随着世界人口和经济的快速增长, 人们对于优质动物产品的需求量与日俱增。预计到2050年, 对于肉和蛋的需求将增加73%, 对奶制品的需求将增加58%[2]。为了满足人们对于优质动物产品的需求, 提高畜禽生产性能和效率是畜牧业发展的重中之重。长期以来, 抗生素不仅能够预防和治疗动物疾病, 还具有促进动物生长的作用[3]。预计2030年, 全球动物养殖使用的抗生素总量将提高到105 596 t, 且仅在亚洲就将达到51 851 t, 占全球使用总量的一半[4]。尽管适量或低剂量的抗生素能够预防疾病或促进动物生产, 但仍会导致抗药性病原菌的出现[5-6]。2019年7月, 中华人民共和国农业农村部发布第194号公告, 饲料无抗已迫在眉睫, 因此寻找合适的抗生素替代品, 成为动物生产的一项重要任务。
益生菌是一类对宿主(人类或动物)有益的活性微生物[7], 包括细菌、真菌(如酵母)等, 能够有效提高动物生产性能[8-9]和对疾病的抵抗力[10], 同时能够抑制耐药性细菌的生长[11-12], 在畜禽生产中具有替代抗生素的潜力。动物消化道中存在着大量的微生物, 包括细菌、原虫、病毒及真菌等[13]。与成年阶段相比, 新生或幼龄动物的消化道微生物更易受日粮等因素的影响, 且该影响作用持续时间更长久[14]。因此, 合理利用益生菌调控消化道微生物和宿主之间建立早期的积极互作, 对于促进其消化道免疫功能的建立、提高生产性能具有长效作用[15]。本文介绍了益生菌的潜在作用机制, 对近10年来利用高通量测序技术研究益生菌调控幼龄畜禽(仔猪、雏鸡和反刍动物)消化道微生物的进展进行了总结和归纳, 并提出了未来研究方向, 包括益生菌如何通过与消化道微生物互作影响其功能, 益生菌对于幼龄畜禽不同健康状态下肠道微生物的影响, 以及宿主因素如何影响益生菌对于幼龄畜禽消化道微生物的作用效果。
1 益生菌的潜在作用机制作为有效的益生菌, 必须保证其自身能够耐受消化道的特殊环境(温度、pH等), 能够附着在消化道上皮细胞上, 在此基础上, 产生抗病原微生物的物质, 并在消化道中发挥持续的益生作用[16]。益生菌在动物消化道中发挥作用的潜在机制主要包括3个方面:首先, 益生菌通过多种途径抑制病原微生物的生长, 具体表现在:(1)益生菌与病原微生物竞争有限的营养源、能量以及消化道结合位点, 从而限制病原微生物生长。青春双歧杆菌(Bifidobacterium adolescentis)S2-1能够与牙龈卟啉单胞菌(Porphyromonas gingivalis)竞争维生素K, 从而抑制其生长[17]。(2)益生菌通过产生抗微生物成分包括细菌素、有机酸、过氧化氢等, 抑制病原微生物生长, 如卷曲乳杆菌(Lactobacillus crispatus) F117和副干酪乳杆菌(Lactobacillus paracasei) F2/F28均能够通过产生大量的过氧化氢来抑制金黄色葡萄球菌(Staphylococcus aureus)的生长[18]。此外, 益生菌产生的乳酸、有机酸能够通过降低消化道pH, 从而抑制沙门氏菌和大肠杆菌等病原微生物的生长[19]。其次, 益生菌能够作用于宿主消化道上皮细胞和树突细胞, 促进上述细胞产生消化道黏液或防御素[20-21], 提高紧密连接以及屏障功能[22], 防止促炎性细胞因子导致的细胞凋亡[23]。主要体现在:乳杆菌[24-25]、链球菌[26]及双歧杆菌[27]等益生菌能够通过纤毛附着在宿主消化道细胞上。在成功黏附后, 肽聚糖(革兰氏阳性菌)和脂多糖(革兰氏阴性菌)等益生菌细胞壁成分与消化道上皮细胞和树突细胞模式识别受体相互作用, 促使后者产生黏液或防御素[28]。此外, 益生菌产生的细菌黏附素如黏液结合蛋白也有利于其结合宿主树突细胞, 从而发挥其吞噬作用[29]。第三, 益生菌可能通过调控宿主特异性免疫功能来发挥作用, 主要表现在乳酸菌对肠淋巴组织的调控作用[30]。乳酸菌的免疫活性不仅体现在调控Toll样受体(TLR)表达、激活树突细胞和自然杀伤细菌等非特异性免疫反应, 还体现在调控调节性T细胞(Treg)应答以及特异性免疫球蛋白A的分泌等免疫反应[31]。例如, 脆弱拟杆菌(Bacteroides fragilis)[32]及梭菌属(Clostridium) IV[33-34]等能够提高无菌小鼠结肠Treg细胞的数量; 婴儿型双歧杆菌(Bifidobacterium infantis)能够促进感染沙门氏菌的小鼠消化道Treg细胞的生成[35]; 健康的人服用婴儿型双歧杆菌35624后, 其Treg细胞表达量提高。此外, 其外周血液中的白介素-10分泌功能增加[36]。尽管在人类和实验动物上的研究一定程度上揭示了益生菌的作用机制, 但上述机制是否同样能够解释益生菌与畜禽消化道微生物的相互作用, 还有待于进一步探索。
2 益生菌调控幼龄畜禽消化道微生物的研究进展基于高通量测序技术研究益生菌调控幼龄畜禽消化道微生物的报道主要集中在近10年, 但总体研究数量仍比较少。在仔猪和雏鸡上研究应用较多的益生菌包括乳杆菌属、肠球菌属及芽孢杆菌属等; 在幼龄反刍动物上研究应用较多的益生菌为酵母。上述益生菌的主要益生作用体现在提高幼龄畜禽消化道有益微生物的丰度, 降低有害微生物的丰度。但在不同动物品种之间, 具体益生作用有较大的区别, 一方面是由于不同动物使用的益生菌菌种/株存在差异; 另一方面是由于不同动物消化道微生物存在差异。因此, 本节将围绕不同益生菌对不同品种的幼龄动物(仔猪、雏鸡及反刍动物)消化道微生物组成的影响作具体阐述。
