K+是植物体内含量最丰富的阳离子,同时钾也是作物生长必需的三大矿质营养元素之一,约占植物干重的2%-10%[1]。与氮、磷两种元素不同,虽然钾不参与任何有机物质的合成,却在植物生长发育过程中起着不可替代的作用,其主要参与植物体内许多重要的生理和生化过程,如(1)促进植物脂肪代谢、水分代谢;(2)促进同化物转化,促进韧皮部的养分运输;(3)控制细胞膜极化;(4)调节气孔运动,促进光合作用;(5)作为酶的激活剂;(6)维持阴阳离子平衡;(7)调节渗透压等[2-5]。同时钾还可以提高植物对高盐、干旱等非生物胁迫及病虫害的耐受性,缺钾会直接影响植物的生长发育,如加速老叶黄化,根系短小且易倒伏等[3]。土壤中所能提供的速效钾的含量很少,约在10-100 μmol/L之间,且土壤内有效钾离子浓度变化巨大,有时相差100倍左右,为了适应不同的钾浓度环境,满足自身对钾营养的需求,保持体内稳态,植物细胞分化出高亲和与低亲和两种钾吸收系统[6]。在外界钾营养匮乏的环境下,植物细胞主要采用高亲和钾吸收系统来进行钾的吸收和转运[5, 7]。本文主要介绍了高亲和钾吸收系统中KUP/HAK/KT家族的基本情况及其分类、高亲和钾离子转运蛋白HAK的系统发育分析及其在植物钾吸收,生长发育,抵抗生物胁迫和非生物胁迫等方面的功能。深入了解钾离子转运蛋白HAK在植物体内的吸收及作用机制对于有效提高钾肥的利用效率,提升作物产量与品质,促进农业发展等方面具有重要的现实意义。
1 钾离子转运载体KUP/HAK/KT家族简介K+在土壤中的流动性较低,大部分被土壤吸附或固定,因此植物根系所能利用的钾浓度很低。在低钾环境(钾浓度低于0.2 mmol/L)下,植物的根系细胞膜进化出高亲和钾转运蛋白从土壤溶液中进行钾的有效吸收与转运,以满足自身对钾的高营养需求[8-9]。根据高亲和K+转运蛋白的序列特征和结构不同,其主要分为4大家族:KUP(K+ uptake)/HAK(High-affinity K+)/KT(K+ transporter)家族、HKT/Trk/Ktr家族、CHX(Cation/hydrogen exchanger)家族以及KEA(K+ efflux anti-porter)家族[10-11]。KUP/HAK/KT家族是植物体内高亲和K+转运家族中最大、成员最多的家族,该家族成员广泛存在于植物的各组织器官中,在细胞膜和液泡膜上行使功能,其成员已在植物、真菌及变形虫中均有发现,但尚未涉及原生生物和动物界 [12]。研究发现水稻(Oryzasativa L.)中存在27个潜在的KUP/HAK/KT家族的转运蛋白,在拟南芥(Arabidopsis thaliana)、玉米(Zea mays L.)、白杨(Populus tomentosa Carr)和番茄(Solanum lycopersicum)中分别存在13、27、31和21个[10, 13-14]。
植物KUP/HAK/KT家族基因基于系统发育关系,保守基序和基因结构分析分为4个主要的系统发育亚族[15]。第I亚族在低钾环境下表达上调,该家族基因编码的转运蛋白均能介导高亲和力钾吸收,也有部分I亚族基因可以为低亲和的钠内流提供通道,如HvHAK1和PhaHAK1等[9, 16-17]。第II亚族参与了植物的生长发育进程,其成员在序列和功能上具有很大的差异,该亚族转运蛋白可介导钾的高亲和与低亲和摄取[16, 18]。部分II亚族成员调控了植物的生长进程,如AtKUP4/TRH1突变会导致拟南芥生长素转运受损、根毛细小;AtKUP2(shy3-1)突变会降低芽细胞扩增等[19-20]。目前,对KUP/HAK/KT第III及第IV亚族转运蛋白生理功能的研究还相对较少[13]。
2 植物钾离子转运蛋白HAK简介及系统发育分析植物HAK钾转运蛋白在介导高亲和K+吸收和转运过程中发挥着关键作用,其转运过程介导H+/K+的同向运输,属于逆化学势梯度,消耗能量的主动运输过程。编码高亲和力K+转运的HAK蛋白在大麦(Hordeum vulgare L.)中首次被克隆出来,后在其他植物中又陆续发现了多个HAK蛋白[21]。在结构方面,HAK钾转运蛋白通常具有12个跨膜区域(Transmembrane segments,TMS),在第2个和第3个TMS之间存在着胞质Loop,C末端和N末端均位于胞内侧[22](图 1)。由于HAK高亲和钾转运蛋白在瓜蟾卵母细胞中不表达,但可补充存在K+吸收缺陷的酵母(Saccharomyce)或细菌突变体中的K+吸收。因此,该蛋白的功能常在酵母和大肠杆菌等异源系统中进行验证[23-24]。
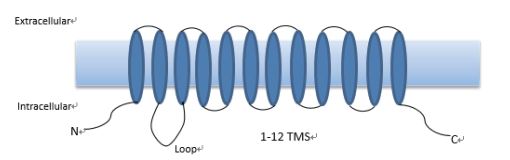
|
| 图 1 植物HAK高亲和钾转运蛋白结构示意图 |
为了研究植物中HAK转运蛋白之间的进化模式和系统发育关系,用MEGA将水稻、拟南芥、玉米、大麦和烟草(Nicotiana tabacum L. / Nicotiana rustica)等13个物种的69个HAK蛋白的氨基酸序列完全比对,通过NJ(Neighbor-Joining,NJ)方法构建了一个综合的系统发育树(图 2)。从系统发育树中,我们发现植物中该转运蛋白的所有成员都被分为7组(A-G),A组包括来自烟草、非洲冰草(Mesembryanthemum crystallinum)、水稻、玉米、海草(Cymodocea nodosa)和芦苇(Phragmites australis Trinius)中的16个基因;B组包括来自水稻、玉米、海草、大麦4个物种中的8个基因;C组包括水稻、玉米中的6个基因;D组包括水稻、玉米、桃(Physcomitrella patens)和芦苇中的9个基因;E组同样包括水稻和玉米中的6个基因;F中包括辣椒(Capsicum chinense)、番茄、拟南芥(Arabidopsis thaliana)、盐芥(Thellungiella halophila)和蓖麻(Ricinus communis)中的5个基因;G组中包括水稻、玉米、芦苇、大麦4个物种中19个基因。该系统发育树中的所有基因都起源于种子植物,表明其可能独立地在种子植物中形成,其中B、C、E和F组均是单子叶植物的基因,其他组既包括单子叶植物的基因,也包括双子叶植物的基因。
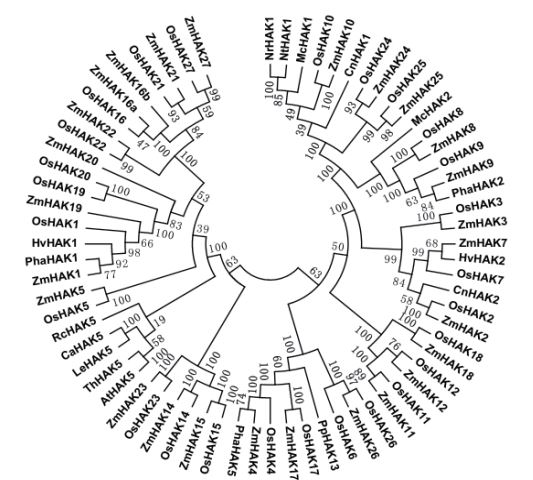
|
| 图 2 植物HAK高亲和钾转运蛋白的系统发育树 |
钾在植物生长发育过程中发挥着不可替代的作用。低钾环境下水稻根和芽中的众多HAK基因均被诱导表达。研究发现OsHAK1在低钾和高钾的酵母环境中均生长良好,OsHAK1被敲除后植株的钾吸收率及转运能力与野生型相比均显著降低,而过表达植株则增强了水稻中钾的积累,这表明OsHAK1是一种能同时介导水稻高亲和与低亲和K+摄取的转运蛋白[13]。OsHAK5能在仅含0.1mmol/L K+的酵母环境中生长,水稻敲除OsHAK5后,其根系净钾流入速率和地上部分的钾转运能力受到严重影响,相反,过表达OsHAK5植株的钾积累量则显著增加[9, 25]。水稻OsHAK16在K+不足时表达上调,OsHAK16被敲除后显著降低了水稻根系净钾吸收速率和生长状态,而过表达OsHAK16仅在0.1 mmol/LK+水平上改善了植物总钾吸收含量及生长状态[26]。OsHAK7、OsHAK10在酵母及细菌突变体中均被验证可介导低亲和钾吸收[27]。当在K+缺陷型酵母中表达时,OsHAK21发挥钾积累功能,弥补了其生长缺陷[28]。这些证据证明HAK转运蛋白在维持水稻K+稳态方面起着至关重要的作用。
