenotypical and molecular responses of Arabidopsis thaliana roots as a result of inoculation with the auxin-producing bacterium Azospirillum brasilense[J]. New Phytologist, 2014, 201: 850-861. DOI:10.1111/nph.12590 |
| [23] |
Lim JH, Kim SD. Synergistic plant growth promotion by the indigenous auxins-producing PGPR Bacillus subtilis AH18 and Bacillus licheniforims K11[J]. Journal of the Korean Society for Applied Biological Chemistry, 2009, 52: 531-538. DOI:10.3839/jksabc.2009.090 |
| [24] |
Zhao Q, Zhang C, Jia Z, et al. Involvement of calmodulin in regulation of primary root elongation by N-3-oxo-hexanoyl homoserine lactone in Arabidopsis thaliana[J]. Frontier in Plant Science, 2015, 5: 1-11. |
| [25] |
Ortiz-Castro R, Diaz-Perez C, Martinez-Trujillo M, et al. Transkingdom signaling based on bacterial cyclodipeptides with auxin activity in plants[J]. Proceedings of the National Academy of Sciences of the United States of America, 2011, 108: 7253-7258. DOI:10.1073/pnas.1006740108 |
| [26] |
Meldau DG, Meldau S, Hoang LH, et al. Dimethyl disulfide produced by the naturally associated bacterium Bacillus sp B55 promotes Nicotiana attenuata growth by enhancing sulfur nutrition[J]. Plant Cell, 2013, 25: 2731-2747. DOI:10.1105/tpc.113.114744 |
| [27] |
Pérez-Flores P, Valencia-Cantero E, Altamirano-Hernandez J, et al. Bacillus methylotrophicus M4-96 isolated from maize(Zea mays) rhizoplane increases growth and auxin content in Arabidopsis thaliana via emission of volatiles[J]. Protoplasma, 2017, 254: 2201-2213. DOI:10.1007/s00709-017-1109-9 |
| [28] |
Raya-González J, Velázquez-Becerra C, Barrera-Ortiz S, et al. N, N-dimethyl hexadecylamine and related amines regulate root morphogenesis via jasmonic acid signaling in Arabidopsis thaliana[J]. Protoplasma, 2017, 254: 1399-1410. DOI:10.1007/s00709-016-1031-6 |
| [29] |
Grieneisen VA, Xu J, Maree AFM, et al. Auxin transport is sufficient to generate a maximum and gradient guiding root growth[J]. Nature, 2007, 449: 1008-1013. DOI:10.1038/nature06215 |
| [30] |
Aida M, Beis D, Heidstra R, et al. The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche[J]. Cell, 2004, 119: 109-120. DOI:10.1016/j.cell.2004.09.018 |
| [31] |
Galinha C, Hofhuis H, Luijten M, et al. PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development[J]. Nature, 2007, 449: 1053-1057. DOI:10.1038/nature06206 |
| [32] |
Verbelen JP, Cnodder TD, Le J, et al. Root apex of Arabidopsis thaliana consists of four distinct zones of growth activities[J]. Plant Signaling & Behavior, 2006, 1: 296-304. |
| [33] |
Moubayidin L, Di Mambro R, Sozzani R, et al. Spatial coordination between stem cell activity and cell differentiation in the root meristem[J]. Development Cell, 2013, 26: 405-415. DOI:10.1016/j.devcel.2013.06.025 |
| [34] |
Dello IR, Nakamura K, Moubayidin L, et al. A genetic framework for the control of cell division and differentiation in the root meristem[J]. Science, 2008, 322: 1380-1384. DOI:10.1126/science.1164147 |
| [35] |
Zhang H, Kim MS, Krishnamachari V, et al. Rhizobacterial volatile emissions regulate auxin homeostasis and cell expansion in Arabidopsis[J]. Planta, 2007, 226: 839-851. DOI:10.1007/s00425-007-0530-2 |
| [36] |
Ortiz-Castro R, Pelagio-Flores R, Mendez-Bravo A, et al. Pyocyanin, a virulence factor produced by Pseudomonas aeruginosa, alters root development through reactive oxygen species and ethylene signaling in Arabidopsis[J]. Molecular Plant-Microbe Interactions, 2014, 27: 364-378. DOI:10.1094/MPMI-08-13-0219-R |