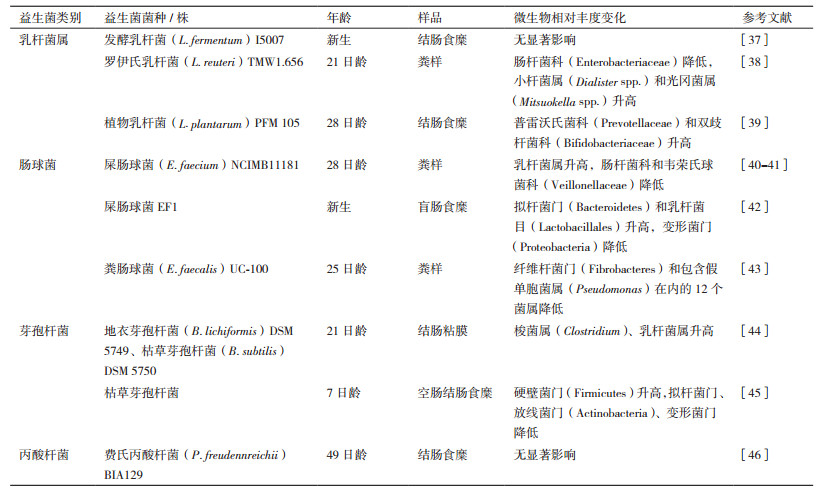
2.1 益生菌调控仔猪消化道微生物的研究在仔猪上研究应用的益生菌主要包括乳杆菌属(Lactobacillus)[37-39]、肠球菌属(Enterococcus)[40-43]、芽孢杆菌属(Bacillus)[44-45]以及丙酸杆菌属(Propionibacterium)[46] (表 1)。上述益生菌调控仔猪消化道微生物的主要作用包括提高了普雷沃氏菌科(Prevotellaceae)[39]、双歧杆菌科(Bifidobacteriaceae)[39]、小杆菌属(Dialister spp.)[38]和光冈菌属(Mitsuokella spp.)[38]、乳杆菌目/属[40, 42, 44]和梭菌属(Clostridium)的相对丰度。其中, 普遍认为普雷沃氏菌是畜禽消化道中的一类有益微生物, 与分解植物源的饲料成分密切相关[47]; 双歧杆菌也是人和动物消化道菌群的重要组成成员之一, 其能够分解碳水化合物生成乳酸, 对于维持消化道健康和菌群稳定具有重要作用[48]; 小杆菌属与消化道短链脂肪酸产量呈正相关, 对于维持消化道免疫功能具有重要作用[49]; 光冈菌属中的M. jalaludinii菌种具有分解植酸磷的作用[50]。除提高有益微生物的相对丰度外, 上述益生菌能够降低肠杆菌科(Enterobacteriaceae)[38]、韦荣氏球菌科(Veillonellaceae)[41]、假单胞菌属(Pseudomonas)[43]、变形菌门(Proteobacteria)和放线菌门(Actinobacteria)的相对丰度。肠杆菌科包含的大肠杆菌(Escherichia coli)存在多个致病性菌株, 如大肠杆菌O157:H7和S102-9, 可引起动物感染性腹泻或肠炎[51-52]; 韦荣氏球菌科可能会引起消化道胆酸浓度过高, 影响消化道健康, 也是炎症性肠炎的标记物之一[53-54]。然而也有研究表明, 添加益生菌对仔猪消化道微生物无显著影响[37, 46], 说明了益生菌和宿主及微生物之间互作的复杂性, 受到益生菌的类型、添加方法、剂量及动物因素(如品种、饲养方式和试验周期)等的影响。
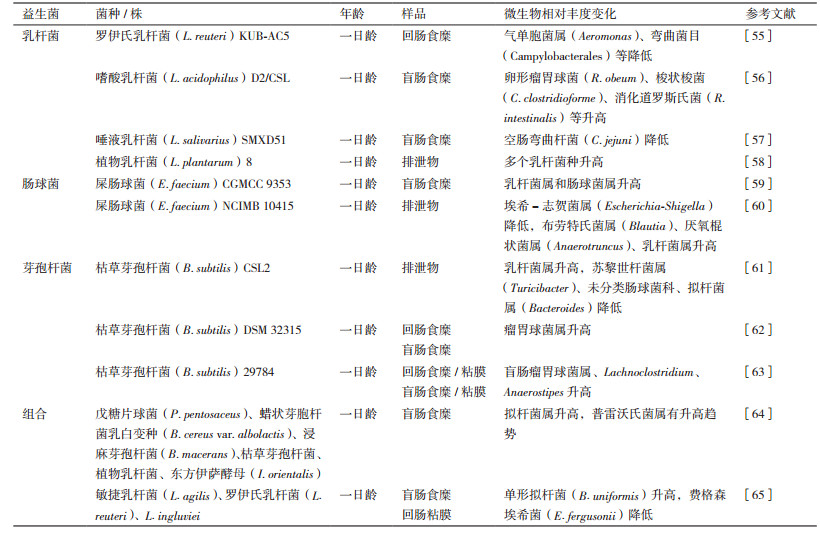
与仔猪上的研究类似, 肉鸡饲粮中常用的益生菌包括乳杆菌[55-58]、肠球菌[59-60]、芽孢杆菌[61-63]及多菌种的组合[64-65] (表 2)。益生菌对雏鸡消化道微生物的影响主要体现在降低了潜在病原微生物的相对丰度, 如罗伊氏乳杆菌KUB-AC5降低了回肠食糜气单胞菌属(Aeromonas)和弯曲杆菌目(Campylobacterales)的相对丰度[55]; 屎肠球菌(E. faecium)NCIMB 10415降低了排泄物中埃希-志贺菌属(Escherichia-Shigella)[60]。气单胞菌属和弯曲菌目是鸡肉中常见的病原微生物[66], 弯曲杆菌属是最常见的食物源性病原微生物[67], 能够引起一系列人类消化道疾病, 而家禽及其产品是重要的弯曲杆菌感染源[68]。此外, 埃希-志贺菌属也包含了大量潜在病原微生物, 如上述致病性大肠杆菌(E. coli O157:H7、E. coli S102-9), 因此, 降低这些潜在病原微生物的相对丰度, 对于降低家禽消化道疾病风险, 提高其免疫性能及生产性能具有重要意义。另一方面, 添加益生菌后, 乳杆菌、瘤胃球菌、肠球菌等丰度升高。上述益生作用可能与益生菌促进消化道免疫功能有关, 如添加乳杆菌[58, 69]、肠球菌[59, 70], 梭菌属[71]或复合益生菌能够提高雏鸡血清免疫球蛋白M、A、G的浓度, 进而降低了致病菌的数量。此外, 添加益生菌能够提高雏鸡空肠黏膜免疫型免疫球蛋白A[72]、杯状细菌数量和中性黏液的产量[73-74], 上述免疫相关物质均能够保护禽类肠黏膜免受有害菌的破坏, 从而保持黏膜稳态[70]。
幼龄反刍动物由于瘤胃尚未完全发育, 其消化道结构类似于单胃动物[75]。研究表明, 犊牛出生后1-3 d内, 瘤胃中已经存在以变形菌门、硬壁菌门和拟杆菌门为主的大量微生物[76], 说明这些微生物可能在瘤胃发育及功能完善的过程中起到重要作用。饲粮因素如益生菌对幼龄反刍动物消化道微生物的研究仍较少, 研究表明, 代乳粉中添加5种益生菌组合, 包括解淀粉芽胞杆菌(B. amyloliquefaciens) H57、小孔芽孢杆菌(B. foraminis)VTM4R85、坚强芽孢杆菌(B. firmus)VTM2R84、地衣芽孢杆菌(B. licheniformis)VTM2R66、地衣芽孢杆菌VTM1R74和腐生葡萄球菌(Staphylococcus saprophyticus bovis) VTM1R96能够提高新生羔羊瘤胃液中普雷沃氏菌科、瘤胃球菌科相对丰度, 降低毛螺菌科的相对丰度[77]; 新生犊牛代乳粉中添加布拉迪酵母(S. boulardii)CNCM I-1079, 瘤胃液中只有瘤胃球菌科UCG-008相对丰度显著降低, 其他微生物相对丰度没有变化。