AtHAK5是拟南芥面对缺钾胁迫时反应的标志基因,在K+饥饿时,拟南芥根中的AtHAK5转录水平增加,T-DNA插入后植株的钾积累量减少,该基因的同源物LeHAK5、ThHAK5、HvHAK1、CaHAK1和OsHAK1均对K+不足有响应[5, 29-30]。通过酵母实验发现,辣椒(Capsicum annuum L.)CaHAK1能介导植株高亲和力K+摄取[31]。虽然番茄LeHAK5不能弥补酵母突变体WD3在0.1 mmol/L钾环境下的生长缺陷,但是LeHAK5-CaHAK1复合体却能补充这种缺陷[32]。从缺钾植株根系中分离得到的盐芥ThHAK5能在0.1 mmol/L K+的酵母环境中生长,介导植物的钾吸收[33]。低钾环境下大麦HvHAK1的转录水平相比于正常钾环境增加了5倍,将大麦HvHAK1 cDNA插入酵母表达载体中发现,HvHAK1能介导酵母在0.1 mmol/L的K+环境中生长[34]。HvHAK4可能有助于大麦吸收K+至生长中的叶片组织中,并维持叶肉K+浓度高于表皮[35]。研究发现,非洲冰草中McHAK1、McHAK2、McHAK4可恢复钾摄取缺陷型酵母菌株在毫摩钾环境下的生长,促进植物钾吸收[36]。獐茅(Aeluropus littoralis)中AlHAK转运蛋白可介导植物的高亲和性钾吸收[37]。芦苇植物中PhaHAK2和PhaHAK5均为高亲和钾转运蛋白,介导植物钾营养积累[17, 38]。海草中CnHAK1过表达后介导了大肠杆菌突变体中的快速K+涌入,这些涌入的浓度依赖性证明CnHAK1为低亲和力K+转运蛋白[39]。烟草NrHAK1在面临低钾胁迫时表达上调,在低钾环境下能增加植株钾离子含量的积累[40]。低钾处理提高了桑椹(Morus alba L.)MaHAK5的转录水平[41]。
研究发现,谷子(Setaria italica)SiHAK1在低钾环境下表达上调显著,促进植物钾吸收,SiHAK2则在K+浓度较高时增加钾营养的积累,通过在酵母中添加不同的钾浓度比较SiHAK1与OsHAK1、OsHAK5和HvHAK1的钾转运能力,发现SiHAK1与其他3种钾运蛋白相比更能适应低钾环境,这些结论证明SiHAK1是一种在低钾环境下进行K+转运的高亲和转运蛋白,且具有更好的钾转运能力[42]。玉米ZmHAK5是一种高亲和的钾转运蛋白,ZmHAK5突变植株钾吸收能力降低,相反,超表达ZmHAK5后则表现出更好的生长状态及钾吸收能力。ZmHAK1在低钾条件下同样表达上调,但其钾转运能力不及ZmHAK5[43]。在甘蔗(Saccharum officinarum)中研究发现,SsHAK1和SsHAK21均具有钾转运活性,可恢复酵母突变菌株的钾离子吸收功能,满足自身的高钾营养需求[44]。以上结果表明钾离子转运蛋白HAK在促进植物钾营养吸收过程中发挥着重要的作用。
3.2 影响植株生长发育钾不仅是植物生长所必需的元素,也是一种重要的细胞溶质,钾的缺失会限制植物细胞扩张,HAK转运蛋白通过吸收钾影响植物的生长发育。利用田间试验发现,OsHAK5超表达水稻与野生型相比株高降低,分蘖增加,该基因敲除后植株株高同样降低,但其分蘖数与野生型相比减少,结实率也大幅下降[45]。水稻OsHAK16敲除后影响了植株的生长,与野生型相比植株根茎干重降低[26]。OsHAK1被敲除后植株蔗糖运输途径受损,根系生长较弱,发育迟缓,花粉活力、生育率与穗肥力均下降,敲除该基因后水稻根部与茎部生长受阻,种子发芽率及谷物产量均小于野生型[13, 41]。这些结果表明在影响水稻的生长发育方面HAK钾转运蛋白发挥着重要作用。通过光学显微镜观察,显示含有CN-HAK1的烟草叶片厚度明显比仅含CN和野生型的更厚(CN为TMV的一种抗性基因),这表明由HAK1引起的钾营养吸收改善了植物的生理状态并影响植物组织的形态[46]。研究发现,在低钾环境下,AtHAK5突变植株根长与野生型相比明显缩短[47]。过表达AtHAK5植株在低钾培养基中生长时,其根毛长度和密度均比野生型增加,这表明AtHAK5可使拟南芥面对低钾胁迫时作出适应性形态改变,促进根毛伸长[48]。在拟南芥AtHAK5突变植株中过表达SiHAK1,发现在低钾条件下SiHAK1弥补了AtHAK5突变后植物的生长缺陷,其植株鲜重,根长度均比AtHAK5突变体显著增加,说明SiHAK1也参与了低钾环境下植物的形态学调整[42]。OsHAK21在拟南芥中的表达同样完全恢复了AtHAK5突变体的低K+敏感表型[28]。因此,HAK家族在钾介导的植物生长发育中发挥着重要的作用。
3.3 增强植物抗旱能力干旱是常见的非生物胁迫,严重限制了作物的产量[49]。植物面对干旱胁迫的一种方式是通过积累K+等溶质改善细胞渗透势来减少水分流失,提高耐旱性。研究表明,水稻敲除OsHAK1后对干旱胁迫的耐受性较低,在营养和生殖阶段均表现为生长发育不良,而OsHAK1过表达幼苗与野生型相比对干旱胁迫表现出更好的耐受性,且比野生型多出35%的粮食产量[50]。过表达NtHAK1可提高烟草对干旱胁迫的耐受性[51]。干旱胁迫下影响桑树MaHAK5的表达水平[41]。由此表明,植物利用HAK转运蛋白增强自身的钾积累,降低细胞渗透势,增强抗旱能力。其次HAK家族可通过降低活性氧(Reactiveoxygen species,ROS)积累量来维持植物体内的水分平衡。干旱胁迫下,植物会积累活性氧,通过有氧代谢产生有害物质,破坏DNA、蛋白质和碳水化合物,引起细胞死亡[52]。与野生型相比,OsHAK1过表达植株具有较低的脂质过氧化水平、较高的抗氧化酶活性(POX、CAT)和较高的脯氨酸积累,说明OsHAK1通过减少ROS的积累来维持植物细胞稳态[50]。以上研究结果表明,高亲和钾转运蛋白HAK在降低细胞渗透势、减少ROS积累及增强植物抗旱能力等方面发挥着重要的作用。