| [37] |
Dubrovsky JG, Sauer M, Napsucialy-Mendivil S, et al. Auxin acts as a local morphogenetic trigger to specify lateral root founder cells[J]. Proceedings of the National Academy of Sciences of the United States of America, 2008, 105: 8790-8794. DOI:10.1073/pnas.0712307105 |
| [38] |
Péret B, De Rybel B, Casimiro L, et al. Arabidopsis lateral root development :an emerging story[J]. Trends in Plant Science, 2009, 14: 399-408. DOI:10.1016/j.tplants.2009.05.002 |
| [39] |
Moreno-Risueno MA, Van Norman JM, Moreno A, et al. Oscillating gene expression determines competence for periodic Arabidopsis root branching[J]. Science, 2010, 329: 1306-1311. DOI:10.1126/science.1191937 |
| [40] |
Xuan W, Audenaert D, Parizot B, et al. Root cap-derived auxin pre-patterns the longitudinal axis of the Arabidopsis root[J]. Current Biology, 2015, 25: 1381-1388. DOI:10.1016/j.cub.2015.03.046 |
| [41] |
Benková E, Michniewicz M, Sauer M, et al. Local, efflux-dependent auxin gradients as a common module for plant organ formation[J]. Cell, 2003, 115: 591-602. DOI:10.1016/S0092-8674(03)00924-3 |
| [42] |
De Rybel B, Vassileva V, Parizot B, et al. A novel Aux/IAA28 signaling cascade activates GATA23-dependent specification of lateral root founder cell identity[J]. Current Biology, 2010, 20: 1697-1706. DOI:10.1016/j.cub.2010.09.007 |
| [43] |
Hofhuis H, Laskowski M, Du YJ, et al. Phyllotaxis and rhizotaxis in Arabidopsis are modified by three PLETHORA transcription factors[J]. Current Biology, 2013, 23: 956-962. DOI:10.1016/j.cub.2013.04.048 |
| [44] |
Du Y, Scheres B. PLETHORA transcription factors orchestrate de novo organ patterning during Arabidopsis lateral root outgrowth[J]. Proceedings of the National Academy of Sciences of the United States of America, 2017, 114: 11709-11714. DOI:10.1073/pnas.1714410114 |
| [45] |
Van Norman JM, Xuan W, Beeckman T, et al. To branch or not to branch :the role of pre-patterning in lateral root formation[J]. Development, 2013, 140: 4301-4310. DOI:10.1242/dev.090548 |
| [46] |
Bailly A, Groenhagen U, Schulz S, et al. The inter-kingdom volatile signal indole promotes root development by interfering with auxin signaling[J]. Plant Journal, 2014, 80: 758-771. DOI:10.1111/tpj.12666 |
| [47] |
Felten J, Legué V, Ditengou FA, et al. Lateral root stimulation in the early interaction between Arabidopsis thaliana and the ectomycorrhizal fungus Laccaria bicolor :is fungal auxin the trigger[J]. Plant Signaling & Behavior, 2009, 5: 864-867. DOI:10.1104/pp.109.147231 |
| [48] |
Splivallo R, Fischer U, Gobel C, et al. Truffles regulate plant root morphogenesis via the production of auxin and ethylene[J]. Plant Physiology, 2009, 150: 2018-2029. DOI:10.1104/pp.109.141325 |
| [49] |
Contreras-Cornejo HA, Lopez-Bucio JS, Mendez-Bravo A, et al. Mitogen-activated protein kinase 6 and ethylene and auxin signaling pathways are involved in Arabidopsis root-system architecture alterations by Trichoderma atroviride[J]. Molecular Plant-Microbe Interactions, 2015, 28: 701-710. DOI:10.1094/MPMI-01-15-0005-R |
| [50] |
D'Angelo-Picard C, Faure D, Penot L, et al. Diversity of N-acyl homoserine lactone-producing and-degrading bacteria in soil and tobacco rhizosphere[J]. Environmental Microbiology, 2005, 7: 1796-1808. DOI:10.1111/j.1462-2920.2005.00886.x |