此外, 布拉迪酵母CNCM I-1079[78]显著降低了盲肠黏膜链球菌属(Streptococcus)的相对丰度, 链球菌属中有多个菌种具有致病性, 如酿脓链球菌(S. pyogenes)和肺炎链球菌(S. pneumoniae); 降低了回肠黏膜艰难梭菌属(Peptoclostridium)的相对丰度, 艰难梭菌属是引起腹泻的一种病原菌[79]; 提高了回肠黏膜纤维杆菌属(Fibrobacter)、罗斯氏菌属(Roseburia)和欧氏菌属(Olsenella)的相对丰度, 罗斯氏菌是一种产丁酸菌, 不仅能够为消化道细胞提供能量, 而且还具有抗炎症的作用, 有利于维持正常免疫功能[80-81]; 欧氏菌是一种乳酸菌, 具有耐胆汁酸的特性且能够利用黏蛋白[82-83]。但也有研究表明, 或布拉迪酵母CNCM I-1079对犊牛粪便微生物无显著影响[84], 这可能是由于粪便微生物并不能够代表整个消化道的微生物变化情况。相比较在仔猪和雏鸡上开展的大量益生菌调控消化道微生物的效果研究, 益生菌调控幼龄反刍动物的研究仍处在起步阶段。基于此, 有必要进一步开展相关研究, 为合理有效使用益生菌促进幼龄反刍动物长期健康和提高其生产性能奠定基础。
3 未来研究方向大量研究表明, 益生菌具有替代抗生素的潜力。但由于目前对于益生菌的作用机制尚未完全明确, 因此在一定程度上限制了益生菌的合理有效利用。为解决这一问题, 今后需要更多研究来阐明益生菌与消化道微生物互作, 以及这种互作如何进一步影响畜禽消化道功能和健康及生产性能。
第一, 除了明确益生菌对消化道微生物组成的影响之外, 还需要进一步明确其对消化道微生物功能的影响, 因为最终在生态系统中起重要作用的是微生物功能, 而不是其组成[85]。事实上, 已有研究表明环境微生物的功能与其组成相比更具有保守性和相关性[86-87]。由此可推断, 益生菌引起的微生物组成上的变化并不一定能够引起或反映功能的变化。因此, 今后需要更多基于宏基因组或宏转录组的研究, 来探索益生益引起的微生物相关基因或转录因子的变化, 从而帮助我们更好地理解益生菌和微生物之间的互作。
第二, 益生菌的益生作用不应仅限于在健康动物上的研究, 还需要研究在非正常状态(如应激/疾病)下的作用效果, 在鸡[88]、猪[89]和犊牛[90]上的研究表明, 益生菌对于健康动物消化道微生物的作用效果并不一定能在应激或疾病状态下表现出来, 这可能是因为在不同健康状态下动物消化道微生物群落也存在差异, 因此需要明确在不同健康状态下动物消化道的核心微生物组成, 从而为精确筛选益生菌菌株, 有效调控消化道微生物, 为治疗疾病或缓解应激奠定基础。
第三, 相比在仔猪和雏鸡上面的研究, 益生菌对幼龄反刍动物消化道微生物的影响仍较为有限。反刍动物拥有独特的消化道结构, 瘤胃微生物能够发酵利用高纤维素含量的饲料[91]; 另一方面, 由于瘤胃微生物的存在, 外源益生菌可能由于竞争或不适应瘤胃特殊的环境(温度、pH)而无法定植[92], 因此有必要充分研究益生菌和瘤胃微生物的互作, 从而开发能够适应瘤胃环境且能有效调控瘤胃发酵功能的益生菌。
第四, 除微生物因素外, 宿主遗传因素以及环境因素也会影响益生菌的作用效果, 在小鼠上的研究表明, 上述因素是造成微生物个体差异的重要原因[93]。近年来研究也表明了宿主遗传因素对牛瘤胃微生物的个体差异存在显著影响[94-95], 在牛上有些可遗传的微生物与SNP(单核苷酸多态性)存在相关性[96]; 当然, 也有研究表明鸡的消化道微生物不受宿主遗传因素(SNP)的影响[97], 造成上述研究结果存在差异的原因仍不明确, 因此还需要继续探索环境(如养殖场和饲粮)和宿主遗传因素(如品种和性别)如何决定个体微生物差异, 以及后者如何影响益生菌调控效果, 从而有针对性地筛选益生菌, 促进动物健康和生产性能。
4 小结益生菌能够有效提高畜禽健康和生长性能, 尽管其调控机制尚未明确, 但益生菌和消化道微生物互作可能很大程度上决定了益生菌的有益作用效果。目前关于益生菌和幼龄畜禽消化道微生物互作的研究非常有限, 在一定程度上限制了益生菌的有效应用。组学技术的发展, 为更深入了解益生菌的作用效果和机制提供了可能, 因此, 今后有必要利用多组学技术, 针对性地研究具体的益生菌菌株、宿主因素等如何影响幼龄畜禽消化道微生物的组成和功能。
| [1] |
Northoff E. 2050 A third more mouths to feed[N]. Food and Agriculture Organization of the United Nations, 2016.
|
| [2] |
McLeod A. World livestock 2011-livestock in food security[N]. Food and Agriculture Organization of the United Nations, 2011.
|
| [3] |
Marshall BM, Levy SB. Food animals and antimicrobials:impacts on human health[J]. Clinical Microbiology Reviews, 2011, 24: 718-733. DOI:10.1128/CMR.00002-11 |
| [4] |