3.4 增强植物耐盐性高盐环境利用膜通透性改变、代谢紊乱、有毒物质积累等途径造成植物生长发育及形态的改变,植物维持胞内K+ /Na+稳态对提高耐盐性,保证自身生长发育等方面发挥着重要的作用[53-54]。盐胁迫环境下,水稻根茎部位OsHAK16表达水平显著上升,该基因的表达增加了植株钾含量但没增加钠积累,OsHAK16显着改善了高盐敏感型酵母突变体的生长,提高植株耐盐性[26]。盐胁迫诱导了水稻根中OsHAK21的表达,OsHAK21突变体中净Na+吸收率明显提高,而净K+吸收率被显著抑制,OsHAK21的破坏扰乱了植株K+ / Na+稳态,导致盐超敏反应[28]。OsHAK21、OsHAK27能在342 mmol/L NaCl的酵母环境下进行生长,具有明显的Na+抗性[25]。水稻OsHAK1具有盐敏感性,高盐环境下能促进植物吸收钾,增强对盐的耐受性[13]。OsHAK5是一种对钠不敏感的钾转运蛋白,通过增加K+吸收、阻止Na+流入来维持细胞稳态,过表达OsHAK5增加了水稻幼苗K+ /Na+比值和对盐胁迫的耐受性,敲除OsHAK5后则导致幼苗对盐胁迫敏感[9, 55]。以上结论证明HAK钾转运家族在增强水稻耐盐性方面发挥着重要作用。
拟南芥中AtHAK5进行F130S点突变后增加了植物的钾吸收,提高了其在酵母中的抗盐能力,同时AtHAK1,AtHAK6,AtHAK2也参与调控了拟南芥的耐盐渠道[56-57]。獐茅原产于沿海地带,其AlHAK转运蛋白具有使植物在高盐环境下保持较高K+ /Na+比的能力[37]。星星草(Puccinellia tenuiflora)是一种优良的盐生草,耐盐性强,其PtHAK5钾转运蛋白在维持植株K+ / Na+比值,增强耐盐性方面发挥重要作用[58]。盐胁迫条件下,芦苇植物中PhaHAK1、PhaHAK2介导的K+持续吸收和保持植株高K+/Na+比是导致芦苇具有高度耐盐性的原因之一[17, 38]。高盐环境下超表达外源基因AlHAK1可维持棉花(Gossypium spp.)中较高的K+ /Na+比,提高了转基因棉花的抗盐能力[59]。海草CnHAK1在高盐环境维持植物K+ /Na+稳态中发挥着重要的作用,在海水中K+浓度为11 mmol/L时进行钾的低亲和转运[39]。烟草NrHAK1插入酵母细胞后能使其在盐胁迫环境下进行生长[40]。面对盐胁迫时OsHAK5提高了烟草BY2细胞的耐盐性,促进其生长[55]。大麦HvHAKI可使K+摄取缺陷型酵母进行高亲和力K+摄取,并介导其进行低亲和力Na+摄取[21]。在酵母实验中谷子SiHAK1表现出高度的Na+不敏感性,甚至能在750 mmol/L的Na +环境中进行生长,且比HvHAK1更能忍受高盐环境 [42]。研究表明,辣椒CaHAK1在盐胁迫下也促进植物K+吸收,具有明显耐盐性[31]。以上结果表明,高亲和钾离子转运蛋白HAK参与了植物对盐分胁迫的反应,在维持植株K+/Na+稳态中起着至关重要的作用。
3.5 增强植物抗病性钾营养充足有利于维持植株良好的生理状态,减轻真菌、细菌及病毒对作物造成的危害,增强植物抗病性[60-62]。研究表明,接种烟草花叶病毒(Tobacco mosaic virus,TMV)的CN-HAK1共表达烟草相比CN转基因烟草和野生型部分抑制了植株HR反应,与野生型相比也没有出现明显的植株矮化,叶片畸形等症状,这表明烟草通过HAK1改善了植物细胞的营养状况,增强了植株对TMV的抵抗力。通过对抗氧化酶(AOEs)SOD、CAT、POD活性、水杨酸(SA)含量及相关抗性基因表达量的检测,证明CN与HAK1在转基因烟草中协同增强了对TMV的抗性,这些结论证明HAK1可能通过参与SA防御途径增强了烟草的TMV抗性,将病毒限制在比较小的坏死斑点中[46]。钾营养充足有助于植物胞内信号传导和免疫,相反钾营养的流失可能触发病原相关分子模式(PAMP)[63-64]。ILK1是一种改变植物生长发育,增强植株对flg22的反应和对高盐高钾的耐受性、提高对细菌病原体抵抗力的体外活性激酶[65-67]。HAK5可与ILK1激酶相互作用,其既可以促使植物在低钾的环境中生长,又能够促进对flg22诱导的响应。植物响应flg22诱导后根系生长受抑制,而HAK5突变体则破坏了这种抑制,HAK5突变细胞和用TEA(K+通道阻止程序)处理后的叶盘均表现出去极化速度显著降低的现象,说明K+转运是flg22诱导的植物生长抑制和PM去极化所必需的。这些证据表明,由HAK5和ILK1介导的钾转运影响了植物的免疫和应激反应,参与植物抗病性[68]。以上研究表明,HAK转运蛋白不仅增强植物钾营养积累,影响植物生长发育,提高抗旱耐盐能力,在植物抗病方面也发挥着十分重要的作用。
4 结论与展望高亲和K+转运蛋白HAK通过提高植物钾离子的吸收,从而在增强植物对于病虫害及高盐干旱等胁迫的耐受性等方面发挥着重要作用。目前关于高亲和K+转运蛋白HAK作用机理的研究虽取得了一定进展,但是仍处于初步阶段。首先该蛋白的许多功能还尚待发掘,目前HAK转运蛋白只解析了当植物面对高盐和干旱两种非生物胁迫的响应过程,对HAK蛋白如何提高植物的抗病性也只停留在初步阶段,当面对病虫害时HAK如何调控植物增强耐受性的作用机理还需要进一步解析。其次HAK如何协作相关基因发挥功能的机理也尚未解析。最后应考虑如何将HAK钾转运蛋白的功能应用到生产实际中,如何应用分子育种和基因工程技术,将能介导植物钾营养吸收,改善植物的生长发育,提高植物的耐盐抗旱及抗病性的HAK基因转入农作物,提高农作物的钾营养吸收能力及对生物和非生物胁迫的耐受性,增加作物产量。
| [1] |
Leigh RA, Wyn Jones RG. A hypothesis relating critical potassium concentrations for growth to the distribution and functions of this ion in the plant cell[J]. New Phytologist, 1984, 97(1): 1-13. DOI:10.1111/j.1469-8137.1984.tb04103.x |
| [2] |
徐赫韩, 侯国媛, 李洋, 等. 植物钾离子通道AKT1的研究进展[J]. 生物技术, 2018, 8(2): 200-204. Xu HH, Hou GY, Li Y, et al. Research progress of plant potassium channel AKT1[J]. Biotechnology, 2018, 8(2): 200-204, 150. |
| [3] |
Zörb C, Senbayram M, Peiter E. Potassium in agriculture-status and perspectives[J]. Journal of Plant Physiology, 2014, 171(9): 656-669. DOI:10.1016/j.jplph.2013.08.008 |
| [4] |
Kollist H, Nuhkat M, Roelfsema MR. Closing gaps :linking elements that control stomatal movement[J]. New Phytologist, 2014, 203(1): 44-62. DOI:10.1111/nph.12832 |
| [5] |