Van Boeckel TP, Brower C, Gilbert M, et al. Global trends in antimicrobial use in food animals[J]. Proceedings of the National Academy of Sciences of the United States of America, 2015, 112: 5649-5654. DOI:10.1073/pnas.1503141112 |
| [5] |
Hao H, Cheng G, Iqbal Z, et al. Benefits and risks of antimicrobial use in food-producing animals[J]. Frontiers in Microbiolology, 2014, 5: 288. |
| [6] |
Lekshmi M, Ammini P, Kumar S, et al. The food production environment and the development of antimicrobial resistance in human pathogens of animal origin[J]. Microorganisms, 2017, 5: 11. DOI:10.3390/microorganisms5010011 |
| [7] |
Hill C, Guarner F, Reid G, et al. Expert consensus document:The international scientific association for probiotics and prebiotics consensus statement on the scope and appropriate use of the term probiotic[J]. Nature Reviews Gastroenterology & Hepatology, 2014, 11: 506-514. |
| [8] |
Timmerman HM, Mulder L, Everts H, et al. Health and growth of veal calves fed milk replacers with or without probiotics[J]. Journal of Dairy Science, 2005, 88: 2154-2165. DOI:10.3168/jds.S0022-0302(05)72891-5 |
| [9] |
Böhmer BM, Kramer W, Roth-Maier DA. Dietary probiotic supplementation and resulting effects on performance, health status, and microbial characteristics of primiparous sows[J]. Journal of Animal Physiology and Animal Nutrtion, 2006, 90: 309-315. DOI:10.1111/j.1439-0396.2005.00601.x |
| [10] |
Mappley LJ, Tchórzewska MA, Nunez A, et al. Oral treatment of chickens with Lactobacillus reuteri LM1 reduces Brachyspira pilosicoli-induced pathology[J]. Journal of Medical Microbiology, 2013, 62: 287-296. DOI:10.1099/jmm.0.051862-0 |
| [11] |
Muñoz-Atienza E, Gómez-Sala B, Araújo C, et al. Antimicrobial activity, antibiotic susceptibility and virulence factors of lactic acid bacteria of aquatic origin intended for use as probiotics in aquaculture[J]. BMC Microbiology, 2013, 13: 15. DOI:10.1186/1471-2180-13-15 |
| [12] |
Varankovich NV, Nickerson MT, Korber DR. Probiotic-based strategies for therapeutic and prophylactic use against multiple gastrointestinal diseases[J]. Frontiers in Microbiology, 2015, 6: 685. |
| [13] |
Taneja V. Microbiome: Impact of gender on function & characteristics of gut microbiome[M]//Principles of GenderSpecific Medicine. Academic Press, 2017: 569-583.