Wang Y, Wu WH. Potassium transport and signaling in higher plants[J]. Annual Review of Plant Biology, 2013, 64: 451-476. DOI:10.1146/annurev-arplant-050312-120153 |
| [6] |
Wang Y, Lü J, Chen D, et al. Genome-wide identification, evolution, and expression analysis of the KT/HAK/KUP family in pear[J]. Genome, 2018, 61(10): 755-765. DOI:10.1139/gen-2017-0254 |
| [7] |
Rubio F, Nieves-Cordones M, Horie T, et al. Doing'business as usual'comes with a cost :evaluating energy cost of maintaining plant intracellular K+ homeostasis under saline conditions[J]. New Phytologist, 2020, 225(3): 1097-1104. DOI:10.1111/nph.15852 |
| [8] |
Véry AA, Nieves-Cordones M, Daly M, et al. Molecular biology of K+ transport across the plant cell membrane :what do we learn from comparison between plant species?[J]. Journal of Plant Physiology, 2014, 171(9): 748-769. DOI:10.1016/j.jplph.2014.01.011 |
| [9] |
Yang T, Zhang S, Hu Y, et al. The role of a potassium transporter OsHAK5 in potassium acquisition and transport from roots to shoots in rice at low potassium supply levels[J]. Plant Physiology, 2014, 166(2): 945-959. |
| [10] |
He C, Cui K, Duan A, et al. Genome-wide and molecular evolution analysis of the Poplar KT/HAK/KUP potassium transporter gene family[J]. Ecology and Evolution, 2012, 2(8): 1996-2004. DOI:10.1002/ece3.299 |
| [11] |
Song Z, Wu X, Gao Y, et al. Genome-wide analysis of the HAK potassium transporter gene family reveals asymmetrical evolution in tobacco(Nicotiana tabacum)[J]. Genome, 2019, 62(4): 267-278. |
| [12] |
Ahn SJ, Shin R, Schachtman DP. Expression of KT/KUP genes in Arabidopsis and the role of root hairs in K+ uptake[J]. Plant Physiology, 2004, 134(3): 1135-1145. |
| [13] |
Chen G, Hu Q, Luo LE, et al. Rice potassium transporter OsHAK1 is essential for maintaining potassium-mediated growth and functions in salt tolerance over low and high potassium concentration ranges[J]. Plant, Cell and Environment, 2015, 38(12): 2747-2765. DOI:10.1111/pce.12585 |
| [14] |
Nieves-Cordones M, Ródenas R, Chavanieu A, et al. Uneven HAK/KUP/KT protein diversity among angiosperms :species distribution and perspectives[J]. Frontiers in Plant Science, 2016, 7: 127. DOI:10.3389/fpls.2016.00127 |
| [15] |
Ou W, Mao X, Huang C, et al. Genome-wide identification and expression analysis of the KUP family under abiotic stress in cassava(Manihot esculenta Crantz)[J]. Frontiers in Physiology, 2018, 9: 17. DOI:10.3389/fphys.2018.00017 |
| [16] |
Horie T, Brodsky DE, Costa A, et al. K+ transport by the OsHKT2; 4 transporter from rice with atypical Na+ transport properties and competition in permeation of K+ over Mg2+ and Ca2+ ions[J]. Plant Physiology, 2011, 156(3): 1493-1507. DOI:10.2307/41435055 |
| [17] |
Takahashi R, Nishio T, Ichizen N, et al. High-affinity K+ transporter PhaHAK5 is expressed only in salt-sensitive reed plants and shows Na+ permeability under NaCl stress[J]. Plant Cell Reports, 2007, 26(9): 1673-1679. DOI:10.1007/s00299-007-0364-1 |
| [18] |
Cheng X, Liu X, Mao W, et al. Genome-wide identification and analysis of HAK/KUP/KT potassium transporters gene family in wheat(Triticum aestivum L.)[J]. International Journal of Molecular Sciences, 2018, 19(12). |
| [19] |
Desbrosses G, Josefsson C, Rigas S, et al. AKT1 and TRH1 are required during root hair elongation in Arabidopsis[J]. Journal of Experimental Botany, 2003, 54(383): 781-788. DOI:10.1093/jxb/erg066 |
| [20] |
Vicente-Agullo F, Rigas S, Desbrosses G, et al. Potassium carrier TRH1 is required for auxin transport in Arabidopsis roots[J]. The Plant Journal, 2004, 40(4): 523-535. DOI:10.1111/j.1365-313X.2004.02230.x |
| [21] |
Santa-María GE, Rubio F, Dubcovsky J, et al. The HAK1 gene of barley is a member of a large gene family and encodes a highaffinity potassium transporter[J]. The Plant Cell, 1997, 9(12): 2281-2289. DOI:10.2307/3870585 |
| [22] |
Very AA, Sentenac H. Molecular mechanisms and regulation of K+ transport in higher plants[J]. Annual Review of Plant Biology, 2003, 54(1): 575-603. DOI:10.1146/annurev.arplant.54.031902.134831 |
| [23] |
Kim EJ, Kwak JM, Uozumi N, et al. AtKUP1 :an Arabidopsis gene encoding high-affinity potassium transport activity[J]. The Plant Cell, 1998, 10(1): 51-62. DOI:10.1105/tpc.10.1.51 |
| [24] |
Li W, Xu G, Alli A, et al. Plant HAK/KUP/KT K+ transporters : function and regulation[C]//Seminars in Cell and Developmental Biology. Academic Press, 2018, 74 : 133-141.
|
| [25] |
Okada T, Yamane S, Yamaguchi M, et al. Characterization of rice KT/HAK/KUP potassium transporters and K+ uptake by HAK1 from Oryza sativa[J]. Plant Biotechnology, 2018, 35: 101-111. DOI:10.5511/plantbiotechnology.18.0308a |
| [26] |
Feng H, Tang Q, Cai J, et al. Rice OsHAK16 functions in potassium uptake and translocation in shoot, maintaining potassium homeostasis and salt tolerance[J]. Planta, 2019, 250(2): 549-561. DOI:10.1007/s00425-019-03194-3 |
| [27] |
Banuelos MA, Garciadeblas B, Cubero B, et al. Inventory and functional characterization of the HAK potassium transporters of rice[J]. Plant Physiology, 2002, 130(2): 784-795. DOI:10.1104/pp.007781 |
| [28] |
Shen Y, Shen L, Shen Z, et al. The potassium transporter OsHAK21 functions in the maintenance of ion homeostasis and tolerance to salt stress in rice[J]. Plant, Cell and Environment, 2015, 38(12): 2766-2779. DOI:10.1111/pce.12586 |
| [29] |
Gierth M, Mäser P, Schroeder JI. The potassium transporter AtHAK5 functions in K+ deprivation-induced high-affinity K+ uptake and AKT1 K+ channel contribution to K+ uptake kinetics in Arabidopsis roots[J]. Plant Physiology, 2005, 137(3): 1105-1114. DOI:10.1104/pp.104.057216 |
| [30] |
Shin R, Schachtman DP. Hydrogen peroxide mediates plant root cell response to nutrient deprivation[J]. Proceedings of the National Academy of Sciences, 2004, 101(23): 8827-8832. DOI:10.1073/pnas.0401707101 |
| [31] |
Martínez-Cordero MA, Martínez V, Rubio F. Cloning and functional characterization of the high-affinity K+ transporter HAK1 of pepper[J]. Plant Molecular Biology, 2004, 56(3): 413-421. DOI:10.1007/s11103-004-3845-4 |