|
| [14] |
Abecia L, Martín-García AI, Martínez G, et al. Nutritional intervention in early life to manipulate rumen microbial colonization and methane output by kid goats postweaning[J]. Journal of Animal Science, 2013, 91: 4832-4840. DOI:10.2527/jas.2012-6142 |
| [15] |
Shen X, Liu L, Peek RM, et al. Supplementation of p40, a Lactobacillus rhamnosus GG-derived protein, in early life promotes epidermal growth factor receptor-dependent intestinal development and long-term health outcomes[J]. Mucosal Immunology, 2018, 11: 1316-1328. DOI:10.1038/s41385-018-0034-3 |
| [16] |
Angelakis E. Weight gain by gut microbiota manipulation in productive animals[J]. Microbial Pathogenesis, 2017, 106: 162-170. DOI:10.1016/j.micpath.2016.11.002 |
| [17] |
Hojo K, Nagaoka S, Murata S, et al. Reduction of vitamin K concentration by salivary Bifidobacterium strains and their possible nutritional competition with Porphyromonas gingivalis[J]. Journal of Applied Microbiology, 2007, 103: 1969-1974. DOI:10.1111/j.1365-2672.2007.03436.x |
| [18] |
Ocaña VS, Pesce de Ruiz Holgado AA, Nader-Macías ME. Selection of vaginal H2O2-generating Lactobacillus species for probiotic use[J]. Current Microbiology, 1999, 38: 279-284. DOI:10.1007/PL00006802 |
| [19] |
Bermudez-Brito M, Muñoz-Quezada S, Gomez-Llorente C, et al. Human intestinal dendritic cells decrease cytokine release against Salmonella infection in the presence of Lactobacillus paracasei upon TLR activation[J]. PLoS One, 2012, 7: e43197. DOI:10.1371/journal.pone.0043197 |
| [20] |
Schlee M, Harder J, Köten B, et al. Probiotic lactobacilli and VSL#3 induce enterocyte β-defensin 2[J]. Clinical and Experimental Immunology, 2008, 151: 528-535. DOI:10.1111/j.1365-2249.2007.03587.x |
| [21] |
Schlee M, Wehkamp J, Altenhoefer A, et al. Induction of human β-defensin 2 by the probiotic Escherichia coli Nissle 1917 is mediated through flagellin[J]. Infection and Immunity, 2007, 75: 2399-2407. DOI:10.1128/IAI.01563-06 |
| [22] |
Seth A, Yan F, Polk DB, et al. Probiotics ameliorate the hydrogen peroxide-induced epithelial barrier disruption by a PKC-and MAP kinase-dependent mechanism[J]. American Journal of Physiology-Gastrointestinal and Liver Physiology, 2008, 294: G1060-G1069. DOI:10.1152/ajpgi.00202.2007 |
| [23] |
Yan F, Cao H, Cover TL, et al. Soluble proteins produced by probiotic bacteria regulate intestinal epithelial cell survival and growth[J]. Gastroenterology, 2007, 132: 562-575. DOI:10.1053/j.gastro.2006.11.022 |
| [24] |
Kankainen M, Paulin L, Tynkkynen S, et al. Comparative genomic analysis of Lactobacillus rhamnosus GG reveals pili containing a human-mucus binding protein[J]. Proceedings of the National Academy of Sciences of the United States of America, 2009, 106: 17193-17198. DOI:10.1073/pnas.0908876106 |
| [25] |
Mack DR, Michail S, Wei S, et al. Probiotics inhibit enteropathogenic E. coli adherence in vitro by inducing intestinal mucin gene expression[J]. American Journal of Physiology-Gastrointestinal and Liver Physiology, 1999, 276: G941-G950. DOI:10.1152/ajpgi.1999.276.4.G941 |
| [26] |
Brittan JL, Nobbs AH. Group B Streptococcus pili mediate adherence to salivary glycoproteins[J]. Microbes and Infection, 2015, 17: 360-368. DOI:10.1016/j.micinf.2014.12.013 |
| [27] |
Turroni F, Serafini F, Mangifesta M, et al. Expression of sortasedependent pili of Bifidobacterium bifidum PRL2010 in response to environmental gut conditions[J]. FEMS Microbiology Letters, 2014, 357: 23-33. DOI:10.1111/1574-6968.12509 |
| [28] |
Lebeer S, Vanderleyden J, De Keersmaecker SCJ. Host interactions of probiotic bacterial surface molecules:comparison with commensals and pathogens[J]. Nature Reviews Microbiology, 2010, 8: 171-184. DOI:10.1038/nrmicro2297 |
| [29] |
Bene KP, Kavanaugh DW, Leclaire C, et al. Lactobacillus reuteri surface mucus adhesins upregulate inflammatory responses through interactions with innate C-type lectin receptors[J]. Frontiers in Microbiology, 2017, 8: 321. |
| [30] |
Famularo G, Moretti S, Marcellini S, et al. Stimulation of immunity by probiotics[M]//Probiotics 2. Dordrecht: Springer, 1997: 133-161.
|
| [31] |
Tsai YT, Cheng PC, Pan TM. The immunomodulatory effects of lactic acid bacteria for improving immune functions and benefits[J]. Applied Microbiology and Biotechnology, 2012, 96: 853-862. DOI:10.1007/s00253-012-4407-3 |
| [32] |
Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota[J]. Proceedings of the National Academy of Sciences of the United States of America, 2010, 107: 12204-12209. DOI:10.1073/pnas.0909122107 |
| [33] |
Atarashi K, Tanoue T, Oshima K, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota[J]. Nature, 2013, 500: 232-236. DOI:10.1038/nature12331 |
| [34] |
Atarashi K, Tanoue T, Shima T, et al. Induction of colonic regulatory T cells by indigenous Clostridium species[J]. Science, 2011, 331: 337-341. DOI:10.1126/science.1198469 |
| [35] |
O'Mahony C, Scully P, O'Mahony D, et al. Commensal-induced regulatory T cells mediate protection against pathogen-stimulated NF-κB activation[J]. PLoS Pathogens, 2008, 4: e1000112. DOI:10.1371/journal.ppat.1000112 |
| [36] |
Konieczna P, Groeger D, Ziegler M, et al. Bifidobacterium infantis 35624 administration induces Foxp3 T regulatory cells in human peripheral blood:potential role for myeloid and plasmacytoid dendritic cells[J]. Gut, 2012, 61: 354-366. DOI:10.1136/gutjnl-2011-300936 |
| [37] |
Liu H, Hou C, Wang G, et al. Lactobacillus reuteri I5007 modulates intestinal host defense peptide expression in the model of IPEC-J2 cells and neonatal piglets[J]. Nutrients, 2017, 9: 559. DOI:10.3390/nu9060559 |
| [38] |
Yang Y, Zhao X, Minh HAL, et al. Reutericyclin producing Lactobacillus reuteri modulates development of fecal microbiota in weanling pigs[J]. Frontiers in Microbiology, 2015, 6: 762. |
| [39] |
Wang T, Teng K, Liu Y, et al. Lactobacillus plantarum PFM 105 promotes intestinal development through modulation of gut microbiota in weaning piglets[J]. Frontiers in Microbiology, 2019, 10: 90. DOI:10.3389/fmicb.2019.00090 |
| [40] |
Pajarillo EAB, Chae JP, Balolong MP, et al. Effects of probiotic Enterococcus faecium NCIMB 11181 administration on swine fecal microbiota diversity and composition using barcoded pyrosequencing[J]. Animal Feed Science and Technology, 2015, 201: 80-88. DOI:10.1016/j.anifeedsci.2015.01.011 |
| [41] |
Chae JP, Pajarillo EAB, Oh JK, et al. Revealing the combined effects of lactulose and probiotic enterococci on the swine fecal microbiota using 454 pyrosequencing[J]. Microbial Biotechnology, 2016, 9: 486-495. DOI:10.1111/1751-7915.12370 |
| [42] |
Wang YB, Du W, Fu AK, et al. Intestinal microbiota and oral administration of Enterococcus faecium associated with the growth performance of new-born piglets[J]. Beneficial Microbes, 2016, 7: 529-538. DOI:10.3920/BM2015.0099 |
| [43] |
Li P, Niu Q, Wei Q, et al. Microbial shifts in the porcine distal gut in response to diets supplemented with Enterococcus faecalis as alternatives to antibiotics[J]. Scientific Reports, 2017, 7: 41395. DOI:10.1038/srep41395 |
| [44] |
Zhang W, Zhu YH, Zhou D, et al. Oral administration of a select mixture of Bacillus probiotics affects the gut microbiota and goblet cell function following Escherichia coli challenge in newly weaned pigs of genotype MUC4 that are supposed to be enterotoxigenic E. coli F4ab/ac receptor negative[J]. Applied and Environmental Microbiology, 2017, 83(3): e02747-16. |
| [45] |
He Y, Mao C, Wen H, et al. Influence of ad libitum feeding of piglets with bacillus subtilis fermented liquid feed on gut flora, luminal contents and health[J]. Scientific Reports, 2017, 7: 44553. DOI:10.1038/srep44553 |
| [46] |
Cousin FJ, Foligne B, Deutsch SM, et al. Assessment of the probiotic potential of a dairy product fermented by Propionibacterium freudenreichii in piglets[J]. Journal of Agricultural and Food Chemistry, 2012, 60: 7917-7927. DOI:10.1021/jf302245m |
| [47] |
Ley RE. Gut microbiota in 2015:Prevotella in the gut:choose carefully[J]. Nature Reviews Gastroenterology & Hepatology, 2016, 13: 69-70. |
| [48] |
Benno Y, Mitsuoka T. Impact of Bifidobacterium longum on human fecal microflora[J]. Microbiology and Immunology, 1992, 36: 683-694. DOI:10.1111/j.1348-0421.1992.tb02071.x |
| [49] |
Umu ÖCO, Fauske AK, Åkesson CP, et al. Gut microbiota profiling in Norwegian weaner pigs reveals potentially beneficial effects of a high-fiber rapeseed diet[J]. PLoS One, 2018, 13: e0209439. DOI:10.1371/journal.pone.0209439 |
| [50] |
Lan GQ, Ho YW, Abdullah N. Mitsuokella jalaludinii sp. nov., from the rumens of cattle in Malaysia[J]. International Journal of Systematic and Evolutionary Microbiology, 2002, 52: 713-718. |
| [51] |
Chanter N, Hall GA, Bland AP, et al. Dysentery in calves caused by an atypical strain of Escherichia coli(S102-9)[J]. Veterinary Microbiology, 1986, 12: 241-253. DOI:10.1016/0378-1135(86)90053-2 |
| [52] |
Gally DL, Stevens MP. Microbe profile:Escherichia coli O157:H7-notorious relative of the microbiologist's workhorse[J]. Microbiology, 2017, 163(1): 1-3. |
| [53] |
Strauss J, Kaplan GG, Beck PL, et al. Invasive potential of gut mucosa-derived Fusobacterium nucleatum positively correlates with IBD status of the host[J]. Inflammatory Bowel Diseases, 2011, 17: 1971-1978. DOI:10.1002/ibd.21606 |
| [54] |