| [32] |
Nieves-Cordones M, Miller AJ, Alemán F, et al. A putative role for the plasma membrane potential in the control of the expression of the gene encoding the tomato high-affinity potassium transporter HAK5[J]. Plant Molecular Biology, 2008, 68(6): 521. DOI:10.1007/s11103-008-9388-3 |
| [33] |
Alemán F, Nieves-Cordones M, Martínez V, et al. Differential regulation of the HAK5 genes encoding the high-affinity K+ transporters of Thellungiella halophila and Arabidopsis thaliana[J]. Environmental and Experimental Botany, 2009, 65(2-3): 263-269. DOI:10.1016/j.envexpbot.2008.09.011 |
| [34] |
Santa-María GE, Rubio F, Dubcovsky J, et al. The HAK1 gene of barley is a member of a large gene family and encodes a highaffinity potassium transporter[J]. The Plant Cell, 1997, 9(12): 2281-2289. DOI:10.2307/3870585 |
| [35] |
Boscari A, Clement M, Volkov V, et al. Potassium channels in barley :cloning, functional characterization and expression analyses in relation to leaf growth and development[J]. Plant, Cell and Environment, 2009, 32(12): 1761-1777. DOI:10.1111/j.1365-3040.2009.02033.x |
| [36] |
Su H, Golldack D, Zhao C, et al. The expression of HAK-type K+ transporters is regulated in response to salinity stress in common ice plant[J]. Plant Physiology, 2002, 129(4): 1482-1493. DOI:10.1104/pp.001149 |
| [37] |
Su Q, Feng S, An L, et al. Cloning and functional expression in Saccharomyces cereviae of a K+ transporter, AlHAK, from the graminaceous halophyte, Aeluropus littoralis[J]. Biotechnology Letters, 2007, 29(12): 1959-1963. DOI:10.1007/s10529-007-9484-5 |
| [38] |
Takahashi R, Nishio T, Ichizen N, et al. Cloning and functional analysis of the K+ transporter, PhaHAK2, from salt-sensitive and salt-tolerant reed plants[J]. Biotechnology Letters, 2007, 29(3): 501-506. DOI:10.1007/s10529-006-9246-9 |
| [39] |
Garciadeblas B, Benito B, Rodríguez-Navarro A. Molecular cloning and functional expression in bacteria of the potassium transporters CnHAK1 and CnHAK2 of the seagrass Cymodocea nodosa[J]. Plant Molecular Biology, 2002, 50(4-5): 623-633. DOI:10.1023/A:1019951023362 |
| [40] |
Guo Z, Yang Q, Wan X, et al. Functional characterization of a potassium transporter gene NrHAK1 in Nicotiana rustica[J]. Journal of Zhejiang University Science B, 2008, 9(12): 944-952. DOI:10.1631/jzus.B0820209 |
| [41] |
Chen G, Zhang Y, Ruan B, et al. OsHAK1 controls the vegetative growth and panicle fertility of rice by its effect on potassiummediated sugar metabolism[J]. Plant Science, 2018, 274: 261-270. DOI:10.1016/j.plantsci.2018.05.034 |
| [42] |
Zhang H, Xiao W, Yu W, et al. Foxtail millet SiHAK1 excites extreme high-affinity K+ uptake to maintain K+ homeostasis under low K+ or salt stress[J]. Plant Cell Reports, 2018, 37(11): 1533-1546. DOI:10.1007/s00299-018-2325-2 |
| [43] |