Gevers D, Kugathasan S, Denson LA, et al. The treatment-naive microbiome in new-onset Crohn's disease[J]. Cell Host & Microbe, 2014, 15: 382-392. |
| [55] |
Nakphaichit M, Thanomwongwattana S, Phraephaisarn C, et al. The effect of including Lactobacillus reuteri KUB-AC5 during post-hatch feeding on the growth and ileum microbiota of broiler chickens[J]. Poultry Science, 2011, 90: 2753-2765. DOI:10.3382/ps.2011-01637 |
| [56] |
De Cesare A, Sirri F, Manfreda G, et al. Effect of dietary supplementation with Lactobacillus acidophilus D2/CSL(CECT 4529)on caecum microbioma and productive performance in broiler chickens[J]. PLoS One, 2017, 12(5): e0176309. DOI:10.1371/journal.pone.0176309 |
| [57] |
Saint-Cyr MJ, Haddad N, Taminiau B, et al. Use of the potential probiotic strain Lactobacillus salivarius SMXD51 to control Campylobacter jejuni in broilers[J]. International Journal of Food Microbiology, 2017, 247: 9-17. DOI:10.1016/j.ijfoodmicro.2016.07.003 |
| [58] |
Gao P, Ma C, Sun Z, et al. Feed-additive probiotics accelerate yet antibiotics delay intestinal microbiota maturation in broiler chicken[J]. Microbiome, 2017, 5: 91. DOI:10.1186/s40168-017-0315-1 |
| [59] |
Zhang L, Li J, Yun TT, et al. Effects of pre-encapsulated and proencapsulated Enterococcus faecalis on growth performance, blood characteristics, and cal microflora in broiler chickens[J]. Poultry Science, 2015, 94: 2821-2830. DOI:10.3382/ps/pev262 |
| [60] |
Beirão BCB, Ingberman M, Fávaro Jr C, et al. Effect of an Enterococcus faecium probiotic on specific IgA following live Salmonella enteritidis vaccination of layer chickens[J]. Avian Pathology, 2018, 47: 325-333. DOI:10.1080/03079457.2018.1450487 |
| [61] |
Oh JK, Pajarillo EAB, Chae JP, et al. Effects of Bacillus subtilis CSL2 on the composition and functional diversity of the faecal microbiota of broiler chickens challenged with Salmonella gallinarum[J]. Journal of Animal Science and Biotechnology, 2017, 8: 1. DOI:10.1186/s40104-016-0130-8 |
| [62] |
Bortoluzzi C, Serpa Vieira B, de Paula Dorigam JC, et al. Bacillus subtilis DSM 32315 supplementation attenuates the effects of Clostridium perfringens challenge on the growth performance and intestinal microbiota of broiler chickens[J]. Microorganisms, 2019, 7: 71. DOI:10.3390/microorganisms7030071 |
| [63] |
Jacquier V, Nelson A, Jlali M, et al. Bacillus subtilis 29784 induces a shift in broiler gut microbiome toward butyrate-producing bacteria and improves intestinal histomorphology and animal performance[J]. Poultry Science, 2019, 98: 2548-2554. DOI:10.3382/ps/pey602 |
| [64] |
Wang Y, Sun J, Zhong H, et al. Effect of probiotics on the meat flavour and gut microbiota of chicken[J]. Scientific Reports, 2017, 7: 6400. DOI:10.1038/s41598-017-06677-z |
| [65] |
Baldwin S, Hughes RJ, Van TTH, et al. At-hatch administration of probiotic to chickens can introduce beneficial changes in gut microbiota[J]. PLoS One, 2018, 13: e0194825. DOI:10.1371/journal.pone.0194825 |
| [66] |
Willis WL, Reid L. Investigating the effects of dietary probiotic feeding regimens on broiler chicken production and Campylobacter jejuni presence[J]. Poultry Science, 2008, 87: 606-611. DOI:10.3382/ps.2006-00458 |
| [67] |
Kilonzo-Nthenge A, Nahashon SN, Chen F, et al. Prevalence and antimicrobial resistance of pathogenic bacteria in chicken and guinea fowl[J]. Poultry Science, 2008, 87: 1841-1848. DOI:10.3382/ps.2007-00156 |
| [68] |
Bull SA, Allen VM, Dominque G, et al. Sources of Campylobacter spp. colonizing housed broiler flocks during rearing[J]. Applied and Environmental Microbiology, 2006, 72: 645-652. DOI:10.1128/AEM.72.1.645-652.2006 |
| [69] |
Koenen ME, Kramer J, Van Der Hulst R, et al. Immunomodulation by probiotic lactobacilli in layer-and meat-type chickens[J]. British Poultry Science, 2004, 45: 355-366. DOI:10.1080/00071660410001730851 |
| [70] |
Zhang Q, Eicher SD, Applegate TJ. Development of intestinal mucin 2, IgA, and polymeric Ig receptor expressions in broiler chickens and Pekin ducks[J]. Poultry Science, 2015, 94: 172-180. DOI:10.3382/ps/peu064 |
| [71] |
Yang CM, Cao GT, Ferket PR, et al. Effects of probiotic, Clostridium butyricum, on growth performance, immune function, and cecal microflora in broiler chickens[J]. Poultry Science, 2012, 91: 2121-2129. DOI:10.3382/ps.2011-02131 |
| [72] |
Peng Q, Zeng XF, Zhu JL, et al. Effects of dietary Lactobacillus plantarum B1 on growth performance, intestinal microbiota, and short chain fatty acid profiles in broiler chickens[J]. Poultry Science, 2016, 95: 893-900. DOI:10.3382/ps/pev435 |
| [73] |
Martínez EA, Babot JD, Lorenzo-Pisarello MJ, et al. Feed supplementation with avian Propionibacterium acidipropionici contributes to mucosa development in early stages of rearing broiler chickens[J]. Beneficial Microbes, 2016, 7: 687-698. DOI:10.3920/BM2016.0077 |
| [74] |
Forte C, Manuali E, Abbate Y, et al. Dietary Lactobacillus acidophilus positively influences growth performance, gut morphology, and gut microbiology in rurally reared chickens[J]. Poultry Science, 2018, 97: 930-936. DOI:10.3382/ps/pex396 |
| [75] |
Baldwin RLVI, McLeod KR, Klotz JL, et al. Rumen development, intestinal growth and hepatic metabolism in the pre-and postweaning ruminant[J]. Journal of Dairy Science, 2004, 87: 55-65. DOI:10.3168/jds.S0022-0302(04)70061-2 |
| [76] |
Jami E, Israel A, Kotser A, et al. Exploring the bovine rumen bacterial community from birth to adulthood[J]. The ISME Journal, 2013, 7: 1069-1079. DOI:10.1038/ismej.2013.2 |
| [77] |
Ishaq SL, Kim CJ, Reis D, et al. Fibrolytic bacteria isolated from the rumen of North American moose(Alces alces)and their use as a probiotic in neonatal lambs[J]. PLoS One, 2015, 10: e0144804. DOI:10.1371/journal.pone.0144804 |
| [78] |
Fomenky BE, Do DN, Talbot G, et al. Direct-fed microbial supplementation influences the bacteria community composition of the gastrointestinal tract of pre-and post-weaned calves[J]. Scientific Reports, 2018, 8: 14147. DOI:10.1038/s41598-018-32375-5 |
| [79] |
Luo Y, Huang C, Ye J, et al. Genome sequence and analysis of Peptoclostridium difficile strain ZJCDC-S82[J]. Evolutionary Bioinformatics, 2016, 12: 41-49. |
| [80] |
Duncan SH, Hold GL, Barcenilla A, et al. Roseburia intestinalis sp. nov., a novel saccharolytic, butyrate-producing bacterium from human faeces[J]. International Journal of Systematic and Evolutionary Microbiology, 2002, 52: 1615-1620. |
| [81] |
Tamanai-Shacoori Z, Smida I, Bousarghin L, et al. Roseburia spp.:a marker of health[J]. Future Microbiology, 2017, 12: 157-170. DOI:10.2217/fmb-2016-0130 |
| [82] |
Kraatz M, Wallace RJ, Svensson L. Olsenella umbonata sp. nov., a microaerotolerant anaerobic lactic acid bacterium from the sheep rumen and pig jejunum, and emended descriptions of Olsenella, Olsenella uli and Olsenella profuse[J]. International Journal of Systematic and Evolutionary Microbiology, 2011, 61: 795-803. DOI:10.1099/ijs.0.022954-0 |
| [83] |
Dewhirst FE, Paster BJ, Tzellas N, et al. Characterization of novel human oral isolates and cloned 16S rDNA sequences that fall in the family Coriobacteriaceae:description of olsenella gen. nov., reclassification of Lactobacillus uli as Olsenella uli comb. nov. and description of Olsenella profusa sp. nov[J]. International Journal of Systematic and Evolutionary Microbiology, 2001, 51: 1797-1804. DOI:10.1099/00207713-51-5-1797 |
| [84] |
Villot C, Ma T, Renaud DL, et al. Saccharomyces cerevisiae boulardii CNCM I-1079 affects health, growth, and fecal microbiota in milk-fed veal calves[J]. Journal of Dairy Science, 2019, 102: 7011-7025. DOI:10.3168/jds.2018-16149 |
| [85] |
Inkpen SA, Douglas GM, Brunet TDP, et al. The coupling of taxonomy and function in microbiomes[J]. Biology & Philosophy, 2017, 32(1225): 1243. |
| [86] |
Franzosa EA, Morgan XC, Segata N, et al. Relating the metatranscriptome and metagenome of the human gut[J]. Proceedings of the National Academy of Sciences of the United States of America, 2014, 111: E2329-E2338. DOI:10.1073/pnas.1319284111 |
| [87] |
Louca S, Parfrey LW, Doebeli M. Decoupling function and taxonomy in the global ocean microbiome[J]. Science, 2016, 353: 1272-1277. DOI:10.1126/science.aaf4507 |
| [88] |
Sohail MU, Ijaz A, Younus M, et al. Effect of supplementation of mannan oligosaccharide and probiotic on growth performance, relative weights of viscera, and population of selected intestinal bacteria in cyclic heat-stressed broilers[J]. Journal of Applied Poultry Research, 2013, 22: 485-491. DOI:10.3382/japr.2012-00682 |
| [89] |
Zhu YH, Li XQ, Zhang W, et al. Dose-dependent effects of Lactobacillus rhamnosus on serum interleukin-17 production and intestinal T-cell responses in pigs challenged with Escherichia coli[J]. Applied and Environmental Microbiology, 2014, 80: 1787-1798. DOI:10.1128/AEM.03668-13 |
| [90] |
Zhang R, Zhou M, Tu Y, et al. Effect of oral administration of probiotics on growth performance, apparent nutrient digestibility and stress-related indicators in Holstein calves[J]. Journal of Animal Physiology and Animal Nutrition, 2016, 100: 33-38. DOI:10.1111/jpn.12338 |
| [91] |
Hungate RE. The rumen and its microbes[M]. New York: Academic Press, 1966.
|
| [92] |
Weimer PJ. Redundancy, resilience, and host specificity of the ruminal microbiota:implications for engineering improved ruminal fermentations[J]. Frontiers in Microbiology, 2015, 6: 296. |
| [93] |
Benson AK, Kelly SA, Legge R, et al. Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors[J]. Proceedings of the National Academy of Sciences of the United States of America, 2010, 107: 18933-18938. DOI:10.1073/pnas.1007028107 |
| [94] |
Zhou M, Peng YJ, Chen Y, et al. Assessment of microbiome changes after rumen transfaunation:implications on improving feed efficiency in beef cattle[J]. Microbiome, 2018, 6: 62. DOI:10.1186/s40168-018-0447-y |
| [95] |
Li F, Hitch TCA, Chen Y, et al. Comparative metagenomic and metatranscriptomic analyses reveal the breed effect on the rumen microbiome and its associations with feed efficiency in beef cattle[J]. Microbiome, 2019, 7: 6. DOI:10.1186/s40168-019-0618-5 |
| [96] |
Li F, Li C, Chen Y, et al. Host genetics influence the rumen microbiota and heritable rumen microbial features associate with feed efficiency in cattle[J]. Microbiome, 2019, 7: 92. DOI:10.1186/s40168-019-0699-1 |
| [97] |
Wen C, Yan W, Sun C, et al. The gut microbiota is largely independent of host genetics in regulating fat deposition in chickens[J]. The ISME Journal, 2019, 13: 1422-1436. DOI:10.1038/s41396-019-0367-2 |