Qin YJ, Wu WH, Wang Y. ZmHAK5 and ZmHAK1 function in K+ uptake and distribution in maize under low K+ conditions[J]. Journal of Integrative Plant Biology, 2019, 61(6): 691-705. DOI:10.1111/jipb.12756 |
| [44] |
Feng X, Wang Y, Zhang N, et al. Genome-wide systematic characterization of the HAK/KUP/KT gene family and its expression profile during plant growth and in response to low-K+ stress in Saccharum[J]. BMC Plant Biology, 2020, 20: 20. DOI:10.1186/s12870-019-2227-7 |
| [45] |
张松.钾离子转运蛋白OsHAK5调控水稻株型机理的探究[D].南京: 南京农业大学, 2015. Zhang S. How does a potassium transporter OsHAK5 affect rice plant architecture[D]. Nanjing : Nanjing Agricultural University, 2015. |
| [46] |
Qin LJ, Zhao D, Zhang Y, et al. Selectable marker-free coexpression of Nicotiana rustica CN and Nicotiana tabacum HAK1 genes improves resistance to tobacco mosaic virus in tobacco[J]. Functional Plant Biology, 2015, 42(8): 802-815. DOI:10.1071/FP14356 |
| [47] |
Qi Z, Hampton CR, Shin R, et al. The high affinity K+ transporter AtHAK5 plays a physiological role in planta at very low K+ concentrations and provides a caesium uptake pathway in Arabidopsis[J]. Journal of Experimental Botany, 2008, 59(3): 595-607. DOI:10.1093/jxb/erm330 |
| [48] |
Zhao S, Zhang ML, Ma TL, et al. Phosphorylation of ARF2 relieves its repression of transcription of the K+ transporter gene HAK5 in response to low potassium stress[J]. The Plant Cell, 2016, 28(12): 3005-3019. DOI:10.1105/tpc.16.00684 |
| [49] |
Zhu JK. Salt and drought stress signal transduction in plants[J]. Annual Review of Plant Biology, 2002, 53(1): 247-273. DOI:10.1146/annurev.arplant.53.091401.143329 |
| [50] |
Chen G, Liu C, Gao Z, et al. OsHAK1, a high-affinity potassium transporter, positively regulates responses to drought stress in rice[J]. Frontiers in Plant Science, 2017, 8: 1885. DOI:10.3389/fpls.2017.01885 |
| [51] |
张祎, 秦利军, 赵丹, 赵德刚. 超量表达NtHAK1基因提高烟草干旱胁迫能力[J]. 植物生理学报, 2017, 53(8): 1444-1452. Zhang Y, Qin LJ, Zhso D, Zhao DG. Improvement of droughtstress in NtHAK1-overexpressing Nicotiana tabacum[J]. Plant Physiology Communications, 2017, 53(8): 1444-1452. |
| [52] |
Jiang Y, Qiu Y, Hu Y, et al. Heterologous expression of AtWRKY57 confers drought tolerance in Oryza sativa[J]. Frontiers in Plant Science, 2016, 7: 145. |
| [53] |
Zhang Z, Zhang J, Chen Y, et al. Genome-wide analysis and identification of HAK potassium transporter gene family in maize(Zeamays L.)[J]. Molecular Biology Reports, 2012, 39(8): 8465-8473. DOI:10.1007/s11033-012-1700-2 |
| [54] |
Assaha DV, Ueda A, Saneoka H, et al. The role of Na+ and K+ transporters in salt stress adaptation in glycophytes[J]. Frontiers in Physiology, 2017, 8: 509. DOI:10.3389/fphys.2017.00509 |
| [55] |
Horie T, Sugawara M, Okada T, et al. Rice sodium-insensitive potassium transporter, OsHAK5, confers increased salt tolerance in tobacco BY2 cells[J]. Journal of Bioscience and Bioengineering, 2011, 111(3): 346-356. DOI:10.1016/j.jbiosc.2010.10.014 |
| [56] |
Alemán F, Caballero F, Ródenas R, et al. The F130S point mutation in the Arabidopsis high-affinity K transporter AtHAK5 increases K over Na and Cs selectivity and confers Na and Cs tolerance to yeast under heterologous expression[J]. Frontiers in Plant Science, 2014, 5: 430. |
| [57] |
Maathuis FJ. The role of monovalent cation transporters in plant responses to salinity[J]. Journal of Experimental Botany, 2005, 57(5): 1137-1147. |
| [58] |
Yang H, Zhang W, Chai W, et al. PtHAK5, a candidate for mediating high-affinity K+ uptake in the halophytic grass, Puccinellia tenuiflora[J]. Frontiers of Agricultural Science and Engineering, 2018, 5(1): 108-117. DOI:10.15302/J-FASE-2018200 |
| [59] |
车文利, 张书玲, 吴立柱, 等. 转AlHAK1基因棉花耐盐能力分析[J]. 中国农业科技导报, 2015, 17(1): 49-56. Che WL, Zhang SL, Wu LZ, et al. Analysis of salt-tolerant ability in transgenic cotton(Gossypium hirsutum L.)with AlHAK1[J]. Journal of Agricultural Science and Technology, 2015, 17(1): 49-56. |
| [60] |
Walters DR, Bingham IJ. Influence of nutrition on disease development caused by fungal pathogens :implications for plant disease control[J]. Annals of Applied Biology, 2007, 151(3): 307-324. DOI:10.1111/j.1744-7348.2007.00176.x |
| [61] |
Römheld V, Kirkby EA. Research on potassium in agriculture :needs and prospects[J]. Plant and Soil, 2010, 335(1-2): 155-180. DOI:10.1007/s11104-010-0520-1 |
| [62] |
Fuchs WH, Grossmann F. Nutrition and resistance of crop plants against pathogens and pests[J]. Handbuch Der Pflanzenernahrung Und Dungung, 1972, 1. |
| [63] |
Jeworutzki E, Roelfsema MR, Anschütz U, et al. Early signaling through the Arabidopsis pattern recognition receptors FLS2 and EFR involves Ca2+-associated opening of plasma membrane anion channels[J]. The Plant Journal, 2010, 62(3): 367-378. DOI:10.1111/j.1365-313X.2010.04155.x |
| [64] |
Atkinson MM, Baker CJ. Alteration of plasmalemma sucrose transport in Phaseolus vulgaris by Pseudomonas syringae pv. syringae and its association with K+ /H+ exchange[J]. Phytopathology, 1987, 77(11): 1573-1578. DOI:10.1094/Phyto-77-1573 |
| [65] |
Magnan F, Ranty B, Charpenteau M, et al. Mutations in AtCML9, a calmodulin-like protein from Arabidopsis thaliana, alter plant responses to abiotic stress and abscisic acid[J]. The Plant Journal, 2008, 56(4): 575-589. DOI:10.1111/j.1365-313X.2008.03622.x |
| [66] |
Leba LJ, Cheval C, Ortiz-Martín I, et al. CML9, an Arabidopsis calmodulin-like protein, contributes to plant innate immunity through a flagellin-dependent signalling pathway[J]. The Plant Journal, 2012, 71(6): 976-989. DOI:10.1111/j.1365-313X.2012.05045.x |
| [67] |
Leba LJ, Perochon A, Cheval C, et al. CML9, a multifunctional Arabidopsis thaliana calmodulin-like protein involved in stress responses and plant growth?[J]. Plant Signaling and Behavior, 2012, 7(9): 1121-1124. DOI:10.4161/psb.21308 |
| [68] |
Brauer EK, Ahsan N, Dale R, et al. The Raf-like kinase ILK1 and the high affinity K+ transporter HAK5 are required for innate immunity and abiotic stress response[J]. Plant Physiology, 2016, 171(2): 1470-1484. |




